
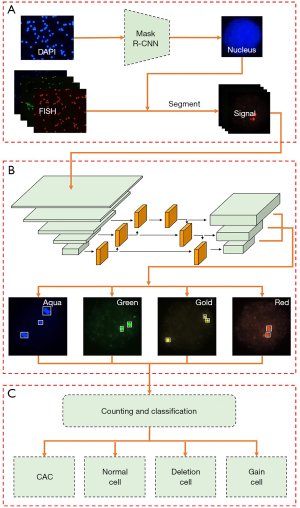
An efficient fluorescence in situ hybridization (FISH)-based circulating genetically abnormal cells (CACs) identification method based on Multi-scale MobileNet-YOLO-V4
Introduction
Quantitation of circulating tumor cells (CTCs) has been reported as a useful tool for prognosis and monitoring of advanced cancers, like breast, colon, and prostate cancer, as well as non-small cell lung cancer (NSCLC). However, several CTCs that extravasate into the bloodstream undergo epithelial-mesenchymal transition remain undetected by the epithelial cell adhesion molecule (EpCAM) based methods. An antigen-independent, 4-color method based on fluorescence in situ hybridization (FISH) was reported to detect CTCs with abnormalities in gene copy numbers—called circulating genetically abnormal cells (CACs). The CACs were thought to be a subset of “sentinel” CTCs found in the blood of patients with nodule-negative low-dose computed tomography (LDCT) scans, 1 to 4 years before the appearance of malignant lung nodules. This suggested that CACs may be markers of lung cancer, which might help in its early diagnosis. Furthermore, CACs in non-malignant nodules are extremely rare in healthy subjects and patients (1-3). Hence, there is an urgent requirement for developing and identifying techniques that would enable the accurate, efficient detection and enumeration of CACs in clinical blood samples, thereby aiding the early diagnosis of cancer.
Several semi-automatic identification systems for the identification of CACs have been developed so far. BioView fluorescence microscope systems are an example of such systems. However, the drawback of such systems is that they require a human operator to classify and count the detected CACs from a series of microscopic images. Normally, CACs are rare in peripheral blood (1–10 cells/mL) (4), a rapid, efficient, and sensitive automatic detection method for CACs is in high demand. Researchers have proposed a few automatic identification methods for CACs utilizing their morphological information. However, the reliability and reproducibility of these methods remain debatable (5-8).
Some studies suggested that the abnormal multiplication of specific genes in CACs indicates the formation of quantifiable standards, implying that it was possible to identify CACs based on the enhancement of fluorescence in two or more probes using the FISH technique (2,3,9,10). This FISH-based CACs identification method had the advantages of distinct and objective interpretation standards (2,3) and has been applied in clinical practice. However, the FISH review and result report are very labor-intense and time-consuming. The cell classification accuracy is often subjected to the reviewer’s experience as well as the degree of fatigue. Therefore, devising an automatic identification of CACs would establish a unified review standard based on large-scale case training to increase review accuracy, thus, enhancing reviewers’ efficiency and reducing review turnaround time.
Deep learning algorithms have undergone significant advancements in the last decade, particularly for biomedical applications such as cell detection (11,12) and cell classification (13,14), which in turn find important applications in the identification of hemocytes and cancer cells (15-17). Therefore, it is possible to develop an automatic identification method for CACs, based on fluorescence signal detection using a deep learning algorithm.
In the present study, an automatic identification method for CACs was developed based on an improved You Only Look Once (YOLO)-V4 (18) algorithm. In the developed detection method, the backbone of YOLO-V4 was replaced with MobileNet-V3 (19) to resolve overfitting and reduce time consumption. Meanwhile, an additional output based on a large-scale feature map was added to avoid misdetection of signal points. The proposed multi-scale MobileNet-YOLO-V4 algorithm achieved an accuracy of 93.86% in a dataset containing >7,000 clinical cell images. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-909/rc).
Methods
Data acquisition
Data was acquired based on methods, namely, Ficoll density separation and 4-color FISH technique (3). Sample preparation and probe hybridization were carried out as described below. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Initially, human full blood samples were enriched using the Ficoll-Hypaque density medium (2). Then, the peripheral blood mononuclear cells (PBMCs) were collected, enumerated, and deposited on glass slides. Next, the cell samples were hybridized with fluorescence probes targeting specific loci on chromosomes. The probe set comprised 3q29 (Spectrum Green), the locus-specific identifier 3p22.1 (Spectrum Red), CEP10 (Spectrum Aqua), and the locus-specific identifier 10q22.3 (Spectrum Gold). Finally, the cells were stained with 4',6-diamidino-2-phenylindole (DAPI; Boehringer Mannheim) and then visualized and digitalized using the BioView Duet-3 instrument.
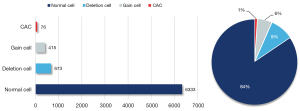
All the images in the dataset were 2,448×2,048 (pixels) in size. The number of cells in each image was >70, and the total number of cells was >7,000. The dataset included 6,333 normal cells, 673 deletion cells, 415 gaining cells and 76 CACs. Figure 1 provided the number of cells for each type and the corresponding proportion.
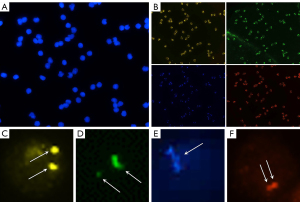
A representative image was shown in Figure 2. In Figure 2, the usual disadvantages of fluorescence signal images, viz., background contaminates (Figure 2C), nonuniformity in signal brightness and size (Figure 2D), the filamentous signal of the aqua channel due to high repetitive sequence content in the centromere region (Figure 2E), and signal overlap (Figure 2F) which lead to difficulties in detection could be observed.
Data preprocessing
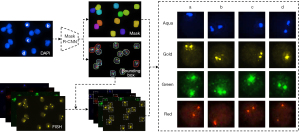
Initially, the nucleus in the DAPI images was detected and segmented using Mask R-CNN (Mask Region Convolutional Neural Networks) (20)—a deep neural network that has been applied widely in medical image segmentation and demonstrated good performance for microscopic images (21,22). Mask R-CNN had ResNet-50 as its backbone network, with critical parameters as follows: non-maximum suppression (NMS) threshold used for postprocessing the region proposal network (RPN) proposals and intersection over union (IOU) between the anchor and the ground truth (GT) box were set to 0.7. For the model, 1,200 images were used for training and 200 for validating. During the training, the size of input images was adjusted to 1,024×1,024, with hyperparameters set as follows: Epoch and batch size were set to 300 and 16, respectively. The initial learning rate was 0.02, which decayed at a rate of 10% per 100 epochs. Simultaneously, a weight decay of 0.0001 and a momentum of 0.9 were adopted. The trained model was tested on the testing sets including 100 images, and 97.67% of dice similarity coefficient (DSC), 100% of independent cell detection rate, and 95.32% of overlapping cell detection rate were obtained. Then, the bounding boxes and masks of the nucleus were obtained. Next, the patch of each nucleus was obtained by cropping the fluorescence signal images in each channel with bounding boxes. The obtained patches were multiplied by the corresponding masks to remove the background noise originating from outside the nucleus. The pipeline for nucleus segmentation was illustrated in Figure 3.
The identification rule of cells
The identification rule of cells, namely clear definition of the outcome that was predicted by the prediction model, was shown in Table 1. A CAC was defined as an intact round or oval cell, having polysomy of at least 2 out of 4 DNA probes per nucleus, which was shown as more than 2 fluorescence signals in 2 or more channels in the image (2,3). The deletion and gain cells were also classified to demonstrate the total abnormality and hybridization efficiency of PBMCs. However, they were not considered as risk factors in later nodule malignancy prediction in the clinical setting. The gain cells were also included as review candidates for human confirmation to avoid any possible misclassification of CACs into a gain cell.
Table 1
| Category | Description |
|---|---|
| Normal cell | 2 signals in all the channels |
| Deletion cell | Less than 2 signals in 1 or more channels |
| Gain cell | More than 2 signals in 1 channel |
| Circulating genetically abnormal cell (CAC) | More than 2 signals in 2 or more channels |
Network architecture for detection of stained signals
YOLO-V4 is utilized frequently for object detection tasks (23-25). It exhibits the characteristics of high precision, high efficiency, flexibility, and generalization (18). Its backbone is constituted of CSPDarknet53 and may be replaced with several other frameworks for different application scenarios (26), giving it additional flexibility for use in various engineering applications. The Multi-scale MobileNet-YOLO-V4 algorithm proposed in the present study presented improvements in the aspects listed below:
- The backbone in YOLO-V4 was replaced with MobileNet-V3, which is lightweight and convenient to train (19). This prevented the issue of overfitting in small-scale datasets and improved the detection efficiency.
- The architecture of YOLO-V4 was optimized by utilizing a new feature map with a larger scale, which enriched more texture and contour information, thereby enabling the detection of small fluorescence signals.
Replacing the backbone
A large quantity of data and labels were required in training, depending on the excessive amount of backbone network parameters, floating-point operations, and the number of layers in the original YOLO-V4 (26). Otherwise, the issue of over-fitting would occur. MobileNet-V3 is a lightweight, low-latency model. In certain visual tasks, such as classification, target detection, semantic segmentation, etc., the model size was significantly reduced, and promising results were obtained (19). YOLO-V4 Tiny is a further lightweight implementation of the YOLO-V4 network. However, the mean average precision (mAP) of YOLO-V4 Tiny (mAP 22%) was inferior to YOLO-V4 (mAP 43.5%) on the MS COCO dataset, although it was faster than YOLO-V4 in detection speed (27). The YOLO-V4 Tiny had been shown in practice to be better at detecting independent objects, but struggled with detecting small objects and overlapping targets (28). For identification of CACs, high accuracy and quick response are both required clinically. Therefore, in the present work, YOLO-V4 was combined with the MobileNet-V3 network. The replaced section is illustrated inside the red dotted box around MobileNet-V3 in Figure 4.
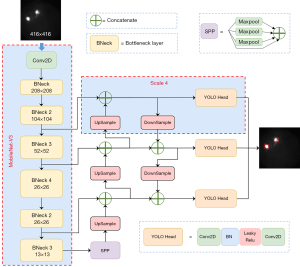
Adding a larger scale
Supposing the size of the input image of YOLO-V4 was 416×416, then three featured output layers, 52×52, 26×26, and 13×13, were selected for upsampling and feature fusion to detect large, medium, and small objects of different sizes. The area corresponding to each grid cell in the input image was related inversely to the size of the feature map; that is, the 13×13 detection layer was suitable for detecting large targets, while the 52×52 detection layer was suitable for detecting small targets. Due to the relatively small size of the signal point, it would have been inappropriate to directly predict the signal points’ position using the 13×13 feature map. Certain fluorescence signals, which were relatively small, such that even a 52×52 feature map failed to detect them, resulted in missed signal detections in the identification of CACs.
To solve the problem of missing small-signal points, a shallow layer feature output was added based on the original layer in the proposed algorithm. To obtain more texture and contour information, the scale 4 was added (the red dotted box named scale 4), which was beneficial for detecting small objects. The minimum scale prediction output was deleted but not abandoned, as it was fused with the feature of the earlier layer through upsampling. The network of the improved Multi-scale MobileNet-YOLO-V4 was illustrated in Figure 4.
Mosaic data augmentation
Mosaic data augmentation (18) was utilized in the training stage, which mixed four images into one image by random flipping, scaling, and cropping, as shown in Figure 5. The utilizing of Mosaic data augmentation enriched the background of the detected objects and enable the model to identify targets smaller than normal size or close targets, thus enhancing the robustness of the model.

Modification of anchor boxes
YOLO-V4 followed the concept of anchor boxes (a set of initial candidate boxes with fixed width and height). The selection of the initial anchor boxes would directly affect the accuracy and speed of detection. In this study, the K-means clustering analysis was used to regain the size of the anchor boxes of the model. Nine different sizes were selected in the 4-color channels, respectively. The smallest of these anchor frames was 10×10, and the largest was 45×37.
Evaluation criterion and parameter setting
The values of accuracy, recall (sensitivity), precision, harmonic mean (also known as F1 score) and mAP were used as the evaluation criteria. These indicators can be defined as follows:
Where true positive (TP), true negative (TN), false positive (FP), and false negative (FN) denote the number of correctly identified signal points, correctly predicted non-signal points, falsely detected signal points, and missed signal points, respectively. In Eq. [5], r and P(r) represent recall and precision respectively. In Eq. [6], n represents the number of detection targets types. In this study, mAP was equal to average precision (AP) because there was only one type of detection target, i.e., fluorescence signal.
The experiment was conducted using Linux with NVIDIA Tesla V100 GPU. Python 3.5.6 was the programming language used in the experiment, based on TensorFlow-GPU 1.14.0 deep learning framework. Epoch, batch size, and initial learning rate were set to 200, 24, and 0.001, respectively, in correspondence with other comparable algorithms. The initialization parameters for training were tabulated in Table 2.
Table 2
| Parameters | Value |
|---|---|
| Input size | 416×416 |
| Batch size | 24 |
| Momentum | 0.9 |
| Initial learning rate | 0.001 |
| Epoch | 200 |
Results
CACs identification
The flowchart for the identification of CACs was presented in Figure 6. In the data preprocessing stage, the mask of the individual nucleus was segmented from the DAPI images by applying the Mask R-CNN. Subsequently, the FISH fluorescence images were also segmented into single-cell images based on the nucleus mask. Next, the segmented single-cell FISH fluorescence images were used as an input for the developed (in this study) algorithm to derive the number and location of the signal points. Hence, the CACs were identified according to the classification rule of cells, as shown in Table 1.
Comparison of four networks
To evaluate the performance of the proposed Multi-scale MobileNet-YOLO-V4 for fluorescence signal detection, the developed approach was compared with the YOLO-V4, YOLO-V4 Tiny, and MobileNet-YOLO-V4. The patches, which were single-cell images of 4-color channels in the dataset, were utilized as the experimental data. K-fold cross-validation (29) was applied for evaluating the overall performance of the model more objectively and comprehensively. In the experiment, five-folds cross-validation were used to randomize the datasets to five parts, and each time, one part was used as testing sets, while the remaining four parts as training sets and validation sets, of which the ratio was 4:1. The final overall performance of the model was presented as the mean of five runs with different data as testing sets. The evaluation criteria have been presented as Eqs. [1-4,6]. In addition, the test time was recorded to confirm the efficiency of the improved network proposed in the present study. The test results for the four networks have been presented in Table 3. Figure 7 showed the precision-recall (P-R) curves of the first test dataset of four detection models in four different color channels based on five-folds cross-validation. The remaining results were shown in Figure S1.
Table 3
| Net | Probe | Accuracy (%) | Recall (%) | Precision (%) | F1 (%) | mAP (%) (IOU =0.5) | Time (s) |
|---|---|---|---|---|---|---|---|
| YOLO-V4 | Aqua | 89.29 | 96.23 | 92.53 | 94.34 | 95.62 | 0.040 |
| YOLO-V4 Tiny | Aqua | 91.96 | 98.25 | 93.49 | 95.81 | 96.71 | 0.017 |
| MobileNet-YOLO-V4 | Aqua | 93.46 | 98.80 | 94.53 | 96.62 | 97.73 | 0.018 |
| Multi-scale MobileNet-YOLO-V4 | Aqua | 97.03 | 99.25 | 97.75 | 98.49 | 99.01 | 0.020 |
| YOLO-V4 | Gold | 94.53 | 98.78 | 95.65 | 97.19 | 99.24 | 0.041 |
| YOLO-V4 Tiny | Gold | 95.10 | 98.96 | 96.06 | 97.49 | 99.39 | 0.016 |
| MobileNet-YOLO-V4 | Gold | 97.57 | 98.98 | 98.55 | 98.77 | 99.77 | 0.018 |
| Multi-scale MobileNet-YOLO-V4 | Gold | 98.20 | 99.29 | 98.89 | 99.09 | 99.86 | 0.018 |
| YOLO-V4 | Green | 96.46 | 99.21 | 97.21 | 98.20 | 99.49 | 0.039 |
| YOLO-V4 Tiny | Green | 97.06 | 99.11 | 97.91 | 98.51 | 99.59 | 0.015 |
| MobileNet-YOLO-V4 | Green | 98.13 | 99.43 | 98.69 | 99.06 | 99.79 | 0.019 |
| Multi-scale MobileNet-YOLO-V4 | Green | 98.93 | 99.56 | 99.36 | 99.46 | 99.91 | 0.019 |
| YOLO-V4 | Red | 87.09 | 98.23 | 88.48 | 93.10 | 97.27 | 0.038 |
| YOLO-V4 Tiny | Red | 94.83 | 98.92 | 95.82 | 97.35 | 98.14 | 0.017 |
| MobileNet-YOLO-V4 | Red | 96.46 | 98.77 | 97.64 | 98.20 | 99.38 | 0.018 |
| Multi-scale MobileNet-YOLO-V4 | Red | 98.13 | 99.29 | 98.82 | 99.06 | 99.83 | 0.019 |
F1, F1-score; mAP, mean average precision; IOU, intersection over union; YOLO, You Only Look Once.
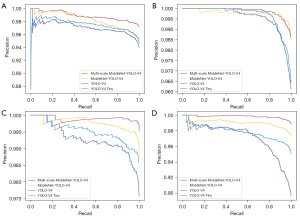
As presented in Table 3, the accuracy of Multi-scale MobileNet-YOLO-V4 was approximately 98%, the highest among all four channels. Moreover, the values of sensitivity, precision, F1-score and mAP were close to 100%. In all these metrics, Multi-scale MobileNet-YOLO-V4 outperformed the other three networks applied in fluorescence signal detection. According to Figure S1, the performance of the proposed method in fluorescence signal detection was superior to that of other networks. It was noteworthy that due to the presence of filamentous signals in the aqua channel (as depicted in Figure 2E), the identification accuracy was consequently lower than the other three channels. However, the accuracy of the aqua channel was improved to 97.03%, with a difference of only approximately 1% compared to the other three channels. Furthermore, in terms of detection time, the last three models using the lightweight network improved the detection speed by 2–3 times when compared to the original YOLO-V4. However, our improved Multi-scale MobileNet-YOLO-V4 achieved the best performance while increasing the detection speed.
Comparison of results with and without the Mosaic data augmentation method
The results of Multi-scale MobileNet-YOLO-V4 in Table 3 were based on the Mosaic data augmentation method. To further describe the influence of Mosaic data augmentation method on the experimental results comprehensively, the Multi-scale MobileNet-YOLO-V4 results with and without Mosaic data augmentation method were compared as shown in Table 4. The results have proved the effectiveness of the Mosaic data enhancement method.
Table 4
| Probe | Mosaic | Accuracy (%) | Recall (%) | Precision (%) | F1 (%) | mAP (%) |
|---|---|---|---|---|---|---|
| Aqua | Yes | 97.03 | 99.25 | 97.75 | 98.49 | 99.01 |
| No | 95.90 | 98.93 | 96.90 | 97.91 | 98.23 | |
| Gold | Yes | 98.20 | 99.29 | 98.89 | 99.09 | 99.86 |
| No | 97.73 | 99.19 | 98.52 | 98.85 | 99.80 | |
| Green | Yes | 98.93 | 99.56 | 99.36 | 99.46 | 99.91 |
| No | 98.43 | 99.49 | 98.93 | 99.21 | 99.85 | |
| Red | Yes | 98.13 | 99.29 | 98.82 | 99.06 | 99.83 |
| No | 97.46 | 99.12 | 98.32 | 98.72 | 99.59 |
F1, F1-score; mAP, mean average precision; YOLO, You Only Look Once.
Results for CACs identification
The fluorescence signals of the 4-color channels detected using Multi-scale MobileNet-YOLO-V4 were marked on the images with red rectangular boxes. The results were then counted, and CACs were detected according to the classification rule of cells. The results have been presented in Figure 8.
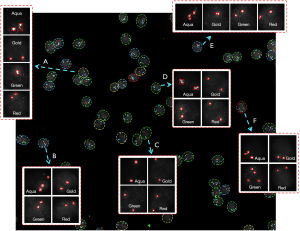
According to the classification rules, it is evident that Figure 8A is a deletion cell, Figure 8B,8E are gain cells, Figure 8C,8D are normal cells, and Figure 8F is a CAC. The evaluation indicators for the identification of CACs were the same as Eqs. [1-4]. Among them, TP indicated the number of correctly identified CACs, FP indicated the number of falsely detected CACs, and FN indicated the number of missed CACs. The test results have been presented in Table 5.
Table 5
| Net | Accuracy (%) | Recall (%) | Precision (%) | F1 (%) | Time (s) |
|---|---|---|---|---|---|
| YOLO-V4 | 87.45 | 74.21 | 72.84 | 73.52 | 0.039 |
| YOLO-V4 Tiny | 89.23 | 77.99 | 74.25 | 76.17 | 0.016 |
| MobileNet-YOLO-V4 | 91.36 | 80.50 | 79.50 | 80.01 | 0.018 |
| Multi-scale MobileNet-YOLO-V4 | 93.86 | 90.57 | 87.80 | 89.16 | 0.019 |
CAC, circulating genetically abnormal cell; F1, F1-score; YOLO, You Only Look Once.
It has been experimentally proven that Multi-scale MobileNet-YOLO-V4 is better than that of the other networks. Using the approach in this study, the accuracy, sensitivity, precision, and F1-score values in the identification of CACs reached 93.86%, 90.57%, 87.80%, and 89.16%, respectively. Further, the detection speed of the method used in this study is advantageous when compared to YOLO-V4. Furthermore, based on the performance of MobileNet-YOLO-V4 and the method used in this study, it could be inferred that adding a larger scale output would indeed be beneficial for the identification CACs.
Discussion
Currently, LDCT scanning is the most extensively used method for the detection of pulmonary nodules. However, LDCT is radiative and unable to distinguish between malignant and non-malignant pulmonary nodules (3). Though CACs act as “liquid biopsy” of cancer and play a critical role in early cancer diagnosis, their detection in the blood poses a challenge. The morphology-based approach for automated CACs identification had three main defects as follows: Firstly, a large amount of training data being used to train the model is necessary for a machine learning classifier based on cell morphology approach (30). For example, the detection accuracy of the model compromised greatly as the CAC quantity was not enough (31). However, CAC is extremely rare cell, and thus, it tends to cause insufficient data and serious data imbalance. Secondly, the morphological analysis of CACs had revealed that they were uneven in morphology—either round or oval, with sizes ranging from 4 to 30 µm (7). Moreover, if the size of CACs was similar to other cells, such as nucleated blood cells in certain prostate cancers (5) and even white blood cells in certain cases (7), the false-positive rate might increase. Therefore, the applicability of methods based on the morphology of CACs was limited. Finally, these methods involved end-to-end prediction, lacking integration with clinically interpretable methods. The blood-based 4-color FISH method for the identification of CACs was possible to achieve undisputed detection results, which identified CACs by analyzing the number of fluorescence signals in the different channels according to the identification rules, as presented in Table 1. The method could accurately identify patients with lung cancer by enabling a reduction in the frequency of non-malignant nodules biopsied, which has been applied in clinical practice (32-36).
From Tables 3,5, it can be observed that the original YOLO-V4 exhibited low accuracy, sensitivity, and precision and also required a long testing time. Numerous high-level semantic features are unessential in fluorescence signal detection as these signals are simple in structure and have a small area. However, extensive texture details and location information are required to be added. Therefore, a deep network structure is unnecessary and may lead to overfitting due to insufficient training data. Improved results obtained using YOLO-V4 Tiny compared to YOLO-V4 proved that there was no requirement for a deep network to detect fluorescence signals. The results were further improved when CSPDarkNet53 was replaced with MobileNet-V3.
As shown in Table 3, the precision and sensitivity of MobileNet-YOLO-V4 were lower than those of Multi-scale MobileNet-YOLO-V4. The MobileNet-YOLO-V4 algorithm missed relatively small signal points, as shown in the experimental results (Figure 9A,9B). In addition, there was false detection to such significantly large noise interference. As depicted in Figure 2, the percentage of signal points in the patches was relatively small. The K-means clustering algorithm was applied to cluster the width and height of the target boxes of the signal points in the dataset. The average overlap (AvgIOU) was used as a measure in the target clustering analysis to cluster the dataset. The average pixel of the signal points was determined as 25×25 in a 416×416 cell image, which was approximately 0.361% in an image. According to the characteristics of the network architecture of YOLO-V4, the small size feature maps at the top level mainly provided deep semantic information, while the large size feature maps at the bottom level provided more information regarding the location of the target. The addition of the 4-fold down-sampling feature map of the output to MobileNet-YOLO-V4 for target detection improved the detection capability of the network for relatively small targets as more information on the size and location of small-signal points was now available. Meanwhile, to address the false detection of noisy points, the output of the smallest scale, i.e., the output of detecting the large targets, was deleted, which enhanced the robustness of the model against interference from noisy points. The results have been presented in Figure 9C,9D.
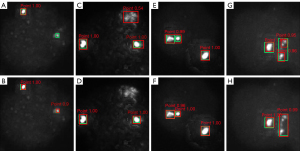
When the distance between two signal points was considerably small or when the points were partially overlapping, as depicted in Figure 9E,9F, they could be detected as one signal point. Meanwhile, the filamentous signal (Figure 9G,9H) of the aqua channel could be easily identified as two or more signal points. Data augmentation for this case was done using the Mosaic data augmentation method (18), which spliced four different images by flipping, zooming, and cropping. Then the background of the detected object was enriched, and the detection of small targets was also enhanced. The experiments conducted in this study have demonstrated that using the Mosaic data augmentation method could enhance the accuracy by approximately 2% in CACs identification. The Mosaic data augmentation method was also effective in detecting overlapping signals.
The accuracy (93.86%) and sensitivity (90.57%) of the results obtained in this study have reached the level of human expert (accuracy 94.2% and sensitivity 89%) compared to the manual counting method using FISH technology (3). Furthermore, the method used in this study is significantly faster than manual identification by a factor of approximately 500. Furthermore, compared to other state-of-the-art methods related to deep learning, used in the identification of CACs, such as NBC (Semi-supervised) (F1 86.9%) (37), Faster R-CNN Deep Detector (F1 85.5%) (38), and improved faster R-CNN based network (F1 84.7%) (39), the F1 of the method used in this study was higher. This indicated the promising overall performance of the method used in this study and hence established it as a highly competitive method that could be used for clinical practice.
Conclusions
In the present study, an efficient automatic approach for the identification of CACs, based on Multi-scale MobileNet-YOLO-V4 was proposed. The Multi-scale MobileNet-YOLO-V4 is a lightweight network for processing fluorescence images. The backbone of YOLO-V4 was replaced with MobileNet-V3 to avoid overfitting and improve detection efficiency. In addition, a larger scale output was added to suitably detect small targets. The experimental results demonstrated that compared to the expert, the proposed approach showed excellent performance accuracy of 93.86% and a processing time of 0.076 s/cell. The present study would provide novel insights into the problems encountered in the identification of CACs.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (grant No. 61971445) and National Key R&D Program of China (grant No. 2019YFF0216302).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-909/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-909/coif). XF, XL and XY are employees of Zhuhai Sanmed Biotech Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brock G, Castellanos-Rizaldos E, Hu L, Coticchia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res 2015;4:280-90.
- Katz RL, He W, Khanna A, Fernandez RL, Zaidi TM, Krebs M, Caraway NP, Zhang HZ, Jiang F, Spitz MR, Blowers DP, Jimenez CA, Mehran RJ, Swisher SG, Roth JA, Morris JS, Etzel CJ, El-Zein R. Genetically abnormal circulating cells in lung cancer patients: an antigen-independent fluorescence in situ hybridization-based case-control study. Clin Cancer Res 2010;16:3976-87. [Crossref] [PubMed]
- Katz RL, Zaidi TM, Pujara D, Shanbhag ND, Truong D, Patil S, Mehran RJ, El-Zein RA, Shete SS, Kuban JD. Identification of circulating tumor cells using 4-color fluorescence in situ hybridization: Validation of a noninvasive aid for ruling out lung cancer in patients with low-dose computed tomography-detected lung nodules. Cancer Cytopathol 2020;128:553-62. [Crossref] [PubMed]
- Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31. [Crossref] [PubMed]
- Ciurte A, Selicean C, Soritau O, Buiga R. Automatic detection of circulating tumor cells in darkfield microscopic images of unstained blood using boosting techniques. PLoS One 2018;13:e0208385. [Crossref] [PubMed]
- Ciurte A, Marita T, Buiga R. Circulating tumor cells classification and characterization in dark field microscopic images of unstained blood. 2015 IEEE International Conference on Intelligent Computer Communication and Processing (ICCP), 3-5 Sept. 2015; Cluj-Napoca, Romania. IEEE, 2015:367-74.
- Esmaeilsabzali H, Beischlag TV, Cox ME, Parameswaran AM, Park EJ. Detection and isolation of circulating tumor cells: principles and methods. Biotechnol Adv 2013;31:1063-84. [Crossref] [PubMed]
- Tsuji K, Lu H, Tan JK, Kim H, Yoneda K, Tanaka F. Detection of Circulating Tumor Cells in Fluorescence Microscopy Images Based on ANN Classifier. Mobile Networks and Applications 2020;25:1042-51. [Crossref]
- Smith AL, Zaidi TM, Shanbhag N, Truong D, Shkedy S, Namer BL, Kuban JD, Katz RL. Circulating Tumor Cell Detection via a Novel FISH Assay Prior to Lung Biopsy Enables Accurate Prediction of Pulmonary Malignancy. Mod Pathol 2017;30:496A.
- Zhang B, He S, Chen C, Chen Y, Li Y, Huang M, Yue D, Huang C, Li C, Xin Y, Zhang J, Wang C. Detection of Circulating Genetically Abnormal Cells Improves the Diagnostic Accuracy in Lung Cancer Presenting with GGNs. J Thorac Oncol 2019;14:S798. [Crossref]
- Xie W, Noble JA, Zisserman A. Microscopy cell counting and detection with fully convolutional regression networks. Comput Methods Biomech Biomed Eng Imaging Vis 2018;6:283-92. [Crossref]
- Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 2019;16:67-70. [Crossref] [PubMed]
- Dürr O, Sick B. Single-Cell Phenotype Classification Using Deep Convolutional Neural Networks. J Biomol Screen 2016;21:998-1003. [Crossref] [PubMed]
- Wolf FA, Angerer P, Theis FJ. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 2018;19:15. [Crossref] [PubMed]
- Wang Q, Bi S, Sun M, Wang Y, Wang D, Yang S. Deep learning approach to peripheral leukocyte recognition. PLoS One 2019;14:e0218808. [Crossref] [PubMed]
- Sirinukunwattana K, Ahmed Raza SE. Yee-Wah Tsang, Snead DR, Cree IA, Rajpoot NM. Locality Sensitive Deep Learning for Detection and Classification of Nuclei in Routine Colon Cancer Histology Images. IEEE Trans Med Imaging 2016;35:1196-206. [Crossref] [PubMed]
- Xu Y, Jia ZP, Ai YQ, Zhang F, Lai MD, Chang EIC. Deep convolutional activation features for large scale Brain Tumor histopathology image classification and segmentation. Proceedings of the 40th IEEE International Conference on Acoustics, Speech, and Signal Processing (ICASSP); 2015 Apr 19-24; Brisbane, Australia. New York: IEEE, 2015.
-
Bochkovskiy A Wang CY Liao HYM YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv:2004.10934v1 [cs.CV]2020 . - Howard A, Sandler M, Chen B, Wang W, Chen LC, Tan M, Chu G, Vasudevan V, Zhu Y, Pang R, Adam H, Le Q. Searching for MobileNetV3. 2019 IEEE/CVF International Conference on Computer Vision (ICCV); Oct 27-Nov 02, 2019; Seoul, South Korea. Los Alamitos: IEEE Computer Soc, 2019.
- He K, Gkioxari G, Dollár P, Girshick R. Mask R-CNN. 2017 IEEE International Conference on Computer Vision (ICCV); 22-29 Oct. 2017; Venice, Italy. New York: IEEE, 2017.
- Johnson JW. Automatic Nucleus Segmentation with Mask-RCNN. Proceedings of the 2019 Computer Vision Conference (CVC); 2019 Apr 25-26; Las Vegas, NV. Cham: Springer International Publishing Ag, 2020.
- Dhieb N, Ghazzai H, Besbes H, Massoud Y. An Automated Blood Cells Counting and Classification Framework using Mask R-CNN Deep Learning Model. 2019 31st International Conference on Microelectronics (ICM); 15-18 Dec. 2019; Cairo, Egypt. New York: IEEE, 2019.
-
Qiao S Pang S Luo G Pan S Wang X Wang M Zhai X Chen T. Automatic Detection of Cardiac Chambers Using an Attention-based YOLOv4 Framework from Four-chamber View of Fetal Echocardiography. arXiv:2011.13096v2 [cs.CV]2020 . - Hu X, Liu Y, Zhao Z, Liu J, Yang X, Sun C, Chen S, Li B, Zhou C. Real-time detection of uneaten feed pellets in underwater images for aquaculture using an improved YOLO-V4 network. Comput Electron Agric 2021;185:106135. [Crossref]
- Yu J, Zhang W. Face Mask Wearing Detection Algorithm Based on Improved YOLO-v4. Sensors (Basel) 2021;21:3263. [Crossref] [PubMed]
- Chen Q, Xiong Q. Garbage Classification Detection Based on Improved YOLOV4. Journal of Computer and Communications 2020;8:285-94. [Crossref]
-
Wang CY Bochkovskiy A Liao HYM Scaled-YOLOv4: Scaling Cross Stage Partial Network. arXiv:2011.08036v2 [cs.CV]2021 .10.1109/CVPR46437.2021.01283 - Kumar A, Kalia A, Sharma A, Kaushal M. A hybrid tiny YOLO v4-SPP module based improved face mask detection vision system. J Ambient Intell Humaniz Comput 2021; Epub ahead of print. [Crossref] [PubMed]
- Wong TT. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognit 2015;48:2839-46. [Crossref]
- Toratani M, Konno M, Asai A, Koseki J, Kawamoto K, Tamari K, Li Z, Sakai D, Kudo T, Satoh T, Sato K, Motooka D, Okuzaki D, Doki Y, Mori M, Ogawa K, Ishii H. A Convolutional Neural Network Uses Microscopic Images to Differentiate between Mouse and Human Cell Lines and Their Radioresistant Clones. Cancer Res 2018;78:6703-7. [Crossref] [PubMed]
- Wang S, Zhou Y, Qin X, Nair S, Huang X, Liu Y. Label-free detection of rare circulating tumor cells by image analysis and machine learning. Sci Rep 2020;10:12226. [Crossref] [PubMed]
- Qiu X, Zhang H, Zhao Y, Zhao J, Wan Y, Li D, Yao Z, Lin D. Application of circulating genetically abnormal cells in the diagnosis of early-stage lung cancer. J Cancer Res Clin Oncol 2022;148:685-95. [Crossref] [PubMed]
- Ye X, Yang XZ, Carbone R, Barshack I, Katz RL. Diagnosis of Non-Small Cell Lung Cancer via Liquid Biopsy Highlighting a Fluorescence-in-situ-Hybridization Circulating Tumor Cell Approach. In: Strumfa I, Bahs G. editors. Pathology: From Classics to Innovations. London: IntechOpen, 2021. doi:
10.5772/intechopen.97631 .10.5772/intechopen.97631 - Liu WR, Zhang B, Chen C, Li Y, Ye X, Tang DJ, Zhang JC, Ma J, Zhou YL, Fan XJ, Yue DS, Li CG, Zhang H, Ma YC, Huo YS, Zhang ZF, He SY, Wang CL. Detection of circulating genetically abnormal cells in peripheral blood for early diagnosis of non-small cell lung cancer. Thorac Cancer 2020;11:3234-42. [Crossref] [PubMed]
- Guber A, Greif J, Rona R, Fireman E, Madi L, Kaplan T, Yemini Z, Gottfried M, Katz RL, Daniely M. Computerized analysis of cytology and fluorescence in situ hybridization (FISH) in induced sputum for lung cancer detection. Cancer Cytopathol 2010;118:269-77. [Crossref] [PubMed]
- Katz R, Zaidi T, Ni X. Liquid Biopsy: Recent Advances in the Detection of Circulating Tumor Cells and Their Clinical Applications. In: Bui MM, Pantanowitz L. editors. Modern Techniques in Cytopathology. Basel: Karger, 2020:43-66.
- Svensson CM, Krusekopf S, Lücke J, Thilo Figge M. Automated detection of circulating tumor cells with naive Bayesian classifiers. Cytometry A 2014;85:501-11. [Crossref] [PubMed]
- Zhang A, Zou Z, Liu Y, Chen Y, Yang Y, Chen Y, Law BNF. Automated Detection of Circulating Tumor Cells Using Faster Region Convolution Neural Network. J Med Imaging Health Inform 2019;9:167-74. [Crossref]
- Liu Y, Si H, Chen Y, Chen Y. Faster R-CNN based Robust Circulating Tumor Cells Detection with Improved Sensitivity. Proceedings of the 2nd International Conference on Big Data Technologies (ICBDT)/3rd International Conference on Business Information Systems Workshop (ICBIS); Aug 28-30, 2019; Jinan, China. New York: Association for Computing Machinery, 2019.


