
Quantification of brown adipose tissue in vivo using synthetic magnetic resonance imaging: an experimental study with mice model
Introduction
Obesity-associated disorders, such as diabetes, cardiovascular diseases, hypertension, strokes, musculoskeletal disorders, and cancer, are the leading cause of mortality in the adult population (1). Obesity is caused by excessive intake of energy stored as fat. Adipose tissues have a vital role in maintaining the body’s energy balance (2). In mammals, the white adipose tissues (WATs) and brown adipose tissues (BATs) are two main types of adipose tissues showing different spatial variations, cell structures, and functional roles. The WATs are distributed in subcutaneous and visceral locations, while BATs are mainly located in the interscapular region in rodents (3). The WATs are composed of large unilocular lipid droplets containing an eccentric nucleus within the cell and a limited amount of cytoplasm, whereas BATs contain small multilocular lipid droplets with an abundance of cytoplasm (3). The BAT is a highly heterogeneous adipose tissue containing dense vascular and iron-rich mitochondria, specifically expressing uncoupling protein 1 (UCP1) (4). Compared to the WAT that stores excess energy as triglycerides, BAT is responsible for releasing energy via thermogenesis (5,6), which utilizes UCP1 in the mitochondria to consume fat and generate heat (7). Our previous study (8) showed that the visceral adipose tissue volume at the level of the 3rd lumbar vertebrae was associated with the incidence of type 2 diabetes. However, the increased quantity or activity of BATs may effectively enhance energy utilization, reduce the storage of WATs, combat the development of obesity and type 2 diabetes, as well as maintain the metabolic balance in the body (6,9). Therefore, BAT is considered a potential target tissue for developing new drugs that might induce the browning of WAT or increase the BAT volume to ameliorate obesity and its related metabolic disorders (10).
Noninvasive imaging approaches are vital for examining BATs in humans, among which positron emission tomography-computed tomography (PET/CT) and magnetic resonance imaging (MRI) are the most popular techniques for distinguishing BAT from WAT and measuring the BAT volume in clinical practice. However, at present, the identification and quantification of BAT in small rodents remain challenging (2).
Imaging using fluorodeoxyglucose F 18 (18F-FDG) PET is the standard method for BAT detection due to its high sensitivity in detecting metabolic alterations. Nonetheless, PET is not suitable for longitudinal studies requiring repeated scans due to its high ionizing radiation (3). In addition, 18F-FDG is mainly taken up by the metabolically active BAT but not by inactive tissues, which can underestimate the total quantity of BATs (2). In contrast, MRI is a valuable tool for quantifying the characteristics of BAT independent of the tissue’s activation state. The lower fat content in BATs and the use of non-ionizing radiation make MRI a particularly attractive method (11,12), especially when assessing BAT alterations in response to pharmacologic stimulation that requires repeated scans. Chemical shift imaging has also been used to differentiate WAT and BAT; yet, BAT shows poor tissue-signal contrast due to the relatively limited imaging spatial resolution in this method (13,14). The use of magnetic resonance spectroscopy (MRS), based on the different endogenous biochemical properties and histological composition of the two adipose tissues, has also been investigated for this purpose (15). However, in practice, MRS is severely limited by spatial resolution (15,16) and magnetic field inhomogeneity, with the latter being commonly present (3).
Synthetic MRI is a novel imaging technique that simultaneously offers conventional weighted images and quantitative maps in one scan, as well as the correction of B1 field inhomogeneity (17,18). The physical properties of tissues like the longitudinal and transverse relaxation times (T1 and T2, respectively, in ms) are obtained from a multiple-dynamic multiple-echo (MDME) sequence. The quantitative relaxation maps obtained using synthetic MRI have been reported as promising candidates for the differential diagnosis of various diseases (19) and have been routinely used for neural (20-22), knee (23), and breast (24) imaging. The T1 and T2 values reflect the tissue compositions (21) and may act as potential quantitative biomarkers for different pathological properties. As the BATs and WATs have differing fat and water contents, with BAT containing a large number of mitochondria, blood vessels, and iron, the relaxation characteristics of BAT and WAT tend to vary. Hamilton et al. (15) used MRS to demonstrate that the T1 relaxation rate of the water component differed between BAT and WAT, and that the water content T1 in the WAT was almost twice that in BAT. However, MRS requires a sufficient tissue volume for accuracy, which may limit spatial resolution. A previous study (2) applied the T2 value to distinguish between BAT and WAT, using a conventional multi-echo spin-echo (MESE) sequence, which was the most commonly used technique for T2 measurement (25); still, the relatively long scanning period hinders its clinical application. In addition, it has been demonstrated that T2-weighted images (T2WIs) can be utilized to estimate the BAT volume in rats (2). The T2WIs obtained using synthetic MRI may be of clinical significance when used in similar applications.
Ouwerkerk et al. (26) showed shorter T2 and lower unsaturated fatty acids (UFAs) in BAT compared to WAT in healthy humans without cold stimulation. Also, previous literature showed that the T1 of water was lower in BAT compared with WAT regardless of the tissue’s activation state (15). Yet, to the best of our knowledge, previous studies have not reported the application of synthetic MRI for distinguishing BAT from WAT and quantifying BAT volumes. Because WAT is mostly fat, the T1 and T2 are close to the T1 and T2 of the most prominent triglyceride signal. In BAT, the water signal is a significantly larger portion of the total signal, and both T1 and T2 are markedly different from the T1 and T2 of triglycerides. In the present study, we hypothesized that the differing tissue composition of the BATs and WATs might lead to different relaxation times, which in turn would allow for differentiation at thermoneutral temperature. Additionally, the synthetic T2-weighted (sT2w) images, obtained simultaneously, may be used for estimating the BAT volume.
Methods
Animal care
A total of 16 C57BL/6 mice, between 20 and 22 weeks of age, were obtained from the Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China). All animals were maintained in an environment of temperature 22±1 °C prior to the MRI scan, relative humidity of 50–60%, and a light/dark cycle of 12/12 h, with free access to a regular diet and water. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (NO. [2017]104) and performed in compliance with institutional guidelines for the care and use of animals.
MRI scanning
All mice were scanned in the prone position using a 3.0 T scanner (SIGNA Pioneer, GE Healthcare, Milwaukee, WI) equipped with a 5-cm diameter receive coil suitable for animals. The chest and abdomen of each mouse were wrapped by a piece of cloth during the examination, which had an auxiliary effect on reducing breathing-induced motion artifacts. A scout localizer scan was performed first, followed by conventional axial T2WI and synthetic MRI examinations. The detailed scan parameters for T2WI were: repetition time (TR), 7,245 ms; echo time (TE), 85 ms; slice thickness, 1 mm; slice spacing, 0 mm; number of slices, 20; field of view, 6×6 cm2; matrix, 352×320; echo-train length, 10; bandwidth, 41.67 kHz. For the synthetic MRI, a two-dimensional MDME pulse sequence was applied to acquire both the axial and sagittal sections. This synthetic MRI sequence used was a multisaturation delay multi-echo fast spin-echo sequence comprising four automatically calculated saturation delays (inversion times, TI) and two TEs (27,28). Hence, the result of each MDME acquisition was 8 (complex) images per slice (4 saturation delays, at 2 TEs). The detailed scan parameters for the MDME sequences were: auto TR range, 4,400 to 4,517 ms; TE, 24.3 and 121.4 ms; slice thickness, 2 mm; slice spacing, 0 mm; number of slices, 20; field of view, 9×9 cm2; matrix, 352×352; echo-train length, 16; bandwidth, 27.78 kHz. The total scanning time of synthetic MRI was 13 minutes and 4 seconds for each model. No respiratory gating of animals was used during the examination. The respiratory rates and body core temperature were not monitored because our experiment was carried out by 3.0 T MRI scanner, which did not provide a technical platform to monitor these indicators.
The quantitative maps (T1 and T2), synthetic T1-weighted (sT1w), and sT2w images were simultaneously generated from the MDME raw data, using an offline post-processing software (synthetic MRI 8.0; Synthetic MR, Linköping, Sweden). Specifically, to retrieve T1 and T2 maps while accounting for B1 inhomogeneity, a least-square fit was performed on the signal intensity S of each pixel of the 8 images per slice according to (18):
where A is an overall intensity scaling factor taking into account the coil sensitivity, the RF chain amplification, and the voxel volume; α is the applied 90 degrees excitation flip angle, and θ is the applied 120 degrees saturation pulse angle. Once T1 and T2 values are calculated, the contrast-weighted images, such as T1w, T2w images, can be synthesized in combination with virtual scanner settings for TE, TR, and TI. The overall processing time is less than one minute.
The values of both BAT and WAT were measured using the means of regions of interest (ROI)-based method (28,29). T2w images showed good contrast compared to the T2 map, so the ROI contours of BAT were manually delineated surrounding the edge of the interscapular BAT (IBAT) in the axial sT2w images slice-by-slice while avoiding the surrounding vessels. Also, comparative ROIs were manually drawn in the dorsal subcutaneous WAT, which was present on the same slices. Thereafter, the mean quantitative values of both IBAT and WAT were automatically calculated across all the pixels in the ROIs using the synthetic MRI software. The delineation of ROIs for each slice was independently conducted by a radiologist with 10 years of experience in MRI, and the ROIs were inspected for accuracy by another radiologist with 15 years of experience in MRI.
To quantify the IBAT volume, the ROIs were manually delineated around the margin of the IBAT on contiguous slices of the axial sT2w images by utilizing the GE post-processing workstation (GE Advanced Workstation 4.7) (Figure 1). Subsequently, the volume of the IBAT was measured by merging all ROIs. The body weight, weight of IBAT gross specimen, and IBAT volume of thirteen mice were analyzed; the remaining three mice were utilized for specimen preparation.
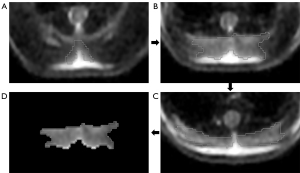
Signal-to-noise ratio (SNR) of BAT and WAT were measured (ITK-SNAP Version 3.8.0) on conventional T2WI using the means of the ROI-based method. The ROI contours of BAT and WAT drawn on conventional T2WI should be as consistent as possible with the ROI contours on the sT2w image and completed by the same two radiologists. The mean pixel value of each ROI was used as the signal intensity of the corresponding tissue, whereas the mean standard deviation of a background ROI placed posterior to the skin surface over the BAT was used as the noise. SNR was determined as the signal intensity of tissue divided by standard deviation of tissue (17).
Dissection and analysis of BAT and WAT ex vivo
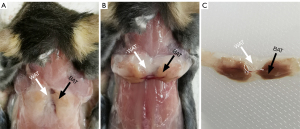
The mice were anesthetized using pentobarbital sodium at a dose of 40 mg/kg of body weight via intraperitoneal (i.p.) injection prior to the experiments. After MRI scanning, the mice were immediately euthanized by administering an overdose of pentobarbital sodium (200 mg/kg, i.p.). A surgical incision was then done in the interscapular region to conduct a visual inspection of the IBAT and the adjacent dorsal subcutaneous WAT layer. The BAT specimens in the interscapular region were excised and weighed, whereas the dorsal subcutaneous WATs were dissected. During BAT dissection, the interference of WAT on BAT was minimized by removing the neighboring WAT that was visible with the naked eye, ensuring the excision of IBAT depots only. The BAT and WAT samples were then stored in 10% normal buffered formalin to perform hematoxylin and eosin (HE), Prussian blue, and UCP1 immunohistochemistry (IHC) staining.
HE, Prussian blue, and IHC staining
The tissues were excised, fixed in 10% formalin for 24 h, embedded into paraffin, serially sectioned to obtain 4 µm sections, deparaffined, and rehydrated. The sections were stained using HE staining according to standard procedures. Prussian blue staining was performed using freshly prepared equal proportions of potassium hexacyanoferrate and hydrochloric acid. After one hour, the sections were counterstained with nuclear fast red, dehydrated, and placed on slides under coverslips.
For performing UCP1 IHC staining, the sections were first dewaxed and then rehydrated. Antigen retrieval was carried out in EDTA buffer (PH 9) by boiling the tissue sections in a microwave oven for 8 min at medium power and then stopping heating for 8 minutes. Finally, samples were heated for 7 minutes at medium and low power. The sections were then incubated with 3% hydrogen peroxide for 25 min to block endogenous peroxidases. Bovine serum albumin (3%) was added to the sections after rinsing them in phosphate-buffered saline. Subsequently, the sections were incubated overnight with primary antibodies at 4 °C; the primary antibodies were rabbit anti-UCP1 (1:500, ab 10983; Abcam, Cambridge, United Kingdom). Finally, the sections were incubated with a horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G secondary antibody (1:200, GB23303, Servicebio, Wuhan, China) for 50 min.
Statistical analysis
All statistical analyses were performed using SPSS Statistics version 25.0 (IBM Corp, NY, USA) and MedCalc Statistical Software version 19.1 (MedCalc Software bv, Ostend, Belgium). The comparisons of both quantitative parameters (T1 and T2) between BAT and WAT were conducted using either a pairwise t-test or Wilcoxon test according to the data distribution. A receiver operating characteristic (ROC) curve was generated for each parameter to assess the parameter’s diagnostic performance in differentiating BAT from WAT using the MedCalc software. The optimal thresholds for differentiation of BAT and WAT, sensitivity, and specificity were determined using Youden’s J statistic. Meanwhile, a ROC curve was plotted to assess the diagnostic performance of SNR in differentiating BAT from WAT using the MedCalc software. The sensitivity and specificity were also determined using Youden’s J statistic. The pairwise comparisons of areas under curves (AUCs) among T1, T2, and SNR were made using the method described by DeLong et al. (30). Spearman correlation analysis was conducted to evaluate the association between the synthetic MRI-derived IBAT volume and the weight of IBAT gross specimen and between the synthetic MRI-derived IBAT volume and the mouse body weights. A value of P<0.05 was considered statistically significant.
Results
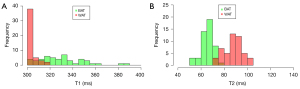
The mean and range of T1 and T2 values of the BAT and WAT are shown in Table 1. The mean T1 values of BAT were significantly higher than those of WAT (pairwise t-test, P<0.001), whereas the mean T2 values of BAT were significantly lower compared to WAT in all mice (pairwise Wilcoxon test, P<0.001). Figure 2 shows the number of overlapping of T1 and T2 in BAT and WAT, respectively.
Table 1
| Parameter (ms) | BAT | WAT | P value | |||
|---|---|---|---|---|---|---|
| Mean ± SD | range | Mean ± SD | range | |||
| T1 | 330.3±19.57 | 301–387 | 304.42±4.14 | 300–319 | <0.001 | |
| T2 | 66.06±5.06 | 54–76 | 88.23±7.68 | 71–102 | <0.001 | |
T1, longitudinal relaxation time; T2, transverse relaxation time; BAT, brown adipose tissue; WAT, white adipose tissue; SD, standard deviation.
The AUC of the T1, T2 values, and SNR for differentiating BAT from WAT was 0.942, 0.995, and 0.915, respectively (Figure 3). The AUC of the T2 values was higher than that of T1 and SNR (P=0.04, P=0.007, respectively), and there was no significant difference between the AUC of T1 and SNR using the DeLong test (P>0.05). The optimal cut-off value for T2 was 76 ms in the differentiation between BAT and WAT (100% sensitivity and 93.7% specificity), and the optimal cut-off value for T1 was 312 ms in the differentiation between BAT and WAT (85.4% sensitivity and 95.8% specificity).
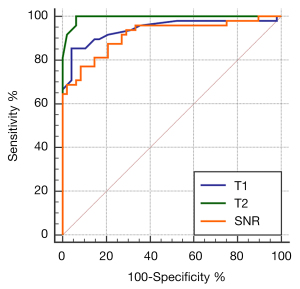
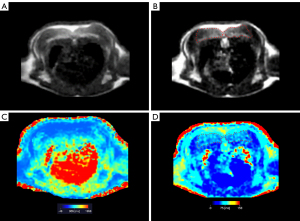
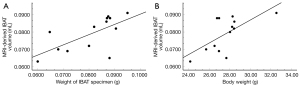
Figure 4 shows the sT1w image, sT2w image, T1 map, and T2 map for the mouse BAT. The synthetic MRI-derived IBAT volume for each mouse was measured based on the sT2w images (Figure 1). A moderate correlation was observed between the synthetic MRI-derived IBAT volume and the weight of the IBAT gross specimen (r=0.662, P=0.014). The synthetic MRI-derived IBAT volume moderately positively correlated with the mouse body weight (r=0.653, P=0.016) (Figure 5). Figure 6 shows that the visualized IBAT and WAT on the MR images were confirmed to be BAT and WAT via autopsy.
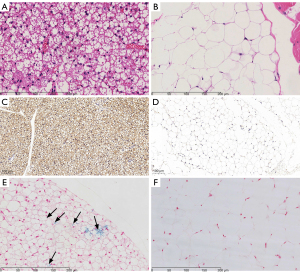
HE, UCP1, and Prussian blue staining were used to identify the BAT and WAT (Figure 7). The BAT was characterized by small multilocular adipocytes containing a large amount of cytoplasm and abundant blood vessels. The WAT exhibited large unilocular lipid droplets containing an eccentric nucleus within the cell and low amounts of cytoplasm. In addition, a positive expression of UCP1 was observed in BAT but not in WAT (Figure 7C,7D). Figure 7E shows the presence of iron particles, which appeared as blue spots in the BAT sections stained with Prussian blue, but not in the WAT (Figure 7F).
Discussion
The WAT is often referred to as “bad fat”, as its excessive accumulation may lead to obesity, type 2 diabetes, cardiovascular disease, and cancer. In contrast, the BAT is referred to as “good fat,” as it consumes energy to generate heat, thereby maintaining body temperature. The BAT is also considered a target tissue for treating metabolic diseases, such as obesity and diabetes. It is therefore of clinical interest to quantify BAT and estimate its volume in vivo.
The T1 and T2 maps are fundamental signal-formation parameters in MRI (21,31,32). The T1 has been associated with tissue fat content (33), free water content (34), the categories and concentration of macromolecules (35), such as myelin (36), and iron content (34,35). The change in T2 has been mainly related to the variation in water (23) and iron contents (37); it has also been associated with macromolecules (23) and tissue fat content (38). T1 and T2 values were also influenced by molecular motion. Small, rapidly rotating molecules (like free water) have long T1 and T2 times. As molecular motion slows (as in proteins and dense solids), T2 shortens, and T1 increases (39).
Synthetic MRI is a novel MRI approach that may be used to simultaneously obtain multiple relaxation maps as well as contrast-weighted images in a single scan (19-24). Importantly, synthetic MRI applies a saturation pulse rather than an inversion pulse, and as the starting position of the T1 relaxation curve is a function of the B1 field, it is possible to measure the local B1 field at the same time. The estimated B1 field can be used to correct the effects of local deviations in flip angle (18,40). Furthermore, synthetic MRI is reportedly accurate (41) and reproducible (41,42), and it has also been applied in several clinical areas. Specifically, Lee et al. (23) evaluated the feasibility and accuracy of synthetic MRI T2 mapping compared with conventional T2 mapping. The phantom study showed an excellent correlation between the T2 values obtained by the two approaches. In the study of Kumar et al. (17), synthetic MRI of the knee was accurate for T1, T2, and proton density quantification. Also, simultaneously generated morphologic MR images had detection rates of structural abnormalities similar to those of conventional MR images, with similar acquisition time. In the present study, the AUC of T2 values for differentiating BAT from WAT was higher than that of SNR on conventional T2WI, and there was no significant difference between the AUC of T1 and SNR. Hence, T2 values obtained from synthetic MRI were superior to conventional T2WI in distinguishing BAT and WAT, and T1 values and conventional T2WI showed similar performance in distinguishing BAT and WAT. It is well-known that breath-induced motion artifact is one of the crucial factors that may influence the accuracy of quantitative MRI mapping techniques. The synthetic MRI showed excellent performance in motionless organs such as knees and brains. Other motional organs, including the liver and heart, were easily affected by the respiratory motion artifacts. The BAT of the mouse is predominantly located in the interscapular region, which is barely influenced by the respiratory movement. Besides, we wrapped the chest and abdomen of each mouse with a piece of cloth during the examination, which had an auxiliary effect on minimizing the possible motion artifacts induced by breathing. However, in order to apply synthetic MRI in the liver or the heart, respiratory or cardiac triggered sequences are needed. In this study, the relaxation maps and contrast-weighted images obtained using synthetic MRI were investigated for estimating the volume of BAT.
It was observed that the mean T1 values in WAT were significantly lower than those in BAT, which indicated a higher fat level in WAT, as the T1 values were negatively correlated with tissue fat content (33). This observation is consistent with previous studies, which have shown that WAT has a higher fat content and stronger resonance strength of fat than BAT (4,5); the T1 values of fat decrease as the fat fraction increases. In addition, the T1 is associated with water content (34). BAT contains intracellular and extracellular water (4), leading to long T1. The T1 is also influenced by the iron content (34,35). The presence of iron in BAT is associated with a low T1 value, and a large amount of water content in BAT corresponds to high T1 values. In our study, the T1 values of BAT were higher than those of WAT. Therefore, we speculated that the water content was the major cause of T1 variation between BAT and WAT. In addition, the differences in the T1 values between BAT and WAT may be attributed to the differences in macromolecule characteristics, such as triglyceride composition and degree of lipid saturation in the two adipose tissues (15,33).
The BAT was also found to show lower T2 values (mean: 66.06±5.06 ms) than WAT (mean: 88.23±7.68 ms). Chen et al. (2) reported that the BAT in rats could be identified with a conventional MESE sequence using a 9.4 T MRI scanner. In their study, T2 values of BAT (mean: 57.76±3.92 ms) were significantly lower than those of WAT (mean: 83.07±2.20 ms). Following are possible explanations for the lower T2 value of BAT: BAT contains large amounts of mitochondria and intracellular iron (2,11), which results in a higher magnetic field susceptibility of the tissue. The presence of iron in the BAT was confirmed via Prussian blue staining. High iron content was associated with a reduced T2 value (6). Next, the rich vasculature in the BAT can lead to a decreased T2 value (2,11). Finally, fat fluidity differences caused by variances in saturated fatty acid content may also cause differences in relaxation time of T1 and T2. A study of excised rodent tissue samples examined with proton MR spectroscopy (15) showed that triglyceride in BAT was more saturated than in WAT, and BAT had a lower proportion of unsaturated triglycerides. Animal studies (43) showed that the triglyceride composition of BAT could be lower in UFA and polyunsaturated fatty acid (PUFA) when deriving fatty acid content in large parts from de novo synthesis from glucose. More saturated fatty acid lowers the fluidity (26), which was also consistent with a lower UFA content in BAT (15,43). Lower fluidity shortens T2 and lengthens T1 (26,44).
Currently, there is no widely accepted standard method for the measurement of the volume of the BAT. In this study, the volume of IBAT was measured via manual segmentation. The obtained results revealed that the volume of IBAT estimated via synthetic MRI was moderately positively correlated with the weight of the excised IBAT specimen, which was similar to that reported by Chen et al. (2). Assuming that the BAT density was approximately equal among the mice used in this study, it can be indirectly inferred that the IBAT volume measured via synthetic MRI may be positively correlated with the actual IBAT volume. Endocrinologists need to establish imaging biomarkers to accurately evaluate the treatment response of a drug in obese or diabetic patients undergoing anti-obesity or anti-diabetic treatments. Using synthetic MRI, the changes in the BAT tissue-relaxation values and the BAT volume may be measured and compared before and after anti-obesity or anti-diabetic treatment, thus allowing for the potential longitudinal assessment of the efficacy of anti-obesity or anti-diabetic drugs.
In the present study, the volume of the IBAT estimated via synthetic MRI was moderately positively correlated with the body weight of the mice, in agreement with the results reported by Hu et al. (45) and Chen et al. (2). However, this result may only be applicable to lean mice, and whether it applies to obese mice needs further validation. In ob/ob obese mice, swollen BAT can be observed due to fat accumulation, while the capillary density, tissue blood flow, the concentration of UCP1 in mitochondria, and the thermogenesis of BAT are all reduced compared to lean mice (46). This evidence suggests that the tissue composition of BAT in obese mice changes, the effective components for heat production decrease, and the fat content increases. However, the relationship between the overall volume of BAT and body weight still needs to be investigated.
The current study has several limitations. Firstly, while the drainage approach is the most commonly used method for measuring the volume of BAT, it was not used in the present study as the density of the BAT is lower than that of water, and the volume of BAT is small, which makes it challenging to avoid artificial measurement errors. Secondly, the inability to completely remove the surrounding WAT and residual blood from the BAT during BAT dissection might have affected the weight of the dissected BAT in a few mice, thus resulting in a higher value than the actual weight. Thirdly, this study lacked a comparison between the values obtained using synthetic MRI and conventional MESE techniques. These values could provide meaningful data if compared with quantitative measures using conventional T1 and T2 quantitative mapping techniques in the same mouse. Lastly, in this study, we did not add conventional sequences to the imaging protocol. So, the analysis of conventional sequences in distinguishing BAT from WAT is lacking. Finally, this study utilized relaxation measures using synthetic MRI to characterize the BAT in the resting state, but did not further explore the quantitative magnetic profiles of BAT in the activated state further, which should be addressed by future studies.
In conclusion, our study demonstrated the feasibility of differentiating BAT from WAT and quantifying BAT volume in vivo via quantitative measurements using synthetic MRI. Synthetic MRI may be an ideal tool to longitudinally evaluate the BAT alteration induced by anti-obesity or anti-diabetic drugs. Future studies are needed to explore further the quantitative magnetic profiles of BAT in the activated state.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81971684, 81771908, 82001882), the Scientific Research Project of Traditional Chinese Medicine Bureau of Guangdong Province (20211173), and the Natural Science Foundation of Guangdong Province (2021A1515011442).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/qims-20-1344). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Sun Yat-sen University (NO. [2017]104) and performed in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lim S, Honek J, Xue Y, Seki T, Cao Z, Andersson P, Yang X, Hosaka K, Cao Y. Cold-induced activation of brown adipose tissue and adipose angiogenesis in mice. Nat Protoc 2012;7:606-15. [Crossref] [PubMed]
- Chen YI, Cypess AM, Sass CA, Brownell AL, Jokivarsi KT, Kahn CR, Kwong KK. Anatomical and functional assessment of brown adipose tissue by magnetic resonance imaging. Obesity (Silver Spring) 2012;20:1519-26. [Crossref] [PubMed]
- Franz D, Syväri J, Weidlich D, Baum T, Rummeny EJ, Karampinos DC. Magnetic Resonance Imaging of Adipose Tissue in Metabolic Dysfunction. Rofo 2018;190:1121-30. [Crossref] [PubMed]
- Verma SK, Nagashima K, Yaligar J, Michael N, Lee SS, Xianfeng T, Gopalan V, Sadananthan SA, Anantharaj R, Velan SS. Differentiating brown and white adipose tissues by high-resolution diffusion NMR spectroscopy. J Lipid Res 2017;58:289-98. [Crossref] [PubMed]
- Hu HH, Perkins TG, Chia JM, Gilsanz V. Characterization of human brown adipose tissue by chemical-shift water-fat MRI. AJR Am J Roentgenol 2013;200:177-83. [Crossref] [PubMed]
- Hu Q, Chen X, Liu J, Di W, Lv S, Tang L, Ding G. Targeted Molecular Magnetic Resonance Imaging Detects Brown Adipose Tissue with Ultrasmall Superparamagnetic Iron Oxide. Biomed Res Int 2018;2018:3619548 [Crossref] [PubMed]
- Cypess AM, Lehman S, Williams G, Tal I, Rodman D, Goldfine AB, Kuo FC, Palmer EL, Tseng YH, Doria A, Kolodny GM, Kahn CR. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 2009;360:1509-17. [Crossref] [PubMed]
- Wang M, Luo Y, Cai H, Xu L, Huang M, Li C, Dong Z, Li ZP, Feng ST. Prediction of type 2 diabetes mellitus using noninvasive MRI quantitation of visceral abdominal adiposity tissue volume. Quant Imaging Med Surg 2019;9:1076-86. [Crossref] [PubMed]
- Villarroya F, Cereijo R, Villarroya J, Giralt M. Brown adipose tissue as a secretory organ. Nat Rev Endocrinol 2017;13:26-35. [Crossref] [PubMed]
- Marzola P, Boschi F, Moneta F, Sbarbati A, Zancanaro C. Preclinical In vivo Imaging for Fat Tissue Identification, Quantification, and Functional Characterization. Front Pharmacol 2016;7:336. [Crossref] [PubMed]
- Hu HH, Hines CD, Smith DL Jr, Reeder SB. Variations in T(2)* and fat content of murine brown and white adipose tissues by chemical-shift MRI. Magn Reson Imaging 2012;30:323-9. [Crossref] [PubMed]
- Huber FA, Del Grande F, Rizzo S, Guglielmi G, Guggenberger R. MRI in the assessment of adipose tissues and muscle composition: how to use it. Quant Imaging Med Surg 2020;10:1636-49. [Crossref] [PubMed]
- Chan XHD, Balasundaram G, Attia ABE, Goggi JL, Ramasamy B, Han W, Olivo M, Sugii S. Multimodal imaging approach to monitor browning of adipose tissue in vivo. J Lipid Res 2018;59:1071-8. [Crossref] [PubMed]
- Guo Y, Li Y, Yang Y, Tang S, Zhang Y, Xiong L. Multiscale Imaging of Brown Adipose Tissue in Living Mice/Rats with Fluorescent Polymer Dots. ACS Appl Mater Interfaces 2018;10:20884-96. [Crossref] [PubMed]
- Hamilton G, Smith DL Jr, Bydder M, Nayak KS, Hu HH. MR properties of brown and white adipose tissues. J Magn Reson Imaging 2011;34:468-73. [Crossref] [PubMed]
- Gifford A, Towse TF, Walker RC, Avison MJ, Welch EB. Characterizing active and inactive brown adipose tissue in adult humans using PET-CT and MR imaging. Am J Physiol Endocrinol Metab 2016;311:E95-E104. [Crossref] [PubMed]
- Kumar NM, Fritz B, Stern SE, Warntjes JBM, Lisa Chuah YM, Fritz J. Synthetic MRI of the Knee: Phantom Validation and Comparison with Conventional MRI. Radiology 2018;289:465-77. [Crossref] [PubMed]
- Hagiwara A, Warntjes M, Hori M, Andica C, Nakazawa M, Kumamaru KK, Abe O, Aoki S. SyMRI of the Brain: Rapid Quantification of Relaxation Rates and Proton Density, With Synthetic MRI, Automatic Brain Segmentation, and Myelin Measurement. Invest Radiol 2017;52:647-57. [Crossref] [PubMed]
- Arita Y, Takahara T, Yoshida S, Kwee TC, Yajima S, Ishii C, Ishii R, Okuda S, Jinzaki M, Fujii Y. Quantitative Assessment of Bone Metastasis in Prostate Cancer Using Synthetic Magnetic Resonance Imaging. Invest Radiol 2019;54:638-44. [Crossref] [PubMed]
- Dean DC 3rd, Hurley SA, Kecskemeti SR, O'Grady JP, Canda C, Davenport-Sis NJ, Carlsson CM, Zetterberg H, Blennow K, Asthana S, Sager MA, Johnson SC, Alexander AL, Bendlin BB. Association of Amyloid Pathology With Myelin Alteration in Preclinical Alzheimer Disease. JAMA Neurol 2017;74:41-9. [Crossref] [PubMed]
- Kang KM, Choi SH, Hwang M, Yoo RE, Yun TJ, Kim JH, Sohn CH. Application of Synthetic MRI for Direct Measurement of Magnetic Resonance Relaxation Time and Tumor Volume at Multiple Time Points after Contrast Administration: Preliminary Results in Patients with Brain Metastasis. Korean J Radiol 2018;19:783-91. [Crossref] [PubMed]
- Andica C, Hagiwara A, Hori M, Haruyama T, Fujita S, Maekawa T, Kamagata K, Yoshida MT, Suzuki M, Sugano H, Arai H, Aoki S. Aberrant myelination in patients with Sturge-Weber syndrome analyzed using synthetic quantitative magnetic resonance imaging. Neuroradiology 2019;61:1055-66. [Crossref] [PubMed]
- Lee SH, Lee YH, Song HT, Suh JS. Quantitative T2 Mapping of Knee Cartilage: Comparison between the Synthetic MR Imaging and the CPMG Sequence. Magn Reson Med Sci 2018;17:344-9. [Crossref] [PubMed]
- Jung Y, Gho SM, Back SN, Ha T, Kang DK, Kim TH. The feasibility of synthetic MRI in breast cancer patients: comparison of T2 relaxation time with multiecho spin echo T2 mapping method. Br J Radiol 2018; Epub ahead of print. [Crossref] [PubMed]
- Poon CS, Henkelman RM. Practical T2 quantitation for clinical applications. J Magn Reson Imaging 1992;2:541-53. [Crossref] [PubMed]
- Ouwerkerk R, Hamimi A, Matta J, Abd-Elmoniem KZ, Eary JF, Abdul Sater Z, Chen KY, Cypess AM, Gharib AM, Proton MR. Spectroscopy Measurements of White and Brown Adipose Tissue in Healthy Humans: Relaxation Parameters and Unsaturated Fatty Acids. Radiology 2021;299:396-406. [Crossref] [PubMed]
- Kang KM, Choi SH, Hwang M, Yun TJ, Kim JH, Sohn CH. T1 Shortening in the Globus Pallidus after Multiple Administrations of Gadobutrol: Assessment with a Multidynamic Multiecho Sequence. Radiology 2018;287:258-66. [Crossref] [PubMed]
- Lee SM, Choi YH, You SK, Lee WK, Kim WH, Kim HJ, Lee SY, Cheon H. Age-Related Changes in Tissue Value Properties in Children: Simultaneous Quantification of Relaxation Times and Proton Density Using Synthetic Magnetic Resonance Imaging. Invest Radiol 2018;53:236-45. [Crossref] [PubMed]
- Cui Y, Han S, Liu M, Wu PY, Zhang W, Zhang J, Li C, Chen M. Diagnosis and Grading of Prostate Cancer by Relaxation Maps From Synthetic MRI. J Magn Reson Imaging 2020;52:552-64. [Crossref] [PubMed]
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Gonçalves FG, Serai SD, Zuccoli G. Synthetic Brain MRI: Review of Current Concepts and Future Directions. Top Magn Reson Imaging 2018;27:387-93. [Crossref] [PubMed]
- Cheng HL, Stikov N, Ghugre NR, Wright GA. Practical medical applications of quantitative MR relaxometry. J Magn Reson Imaging 2012;36:805-24. [Crossref] [PubMed]
- Garnov N, Linder N, Schaudinn A, Blüher M, Karlas T, Schütz T, Dietrich A, Kahn T, Busse H. Comparison of T1 relaxation times in adipose tissue of severely obese patients and healthy lean subjects measured by 1.5 T MRI. NMR Biomed 2014;27:1123-8. [Crossref] [PubMed]
- Gelman N, Ewing JR, Gorell JM, Spickler EM, Solomon EG. Interregional variation of longitudinal relaxation rates in human brain at 3.0 T: relation to estimated iron and water contents. Magn Reson Med 2001;45:71-9. [Crossref] [PubMed]
- Rooney WD, Johnson G, Li X, Cohen ER, Kim SG, Ugurbil K, Springer CS Jr. Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med 2007;57:308-18. [Crossref] [PubMed]
- Lutti A, Dick F, Sereno MI, Weiskopf N. Using high-resolution quantitative mapping of R1 as an index of cortical myelination. Neuroimage 2014;93:176-88. [Crossref] [PubMed]
- Gelman N, Gorell JM, Barker PB, Savage RM, Spickler EM, Windham JP, Knight RA. MR imaging of human brain at 3.0 T: preliminary report on transverse relaxation rates and relation to estimated iron content. Radiology 1999;210:759-67. [Crossref] [PubMed]
- Chaland B, Mariette F, Marchal P, De Certaines J. 1H nuclear magnetic resonance relaxometric characterization of fat and water states in soft and hard cheese. J Dairy Res 2000;67:609-18. [Crossref] [PubMed]
- Bottomley PA, Foster TH, Argersinger RE, Pfeifer LM. A review of normal tissue hydrogen NMR relaxation times and relaxation mechanisms from 1-100 MHz: dependence on tissue type, NMR frequency, temperature, species, excision, and age. Med Phys 1984;11:425-48. [Crossref] [PubMed]
- Warntjes JB, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn Reson Med 2008;60:320-9. [Crossref] [PubMed]
- Krauss W, Gunnarsson M, Andersson T, Thunberg P. Accuracy and reproducibility of a quantitative magnetic resonance imaging method for concurrent measurements of tissue relaxation times and proton density. Magn Reson Imaging 2015;33:584-91. [Crossref] [PubMed]
- Hagiwara A, Hori M, Cohen-Adad J, Nakazawa M, Suzuki Y, Kasahara A, Horita M, Haruyama T, Andica C, Maekawa T, Kamagata K, Kumamaru KK, Abe O, Aoki S. Linearity, Bias, Intrascanner Repeatability, and Interscanner Reproducibility of Quantitative Multidynamic Multiecho Sequence for Rapid Simultaneous Relaxometry at 3 T: A Validation Study With a Standardized Phantom and Healthy Controls. Invest Radiol 2019;54:39-47. [Crossref] [PubMed]
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004;84:277-359. [Crossref] [PubMed]
- Robinson MD, Cistola DP. Nanofluidity of fatty acid hydrocarbon chains as monitored by benchtop time-domain nuclear magnetic resonance. Biochemistry 2014;53:7515-22. [Crossref] [PubMed]
- Hu HH, Smith DL Jr, Nayak KS, Goran MI, Nagy TR. Identification of brown adipose tissue in mice with fat-water IDEAL-MRI. J Magn Reson Imaging 2010;31:1195-202. [Crossref] [PubMed]
- Branca RT, He T, Zhang L, Floyd CS, Freeman M, White C, Burant A. Detection of brown adipose tissue and thermogenic activity in mice by hyperpolarized xenon MRI. Proc Natl Acad Sci U S A 2014;111:18001-6. [Crossref] [PubMed]


