
Applying multisequence MRI radiomics of the primary tumor and lymph node to predict HPV-related p16 status in patients with oropharyngeal squamous cell carcinoma
Introduction
Over the last decade, the prevalence of human papillomavirus (HPV)-positive oropharyngeal squamous cell carcinoma (OPSCC) has rapidly increased, especially in the Western world (1). This is in contrast with the prevalence of HPV in China, where for a long time the prevalence has been much lower. However, the relative percentage of HPV-related OPSCC in China was recently reported to be close to that of many Western countries, and a strong predilection for the tonsillar lesion was discovered (2). Patients with HPV-positive OPSCC have demonstrated superior survival and better treatment response compared with HPV-negative patients (3). The evaluation of HPV status is essential for making treatment plans and predicting prognosis. Meanwhile, HPV status strongly correlates with p16 expression as detected by immunochemistry (IHC) (4). Therefore, p16 expression is always used to evaluate HPV status.
Conventional and advanced imaging methods provide useful information for the diagnosis, assessment of treatment response, and surveillance of patients with OPSCC (5-7). However, in traditional imaging analysis, the characteristics of whole-tumor imaging findings are based on a radiologist’s diagnostic experience, and intratumor heterogeneity has not been analyzed using quantitative imaging methods. Radiomics, which provides high-throughput mining of large amounts of quantitative features derived from medical imaging, is a promising tool in the decision support systems of precision medicine (8-11). Evidence from previous studies shows that radiomics features could be helpful for personalized risk stratification (12) and individual treatment decisions (13) and may also serve as prognostic indicators (14) of OPSCC.
Therefore, quantitative imaging analysis based on radiomics could provide a noninvasive approach for assessing the HPV-related p16 status of patients with OPSCC. In previously reported studies, researchers developed different models to decode the imaging phenotypes stratified by p16, based on computer tomography (CT), positron emission tomography (PET)/CT, or single-sequence magnetic resonance imaging (MRI) (15-17). However, the predictive power of these radiomics models needs further improvement. Many studies of other tumors have demonstrated that the multisequence prediction model has a better classification performance compared with the single-sequence model (18-21). We hypothesized that the radiomics model based on multisequence MRI may improve the predictive accuracy of p16 status in OPSCC. Moreover, previous studies have suggested that primary tumor (PT) and lymph node (LN) imaging features may play complementary roles for each other (22); for instance, the fusion of PT and LN features was found to outperform PT or LN features alone in the prediction of prognosis or treatment response in squamous cell cancer (23,24). Thus, in the current study, we aimed to evaluate the predictive values of radiomics features derived from PT and LN images based on multisequence MRI for the HPV-related p16 status of OPSCC patients. We present the following article in accordance with the STARD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-819/rc).
Methods
Patients
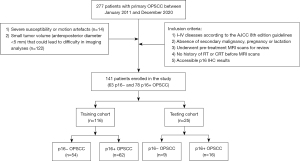
We employed a multicenter, retrospective design for this study. Patients were retrospectively and consecutively enrolled from Fudan University Shanghai Cancer Center (FUSCC) and Shanghai Ninth People’s Hospital between January 2011 and December 2020. In this period, the complete imaging, pathology, and clinical data of patients were collected. Patients with histopathologically proven primary OPSCC who met the following inclusion criteria were included in our analyses: (I) presence of stages I–IV OPSCC, restaged according to the American Joint Commission on Cancer (AJCC; eighth edition staging manual) guidelines; (II) absence of secondary malignancy, pregnancy, or lactation; (III) availability of pretreatment MRI scans for review; (IV) no history of radiotherapy (RT) or chemoradiotherapy (CRT) before the MRI scans; and (V) accessible p16 IHC results. The exclusion criteria were as follows: (I) severe susceptibility or motion artefacts (n=14) or (II) small tumor volume (anteroposterior diameter <5 mm) that could lead to difficulty in imaging analyses (n=122). The time interval between pathological examination and MRI was within 1 week. A total of 141 patients with primary OPSCC who met the criteria were finally identified. The training cohort consisted of 116 patients from the FUSCC and the testing cohort consisted of 25 patients from Shanghai Ninth People’s Hospital. Baseline clinical variables were collected, including age, gender, TNM stage, smoking history, treatment methods, and the location of lesions. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics board of the FUSCC, and individual consent for this retrospective analysis was waived. The workflow of this study is displayed in Figure 1. The study protocol can be obtained from the corresponding author.
p16 expression assessment
A commercial antibody of p16 (25) was used to detect the p16 expression of all patients. p16 with a 70% nuclear and cytoplasmic staining in IHC was employed as the cutoff. IHC was performed on 4-µm sections of paraffin-embedded tissues to determine the expression level of p16 protein. In brief, the slides were incubated in p16 antibody (M78710, Dako Products, Agilent Technologies) diluted 1:200 at 4 ℃ overnight and incubated in a second antibody (Dako Products, Agilent Technologies) at 37 ℃ for 40 minutes. Then, the slides were stained with the avidin–biotin peroxidase method using diaminobenzidine (DAB) and then counter-stained with hematoxylin. Before and after each step mentioned above, a wash was completed 3 times with phosphate-buffered saline (PBS). The testing of p16 expression were performed using the EnVision FLEX high-pH visualization system (Dako Products, Agilent Technologies) according to the manufacturer’s instructions. Slides were examined under a light microscope for evaluation.
MR imaging protocol
The MR imaging was performed using 3.0 T scanners (Siemens Healthineers; GE Healthcare) with a dedicated 16-channel head-neck synergic coil. The MR imaging protocol included axial T1-weighted imaging (T1WI) and T2-weighted imaging (T2WI), coronal short tau inversion recovery (STIR) imaging, and contrast-enhanced T1WI (CE-T1WI). A rapid bolus of gadolinium contrast agent (Magnevist; Bayer HealthCare Pharmaceuticals Inc.) was injected intravenously at a dose of 0.1 mmol/kg of body weight. Axial, coronal, and sagittal CE-T1WIs were obtained. Axial T2WI and CE-T1WI were chosen for further imaging analysis. The following imaging parameters for the Siemens scanner were applied: axial T2WI [repetition time (TR), 2,500 ms; time to echo (TE), 78 ms; slice thickness, 5 mm; matrix, 384×324; pixel spacing, 0.7 mm × 0.7 mm] and axial CE-T1WI (TR, 4.2 ms; TE, 1.5 ms; slice thickness, 3 mm; matrix, 384×324; pixel spacing, 0.7 mm × 0.7 mm). The imaging parameters for the GE scanner were as follows: axial T2WI (TR, 3,100 ms; TE, 86.7 ms; slice thickness, 6 mm; matrix, 320×160; pixel spacing, 0.84 mm × 0.84 mm) and axial CE-T1WI (TR, 220 ms; TE, 2.5 ms; slice thickness, 6 mm; matrix, 256×160; pixel spacing, 0.84 mm × 0.84 mm). In the training cohort, 65 patient scans were acquired using the Siemens scanner and 51 were acquired using the GE scanner. All 25 patients in the testing cohort were scanned with the Siemens device.
Image analysis
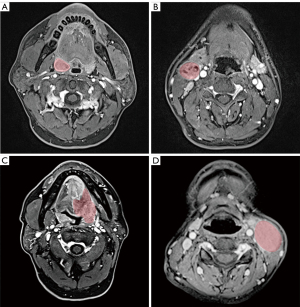
Two radiologists with 9 years’ and 5 years’ experience, respectively, in head and neck radiology assessed all of the images taken for each patient and staged the tumors according to the established staging system by consensus. If there was an inconsistent interpretation, a third radiologist would review the scans to reach a consensus. The regions of interest (ROIs) were manually contoured by one radiologist, and all segmentations were reviewed by another radiologist. All ROIs of PT and LN were delineated on both T2WI and CE-T1WI. When delineating the certain ROIs on an image, the radiologists were allowed to review other MR sequences to improve their delineations. Segmentation of the PT and maximum LN were separately contoured using ITK-SNAP software v.3.6 (http://www.itksnap.org/pmwiki/pmwiki.phpn=Downloads.SNAP3) with a semiautomated graphical user interface. All metastatic LNs were evaluated on axial, coronal, and sagittal images. The LN with the largest short-axis diameter was chosen as the maximum LN. Figure 2 shows the representative cases of HPV-positive and HPV-negative OPSCC with a manual ROI. In order to ensure all readers were blinded to the patients, all images were anonymous.
Radiomics model development
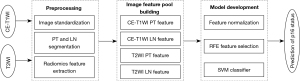
Since the MR images were scanned using different scanners, the different distributions of image spacing and gray values might have influenced the uniformity of tumor imaging features. Thus, the 3D B-spline interpolation algorithm was first used to resample the CE-T1W and T2W images with a new image resolution of 1 mm × 1 mm × 7 mm. The gray values of the MR image were also normalized with a scale of 100 by centering it at the mean with standard deviation to ensure the same sequence in different scanners used the same gray scale.
After MR image standardization, the original images were generated. The Laplacian of Gaussian (LoG) and wavelet images were generated from original images using a 3D LoG image filter and 3D wavelet image filter. Then, the LoG features and wavelet features were computed based on the LoG and wavelet images, respectively. Therefore, the following 3 types of image features were acquired: original features, LoG features, and wavelet features. Each type of image feature involved a shape feature, histogram feature, and texture feature. Moreover, the texture features were extracted from the gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level dependence matrix (GLDM), and gray-level size zone matrix (GLSZM). Finally, a total of 2,092 radiomics features were extracted from the PT and LN based on multisequence imaging consisting of 200 original features, 516 LoG features, and 1,376 wavelet features. The publicly available Pyradiomics (https://aim.hms.harvard.edu/pyradiomics) and SimpleITK (https://simpleitk.org/) libraries were used for the imaging preprocessing and feature extraction.
A standard scaler was used to normalize each type of image feature by removing the mean and scaling to the unit variance. Then, the recursive feature elimination (RFE) feature selector configured with the linear support vector machine (SVM) classifier was applied to remove redundancy features and select the optimal image features. Finally, the SVM classifier was employed to build the machine learning-based classification models depending on the p16 status. After the radiomics models were built based on the training data set, independent testing data sets were used to assess the performance of the models. Figure 3 shows the workflow of the proposed radiomics model. The Python v.3.7 Scikit-learn v.0.21 (http://scikit-learn.org/) package was used for feature normalization, feature selection, and SVM implementation.
Statistical analysis
The continuous variables are expressed as medians, and the categorical variables are expressed as percentages. The Mann-Whitney test, independent t-test, chi-squared test, and Fisher exact test were used, as appropriate, for the univariate analyses. The area under the receiver operation characteristic (ROC) curve (AUC) value and the corresponding 95% confidence interval (CI) were computed to evaluate and compare the prediction performance generated with the different MRI features. Meanwhile, other quantitative evaluation indices involving accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and odds ratio (OR) were also computed to evaluate the predictive performance. Differences in the AUC values between different models were estimated using the Delong test, and false discovery rate (FDR) correction was applied to multiple testing for corrections of cohousing effects. Moreover, the F1 score and precision were used to evaluate the performance of the model in the testing cohort for class imbalance. The pathological results and imaging predictions were compared and analyzed according to the optimal PT-LN fused model based on multisequence imaging. The statistical analyses were performed using the Fisher exact test.
All the processes of model development and statistical data analysis were performed using Python v.3.7 programming software (https://www.python.org/) on a computer with an Intel Core i7-8700 central processing unit (CPU) with 3.2 GHz ×2, 16 gigabyte (GB) random access memory (RAM), and a NVIDIA GeForce GTX 1,070 graphics processing unit. Our Python scripts are available in the GitHub repository (https://github.com/GongJingUSST/OCCp16StatusPrediction).
Results
Clinical characteristics
Among the 141 examined patients with OPSCC, 63 patients were p16− and 78 patients were p16+. In the training cohort, 54 patients were p16− and 62 patients were p16+, and in the testing cohort, 9 patients were p16− and 16 patients were p16+. There were no significant differences in the P16+ or P16− distribution between the training and testing cohorts (P>0.05).
Compared with the p16+ patients, the p16− patients were older and more likely to have a history of heavy smoking consumption in both the training cohort and the testing cohort. The p16+ patients with OPSCC were more likely to have tonsillar but not nontonsillar lesions than were the p16− patients with OPSCC in both the training and testing cohorts. Due to the different staging principles for HPV-positive and HPV-negative OPSCC, the chi-squared test was not performed to compare the differences in the TNM stage between the 2 groups. Moreover, the p16− patients tended to be of advanced nodal stage. No statistically significant differences were identified for sex or treatment method based on p16 status (P>0.05). Patient characteristics stratified by p16 status are shown in Table 1.
Table 1
| Characteristics | Training cohort | Testing cohort | All patients | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All, N=116 | P16−, N=54 | P16+, N=62 | P value | All, N=25 | P16−, N=9 | P16+, N=16 | P value | All, N=141 | P16−, N=63 | P16+, N=78 | P value | |||
| Sex, n (%) | 0.794 | 0.280 | 0.347 | |||||||||||
| Male | 99 (85.3) | 47 (87.0) | 52 (83.9) | 22 (88.0) | 9 (100.0) | 13 (81.3) | 121 (85.8) | 56 (88.9) | 65 (83.3) | |||||
| Female | 17 (14.7) | 7 (13.0) | 10 (16.1) | 3 (12.0) | 0 (0.0) | 3 (18.7) | 20 (14.2) | 7 (11.1) | 13 (16.7) | |||||
| Age, median [interquartile range] | 58 [50–62] | 59 [54–67] | 56 [48–61] | 0.028 | 61 [52–65] | 65 [56–67] | 56 [48–64] | 0.042 | 58 [50–63] | 60 [54–67] | 56 [48–61] | 0.007 | ||
| Smoking, n (%) | 0.047 | 0.078 | 0.009 | |||||||||||
| Never | 47 (40.5) | 19 (35.1) | 28 (45.2) | 12 (48.0) | 2 (22.2) | 10 (62.5) | 59 (41.9) | 21 (33.3) | 38 (48.7) | |||||
| <10 pack/year | 13 (11.2) | 3 (5.6) | 10 (16.1) | 1 (4.0) | 0 (0.0) | 1 (6.3) | 14 (9.9) | 3 (4.8) | 11 (14.1) | |||||
| >10 pack/year | 56 (48.3) | 32 (59.3) | 24 (38.7) | 12 (48.0) | 7 (77.8) | 5 (31.2) | 68 (48.2) | 39 (61.9) | 29 (37.2) | |||||
| Location, n (%) | 0.002 | 0.030 | <0.001 | |||||||||||
| Tonsil | 76 (65.5) | 26 (48.1) | 50 (80.6) | 14 (56.0) | 2 (22.2) | 12 (75.0) | 90 (63.8) | 28 (44.4) | 62 (79.5) | |||||
| Base of tongue | 29 (25.0) | 19 (35.2) | 10 (16.1) | 7 (28.0) | 5 (55.6) | 2 (12.5) | 36 (25.6) | 24 (38.1) | 12 (15.4) | |||||
| Soft palate | 9 (7.8) | 7 (13.0) | 2 (3.2) | 4 (16.0) | 2 (22.2) | 2 (12.5) | 13 (9.2) | 9 (14.3) | 4 (5.1) | |||||
| Posterior wall | 2 (1.7) | 2 (3.7) | 0 (0.0) | 2 (1.4) | 2 (3.2) | 0 (0.0) | ||||||||
| Treatment, n (%) | 0.087 | 0.671 | >0.99 | |||||||||||
| RT/CRT | 102 (87.9) | 48 (88.9) | 54 (87.1) | 16 (64.0) | 5 (55.6) | 11 (68.8) | 118 (83.7) | 53 (84.1) | 65 (83.3) | |||||
| S ± RT/CRT | 14 (12.1) | 6 (11.1) | 8 (12.9) | 9 (36.0) | 4 (44.4) | 5 (31.2) | 23 (16.3) | 10 (15.9) | 13 (16.7) | |||||
| T stage, n (%) | ||||||||||||||
| T1 | 14 (12.0) | 6 (11.1) | 8 (12.9) | 3 (12.0) | 2 (22.2) | 1 (6.3) | 17 (12.1) | 8 (12.7) | 9 (11.5) | |||||
| T2 | 48 (41.4) | 21 (38.9) | 27 (43.6) | 14 (56.0) | 7 (77.8) | 7 (43.7) | 62 (44.0) | 28 (44.4) | 34 (43.6) | |||||
| T3 | 43 (37.1) | 19 (35.2) | 24 (38.7) | 5 (20.0) | 0 (0.0) | 5 (31.2) | 48 (34.0) | 19 (30.2) | 29 (37.2) | |||||
| T4 | 11 (9.5) | 8 (14.8) | 3 (4.8) | 3 (12.0) | 0 (0.0) | 3 (18.8) | 14 (9.9) | 8 (12.7) | 6 (7.7) | |||||
| N stage, n (%) | ||||||||||||||
| N0 | 2 (1.7) | 2 (3.7) | 0 (0.0) | 2 (8.0) | 0 (0.0) | 2 (12.5) | 4 (2.8) | 2 (3.2) | 2 (2.6) | |||||
| N1 | 41 (35.3) | 8 (14.8) | 33 (53.2) | 15 (60.0) | 7 (77.8) | 8 (50.0) | 56 (39.7) | 15 (23.8) | 41 (52.5) | |||||
| N2 | 25 (21.6) | 8 (14.8) | 17 (27.4) | 1 (4.0) | 0 (0.0) | 1 (6.3) | 26 (18.5) | 8 (12.7) | 18 (23.1) | |||||
| N3 | 48 (41.4) | 36 (66.7) | 12 (19.4) | 7 (28.0) | 2 (22.2) | 5 (31.2) | 55 (39.0) | 38 (60.3) | 17 (21.8) | |||||
| Overall stage, n (%) | ||||||||||||||
| I | 18 (15.5) | 1 (1.9) | 17 (27.4) | 6 (24.0) | 0 (0.0) | 6 (37.5) | 24 (17.0) | 1 (1.6) | 23 (29.5) | |||||
| II | 31 (26.8) | 1 (1.9) | 30 (48.4) | 4 (16.0) | 0 (0.0) | 4 (25.0) | 35 (24.8) | 1 (1.6) | 34 (43.6) | |||||
| III | 21 (18.1) | 6 (11.1) | 15 (24.2) | 13 (52.0) | 7 (77.8) | 6 (37.5) | 34 (24.1) | 13 (20.6) | 21 (26.9) | |||||
| IVa | 10 (8.6) | 10 (18.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (7.1) | 10 (15.9) | 0 (0.0) | |||||
| IVb | 36 (31.0) | 36 (66.6) | 0 (0.0) | 2 (8.0) | 2 (22.2) | 0 (0.0) | 38 (27.0) | 38 (60.3) | 0 (0.0) | |||||
OPSCC, oropharyngeal squamous cell carcinoma; CRT, chemoradiotherapy; RT, radiotherapy; S, surgery.
Radiomics models for P16 prediction
In total, 9 prediction models were built based on the training data set, as follows: a sole PT model, a sole LN model, and a PT-LN fusion model based on T2WI; a sole PT model, a sole LN model, and a PT-LN fusion model based on CE-T1WI; and a sole PT model, a sole LN model, and a PT-LN fusion model based on multisequence imaging (T2WI and CE-T1WI). In the T2WI-based training data set, the PT-LN fusion model yielded a statistically significant (P<0.05) better performance compared to the models based on solely PT or LN (0.91 vs. 0.52/0.78, respectively). In the CE-T1WI data set, the AUC values of the PT, LN, and PT-LN fusion models were 0.79, 0.77, and 0.74, respectively. No statistically significant differences were identified between these 3 models (P>0.05). The sole PT model based on multisequence imaging outperformed the single CE-T1WI- and T2WI-based models (0.89 vs. 0.79/0.52, respectively) with statistical significance (P<0.05). Based on the LN features alone, the multisequence and single-sequence models yielded a similar classification performance (0.70 vs. 0.77/0.78, respectively) with no statistically significant differences. Finally, the PT-LN fusion model based on multisequence imaging yielded a satisfactory classification performance with an AUC of 0.90 for the prediction of p16 expression. In terms of other evaluation metrics, including accuracy, sensitivity, specificity, PPV, NPV, and OR, the final fusion model showed better classification performance than did the other 8 models. Table 2 and Figure 4A present the performance of the models in the training cohort.
Table 2
| Models | AUC | 95% CI | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | OR |
|---|---|---|---|---|---|---|---|---|
| CE-T1WI-based | ||||||||
| Sole PT | 0.79±0.04 | 0.70–0.86 | 80.6 | 64.8 | 72.5 | 74.5 | 73.3 | 7.7 |
| Sole LN | 0.77±0.04 | 0.68–0.85 | 67.7 | 68.5 | 71.1 | 64.9 | 68.1 | 4.6 |
| PT-LN fusion | 0.74±0.05 | 0.64–0.82 | 64.5 | 68.5 | 70.2 | 62.7 | 66.4 | 3.96 |
| T2WI-based | ||||||||
| Sole PT | 0.52±0.05 | 0.41–0.62 | 58.1 | 42.6 | 53.7 | 46.9 | 50.1 | 1.0 |
| Sole LN | 0.78±0.04 | 0.69–0.86 | 74.2 | 64.8 | 70.8 | 68.6 | 69.8 | 5.3 |
| PT-LN fusion | 0.91±0.03 | 0.85–0.95 | 85.4 | 83.3 | 85.4 | 83.3 | 84.5 | 29.4 |
| Multisequences-based | ||||||||
| Sole PT | 0.89±0.03 | 0.82–0.94 | 83.9 | 74.1 | 78.8 | 80.0 | 79.3 | 14.9 |
| Sole LN | 0.70±0.05 | 0.55–0.85 | 67.7 | 50.0 | 60.9 | 57.4 | 59.4 | 2.1 |
| PT-LN fusion | 0.90±0.03 | 0.82–0.95 | 87.1 | 83.3 | 85.7 | 84.9 | 85.3 | 33.8 |
AUCs are presented as mean ± standard deviation. CE, contrast-enhanced; T1WI, T1-weighted imaging; T2WI, T2-weighted imaging; PT, primary tumor; LN, lymph node; AUC, area under curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; OR, odds ratio.
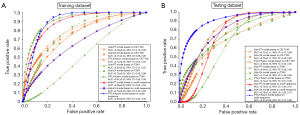
The radiomics models based on the testing data set showed comparable performance in the training data set. The fusion models yielded better performance compared to models based on solely PT or LN (CE-T1WI: 0.80 vs. 0.71/0.73; T2WI: 0.74 vs. 0.64/0.71). Models combining multisequences outperformed the single CE-T1WI and T2WI models (PT: 0.74 vs. 0.71/0.64; LN: 0.78 vs. 0.73/0.71). Finally, the PT-LN fused model based on multisequence imaging yielded the best classification performance with the highest AUC value of 0.91 for the prediction of p16 expression. The differences between the final models and other models were significant (P<0.05). In terms of other evaluation metrics, including accuracy, sensitivity, specificity, PPV, NPV, and OR, the final fusion model also showed better classification performance compared to the other models. The F1 score and precision suggested that the prediction models in the testing cohort achieved a fine performance. Particularly, the PT-LN fused model based on multisequence imaging yielded the best classification performance with the highest precision of 0.92 and a comparable F1 score of 0.83. Table 3 and Figure 4B present the performance of models in the testing cohort.
Table 3
| Models | AUC | 95% CI | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) | F1 score | Precision | OR |
|---|---|---|---|---|---|---|---|---|---|---|
| CE-T1WI-based | ||||||||||
| Sole PT | 0.71±0.11 | 0.47–0.88 | 43.8 | 88.9 | 87.5 | 47.1 | 60.0 | 0.76 | 0.875 | 6.2 |
| Sole LN | 0.73±0.11 | 0.48–0.90 | 62.5 | 77.8 | 83.3 | 53.8 | 68.0 | 0.71 | 0.83 | 5.8 |
| PT-LN fusion | 0.80±0.10 | 0.55–0.94 | 62.5 | 77.8 | 83.3 | 53.8 | 68.0 | 0.82 | 0.83 | 5.8 |
| T2WI-based | ||||||||||
| Sole PT | 0.64±0.13 | 0.38–0.85 | 50.0 | 67.7 | 72.7 | 42.9 | 56.0 | 0.79 | 0.72 | 2.0 |
| Sole LN | 0.71±0.10 | 0.48–0.88 | 56.2 | 77.8 | 81.8 | 50.0 | 64.0 | 0.81 | 0.82 | 4.5 |
| PT-LN fusion | 0.74±0.10 | 0.51–0.95 | 56.2 | 66.7 | 75.0 | 46.2 | 60.0 | 0.85 | 0.85 | 2.6 |
| Multisequence-based | ||||||||||
| Sole PT | 0.74±0.12 | 0.46–0.91 | 75.0 | 55.6 | 75.0 | 55.6 | 68.0 | 0.81 | 0.78 | 3.8 |
| Sole LN | 0.78±0.10 | 0.55–0.75 | 87.5 | 66.7 | 82.3 | 75.0 | 80.0 | 0.84 | 0.82 | 14 |
| PT-LN fusion | 0.91±0.06 | 0.72–0.98 | 75.0 | 88.9 | 92.3 | 66.7 | 80.0 | 0.83 | 0.92 | 24 |
AUCs are presented as mean±standard deviation. CE, contrast-enhanced; PT, primary tumor; LN, lymph node; AUC, area under curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; OR, odds ratio.
In the final fusion model based on multisequence radiomics features, the following 6 radiomics features were included: 1 CE-T1WI PT wavelet feature (LHH first-order kurtosis), 1 T2WI PT LoG feature (GLSZM size zone nonuniformity normalized (δ =10), 3 T2WI PT wavelet features (LHL GLSZM size zone nonuniformity normalized, LHH GLCM Id, and LHH GLSZM small area emphasis), and 1 T2WI LN wavelet feature (LLH GLDM dependence entropy). It can be seen that features based on the PT were critical to predicting the p16 status of OPSCC. Supplementary Table 1 lists the features included in each radiomics model.
The results of the comparison of pathology and imaging prediction in the optimal PT-LN fused model based on multisequence imaging are shown in Table 4. The total diagnostic accuracy of imaging prediction was 80.0%. The accuracy for detecting P16− was 88.9%, and the accuracy for detecting P16+ was 75.0%. There were significant differences between the P16− and P16+ groups (P=0.004).
Table 4
| Category | Imaging prediction | Total | Statistical data | |
|---|---|---|---|---|
| Wrong | Right | |||
| Pathological results, n (%) | χ2=9.420; P=0.004 | |||
| P16− | 1 (11.1) | 8 (88.9) | 9 | |
| P16+ | 4 (25.0) | 12 (75.0) | 16 | |
| Total | 5 | 20 | 25 | |
PT, primary tumor; LN, lymph node.
Discussion
HPV-positive and HPV-negative OPSCC have different carcinogenic mechanisms, pathogeneses, and prognoses. In this study, we used machine learning classification methods to build a 6-feature-based radiomics model, which was validated as a significant predictor of p16 status in patients with OPSSCC. Compared with a sole PT or LN model or models based on single-sequence imaging, the final PT-LN fusion model based on multisequence imaging had significantly superior predictive performance.
In our study, the PT-LN fusion model based on multisequence imaging was able to distinguish HPV-positive from HPV-negative patients. On routine imaging, HPV-positive and HPV-negative OPSCC exhibit different imaging characteristics. HPV-positive OPSCC is more likely to consist of enhanced tumors with well-defined margins and cystic nodal metastases, whereas HPV-negative OPSCC is more often composed of tumors with poorly defined borders and invasion of the adjacent muscle (26). The distinct imaging entities may be explained by histopathology. The HPV-negative tumor exhibits an infiltrative growth pattern with a pronounced stromal desmoplastic reaction (27). The phenomenon of cystic nodal metastasis may result from the spontaneous degradation of keratin in HPV-positive tumors (28). However, these imaging characteristics are not very specific. Some HPV-negative tumors can also show intranodal cystic changes and unclear margins (6,26,28), so a more accurate method is needed to distinguish HPV-positive from HPV-negative OPSCC. In recent years, radiomics has been proposed as a potential tool for predicting HPV or p16 status. The imaging features based on radiomics can help explore the microscopic structural characteristics and heterogeneity of tumors better than can the conventional morphological characteristics (29,30). In our study, the radiomics features (LHH first-order kurtosis, GLSZM size zone non uniformity normalized, LHL GLSZM size zone non uniformity normalized, LHH GLCM Id, LHH GLSZM small area emphasis, and LLH GLDM dependence entropy) involved in the final fusion model consistently reflected the uniformity and heterogeneity of the tumor. These imaging features exhibited less-prominent irregularities and more homogenous distribution in HPV-positive tumors than in HPV-negative tumors.
Our study proved that the combination of multisequence radiomics features could improve the predictive performance compared with that of single-sequence features, which is consistent with previous radiomics studies (18-21,31). This finding suggests that the radiomics features extracted from each sequence contain complementary information needed for the prediction of p16 status, thus providing more valuable information and achieving increased performance when combined. Lesions on postcontrast imaging showed more obvious margins. Cystic changes and necrosis of the PT or LN appeared as high intensity on T2WI, which made these changes easier to identify. Therefore, the combination of CE-T1WI and T2WI may guarantee the more accurate recognition of lesion extent and intralesion intensity.
In the current study, PT-LN fusion radiomics models improved the classification performance compared with the use of PT or LN alone for the prediction of p16 status. Haider et al. (17) also suggested the potential benefit of combining tumoral and nodal radiomics features in the prediction of HPV status based on fludeoxyglucose-positron emission tomography (FDG-PET). Similarly, another study reported that the combined evaluation of dynamic contrast-enhanced (DCE)- and diffusion-weighted imaging (DWI)-derived parameters from both PTs and LNs could predict response to chemoradiation in squamous cell carcinomas of the head and neck, whereas each parameter alone could not predict the response to (24). Differences in PT- and LN-related imaging prognosticators reflect the different intrinsic biologic characteristics of the respective sites (5); therefore, the combination of PT and LN radiomics features can mutually complement the information in the respective prediction models.
Multiple studies have proven that radiomics can be applied to identify the HPV status of OPSCC based on CT (AUC =0.7–0.8, AUC =0.834) (16,32). Haider et al. (17) reported that PET-based radiomics signatures yielded a similar classification performance to CT-based models for the prediction of HPV status; however, models combining PET and CT features outperformed the single-imaging modality model, with an AUC value of 0.78. However, our model produced results superior to these findings, which may be attributable to the better soft-tissue contrast of MRI in this anatomically challenging region. The margin of the primary tumor, intratumor necrosis, and cystic changes can be demonstrated more clearly on MRI. Bos et al. (15) extracted 77 radiomics features from CE-T1WI MRI to build a radiomics model predictive of HPV status (AUC =0.76). Our study yielded a higher performance (AUC =0.91), not only for the fusion of PT and LN but also for the combination of CE-T1WI and T2WI radiomics features, with both of these potentially adding more valuable information to the predictive models, as mentioned above. Two studies have also built radiomics models to predict HPV status based on multisequence MRI radiomics (33,34). However, unlike our study, both of these studies were performed in a single institution, and the predictive models were not validated by an external testing cohort. Therefore, the generalizability of these predictive models is not very satisfactory. Beyond just predicting P16 status, the radiomics model has implications for providing a better understanding of the whole tumor histology in the living patient and has the potential to predict the treatment response, prognosis, and immune state noninvasively in further studies, potentially assisting oncologists with their clinical decision-making.
For model development and validation, there are some issues to be addressed. SVM consistently requires less computation and achieves good accuracy even in a small sample size. Because of the relatively small data set enrolled in our study, an overfitting problem due to the high-dimensionality data sets might have occurred in the machine learning algorithm. Additionally, other methods were used for model development, including logistic regression, random forest, etc., and the SVM was preferred over these other classification algorithms. Therefore, the SVM was more suitable for the current study. Moreover, the leave-one-out cross-validation (LOOCV) method, which is a special K-fold validation method, was used to train and test the classification models. By using LOOCV, the final fusion model achieved satisfactory performance in terms of AUC (0.92±0.03; 95% CI: 0.87–0.96), accuracy (85.34%), sensitivity (83.9%), specificity (87.0%), PPV (88.1%), NPV (82.5%), and OR (34.9), which were similar to the current results.
In this study, we not only fused the radiomics features from the PT and LN, but also combined CE-T1WI- and T2WI-derived features, all of which added valuable information to each other and improved the classification performance of the predictive models. Moreover, our results, which were based on a multi-institutional cohort, confirmed the generalizability of the radiomics models and their potential to be used as a noninvasive biomarker for the molecular phenotyping of OPSCC.
Despite the promising results, this study also had several limitations. First, our sample size was quite limited compared to previous CT-based studies. Second, the proportion of p16+ to p16− patients was imbalanced in the testing cohort, and thus more cases will be collected in future studies to eliminate the data imbalance. Third, we presume that additional improvement predicting HPV status may be achieved when combining radiomics with clinical characteristics; thus, the inclusion of demographic information should be collected for more in-depth research. Fourth, advanced MRI techniques, such as DCE and DWI, were not performed in this study, and the quantitative parameters derived from these sequences could be used to evaluate the blood supply and cell density of a tumor. Finally, the exclusion of patients with small tumor volumes limited the generalizability of the study since using MRI imaging to determine P16 status would be helpful for smaller tumors which are not amenable to biopsy and do not have a notable nodal component; therefore thin-slice, high-resolution MRI scanning will be applied to demonstrate small tumors more clearly in further research.
In future research, a larger sample size from a greater number of institutions should be collected, and variable sequences with quantitative imaging metrics should be employed. Demographic information may be needed to build the clinical–radiomics combined model for the prediction of HPV status and prognosis in patients with OPSCC.
Conclusions
In conclusion, this study found that in patients with OPSCC, the PT-LN fusion radiomics models improved the classification performance of PT or LN features alone for the prediction of p16 status. The radiomics models based on multisequence imaging outperformed single-sequence imaging models in predicting the p16 status. The PT-LN fusion model based on multisequence MRI radiomics features could serve as a noninvasive method for reflecting the molecular information of OPSCC, potentially assisting oncologists with their clinical decision-making.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-819/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-819/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chaturvedi AK, Anderson WF, Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS, Bray F, Gillison ML. Worldwide trends in incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol 2013;31:4550-9. [Crossref] [PubMed]
- Xu T, Shen C, Wei Y, Hu C, Wang Y, Xiang J, Sun GH, Su F, Wang Q, Lu X. Human papillomavirus (HPV) in Chinese oropharyngeal squamous cell carcinoma (OPSCC): A strong predilection for the tonsil. Cancer Med 2020;9:6556-64. [Crossref] [PubMed]
- Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, Westra WH, Chung CH, Jordan RC, Lu C, Kim H, Axelrod R, Silverman CC, Redmond KP, Gillison ML. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med 2010;363:24-35. [Crossref] [PubMed]
- Fakhry C, Lacchetti C, Rooper LM, Jordan RC, Rischin D, Sturgis EM, Bell D, Lingen MW, Harichand-Herdt S, Thibo J, Zevallos J, Perez-Ordonez B. Human Papillomavirus Testing in Head and Neck Carcinomas: ASCO Clinical Practice Guideline Endorsement of the College of American Pathologists Guideline. J Clin Oncol 2018;36:3152-61. [Crossref] [PubMed]
- Ng SH, Liao CT, Lin CY, Chan SC, Lin YC, Yen TC, Chang JT, Ko SF, Fan KH, Wang HM, Yang LY, Wang JJ. Dynamic contrast-enhanced MRI, diffusion-weighted MRI and (18)F-FDG PET/CT for the prediction of survival in oropharyngeal or hypopharyngeal squamous cell carcinoma treated with chemoradiation. Eur Radiol 2016;26:4162-72. [Crossref] [PubMed]
- Cantrell SC, Peck BW, Li G, Wei Q, Sturgis EM, Ginsberg LE. Differences in imaging characteristics of HPV-positive and HPV-Negative oropharyngeal cancers: a blinded matched-pair analysis. AJNR Am J Neuroradiol 2013;34:2005-9. [Crossref] [PubMed]
- Brenet E, Barbe C, Hoeffel C, Dubernard X, Merol JC, Fath L, Servagi-Vernat S, Labrousse M. Predictive Value of Early Post-Treatment Diffusion-Weighted MRI for Recurrence or Tumor Progression of Head and Neck Squamous Cell Carcinoma Treated with Chemo-Radiotherapy. Cancers (Basel) 2020.
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Chu CS, Lee NP, Adeoye J, Thomson P, Choi SW. Machine learning and treatment outcome prediction for oral cancer. J Oral Pathol Med 2020;49:977-85. [Crossref] [PubMed]
- Morgan HE, Wang K, Dohopolski M, Liang X, Folkert MR, Sher DJ, Wang J. Exploratory ensemble interpretable model for predicting local failure in head and neck cancer: the additive benefit of CT and intra-treatment cone-beam computed tomography features. Quant Imaging Med Surg 2021;11:4781-96. [Crossref] [PubMed]
- Yu K, Zhang Y, Yu Y, Huang C, Liu R, Li T, Yang L, Morris JS, Baladandayuthapani V, Zhu H. Radiomic analysis in prediction of Human Papilloma Virus status. Clin Transl Radiat Oncol 2017;7:49-54. [Crossref] [PubMed]
- Rich B, Huang J, Yang Y, Jin W, Johnson P, Wang L, Yang F. Radiomics Predicts for Distant Metastasis in Locally Advanced Human Papillomavirus-Positive Oropharyngeal Squamous Cell Carcinoma. Cancers (Basel) 2021.
- Alabi RO, Elmusrati M, Sawazaki-Calone I, Kowalski LP, Haglund C, Coletta RD, Mäkitie AA, Salo T, Almangush A, Leivo I. Comparison of supervised machine learning classification techniques in prediction of locoregional recurrences in early oral tongue cancer. Int J Med Inform 2020;136:104068. [Crossref] [PubMed]
- Giannitto C, Marvaso G, Botta F, Raimondi S, Alterio D, Ciardo D, et al. Association of quantitative MRI-based radiomic features with prognostic factors and recurrence rate in oropharyngeal squamous cell carcinoma. Neoplasma 2020;67:1437-46. [Crossref] [PubMed]
- Bos P, van den Brekel MWM, Gouw ZAR, Al-Mamgani A, Waktola S, Aerts HJWL, Beets-Tan RGH, Castelijns JA, Jasperse B. Clinical variables and magnetic resonance imaging-based radiomics predict human papillomavirus status of oropharyngeal cancer. Head Neck 2021;43:485-95. [Crossref] [PubMed]
- Leijenaar RT, Bogowicz M, Jochems A, Hoebers FJ, Wesseling FW, Huang SH, Chan B, Waldron JN, O'Sullivan B, Rietveld D, Leemans CR, Brakenhoff RH, Riesterer O, Tanadini-Lang S, Guckenberger M, Ikenberg K, Lambin P. Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br J Radiol 2018;91:20170498. [Crossref] [PubMed]
- Haider SP, Mahajan A, Zeevi T, Baumeister P, Reichel C, Sharaf K, Forghani R, Kucukkaya AS, Kann BH, Judson BL, Prasad ML, Burtness B, Payabvash S. PET/CT radiomics signature of human papilloma virus association in oropharyngeal squamous cell carcinoma. Eur J Nucl Med Mol Imaging 2020;47:2978-91. [Crossref] [PubMed]
- Wang Y, Wan Q, Xia X, Hu J, Liao Y, Wang P, Peng Y, Liu H, Li X. Value of radiomics model based on multi-parametric magnetic resonance imaging in predicting epidermal growth factor receptor mutation status in patients with lung adenocarcinoma. J Thorac Dis 2021;13:3497-508. [Crossref] [PubMed]
- Li Z, Chen F, Zhang S, Ma X, Xia Y, Shen F, Lu Y, Shao C. The feasibility of MRI-based radiomics model in presurgical evaluation of tumor budding in locally advanced rectal cancer. Abdom Radiol (NY) 2022;47:56-65. [Crossref] [PubMed]
- Xue K, Liu L, Liu Y, Guo Y, Zhu Y, Zhang M. Radiomics model based on multi-sequence MR images for predicting preoperative immunoscore in rectal cancer. Radiol Med 2022;127:702-13. [Crossref] [PubMed]
- Wang S, Jiang T, Hu X, Hu H, Zhou X, Wei Y, Mao X, Zhao Z. Can the combination of DWI and T2WI radiomics improve the diagnostic efficiency of cervical squamous cell carcinoma? Magn Reson Imaging 2022;92:197-202. [Crossref] [PubMed]
- Park SH, Hahm MH, Bae BK, Chong GO, Jeong SY, Na S, Jeong S, Kim JC. Magnetic resonance imaging features of tumor and lymph node to predict clinical outcome in node-positive cervical cancer: a retrospective analysis. Radiat Oncol 2020;15:86. [Crossref] [PubMed]
- Lu N, Zhang WJ, Dong L, Chen JY, Zhu YL, Zhang SH, Fu JH, Yin SH, Li ZC, Xie CM. Dual-region radiomics signature: Integrating primary tumor and lymph node computed tomography features improves survival prediction in esophageal squamous cell cancer. Comput Methods Programs Biomed 2021;208:106287. [Crossref] [PubMed]
- Chawla S, Kim S, Dougherty L, Wang S, Loevner LA, Quon H, Poptani H. Pretreatment diffusion-weighted and dynamic contrast-enhanced MRI for prediction of local treatment response in squamous cell carcinomas of the head and neck. AJR Am J Roentgenol 2013;200:35-43. [Crossref] [PubMed]
- Brcic I, Gallob M, Schwantzer G, Zrnc T, Weiland T, Thurnher D, Wolf A, Brcic L. Concordance of tumor infiltrating lymphocytes, PD-L1 and p16 expression in small biopsies, resection and lymph node metastases of oropharyngeal squamous cell carcinoma. Oral Oncol 2020;106:104719. [Crossref] [PubMed]
- Corey AS, Hudgins PA. Radiographic imaging of human papillomavirus related carcinomas of the oropharynx. Head Neck Pathol 2012;6:S25-40. [Crossref] [PubMed]
- El-Mofty SK, Zhang MQ, Davila RM. Histologic identification of human papillomavirus (HPV)-related squamous cell carcinoma in cervical lymph nodes: a reliable predictor of the site of an occult head and neck primary carcinoma. Head Neck Pathol 2008;2:163-8. [Crossref] [PubMed]
- Goldenberg D, Begum S, Westra WH, Khan Z, Sciubba J, Pai SI, Califano JA, Tufano RP, Koch WM. Cystic lymph node metastasis in patients with head and neck cancer: An HPV-associated phenomenon. Head Neck 2008;30:898-903. [Crossref] [PubMed]
- Lee JY, Han M, Kim KS, Shin SJ, Choi JW, Ha EJ. Discrimination of HPV status using CT texture analysis: tumour heterogeneity in oropharyngeal squamous cell carcinomas. Neuroradiology 2019;61:1415-24. [Crossref] [PubMed]
- Mungai F, Verrone GB, Pietragalla M, Berti V, Addeo G, Desideri I, Bonasera L, Miele V. CT assessment of tumor heterogeneity and the potential for the prediction of human papillomavirus status in oropharyngeal squamous cell carcinoma. Radiol Med 2019;124:804-11. [Crossref] [PubMed]
- Nebbia G, Zhang Q, Arefan D, Zhao X, Wu S. Pre-operative Microvascular Invasion Prediction Using Multi-parametric Liver MRI Radiomics. J Digit Imaging 2020;33:1376-86. [Crossref] [PubMed]
- Choi Y, Nam Y, Jang J, Shin NY, Ahn KJ, Kim BS, Lee YS, Kim MS. Prediction of Human Papillomavirus Status and Overall Survival in Patients with Untreated Oropharyngeal Squamous Cell Carcinoma: Development and Validation of CT-Based Radiomics. AJNR Am J Neuroradiol 2020;41:1897-904. [Crossref] [PubMed]
- Sohn B, Choi YS, Ahn SS, Kim H, Han K, Lee SK, Kim J. Machine Learning Based Radiomic HPV Phenotyping of Oropharyngeal SCC: A Feasibility Study Using MRI. Laryngoscope 2021;131:E851-6. [Crossref] [PubMed]
- Suh CH, Lee KH, Choi YJ, Chung SR, Baek JH, Lee JH, Yun J, Ham S, Kim N. Oropharyngeal squamous cell carcinoma: radiomic machine-learning classifiers from multiparametric MR images for determination of HPV infection status. Sci Rep 2020;10:17525. [Crossref] [PubMed]


