
Deep learning-based segmentation of epithelial ovarian cancer on T2-weighted magnetic resonance images
Introduction
Ovarian cancer has the second highest incidence and the highest mortality rates in gynecological malignancy (1,2). Epithelial ovarian cancer (EOC) represents 90–95% of all ovarian cancers, accounting for more than 70% of ovarian cancer deaths (3). The standard treatment strategy for EOC consists of operative tumor debulking and administration of intravenous chemotherapy (4). Assessment of tumor prior to surgery is of paramount importance in guiding treatment decisions and ultimately improving patient outcomes (5). Magnetic resonance imaging (MRI) is widely used in the detection and characterization of tumors owing to the superior soft tissue contrast and tissue characterization (6). Recent radiomics studies based on MRI have demonstrated many encouraging advances. In our previous studies, we developed an assessment model built for differentiating borderline epithelial ovarian tumors from malignant epithelial ovarian tumors (7), which was praised by Dr. Araki at Harvard Medical School as “may significantly change clinical management and outcome” (8). We also developed a radiomics diagnostic model for differentiating type I and II EOC (9). Moreover, a radiomics study has demonstrated the feasibility of EOC prognosis prediction (10).
Recognizing and delineating tumor regions is a prerequisite for quantitative radiomics analysis. In addition, it helps simplify target volume delineation in radiotherapy as well as subsequent efficacy evaluation. Nevertheless, EOC segmentation is commonly conducted in a slice-by-slice manner by clinicians in clinical practices, which is both time-consuming and labor-intensive. Hence, a solution to this problem is urgently required.
Dastidar et al. (11) applied an automatic segmentation software named Anatomatic for ovarian tumor segmentation on MRI images. The software easily achieved accurate volumetric estimations of tumors on patients with ovarian tumor without ascites by combining four different methods: image enhancement, amplitude segmentation, region growing and decision trees (IARD). However, for patients with ascites, this software tends to misinterpret ascites as tumor cyst fluid, thus limiting its clinical application. Currently, deep learning (DL), represented by convolutional neural networks (CNNs) and transformers, has achieved enormous success in the field of medical image segmentation. CNN is a classical DL architecture that extracts hierarchical features from an image via convolution operation to identify the region of interest pixel by pixel, and is widely used in the segmentations on MR images of brain tumor (12), breast tumor (13), colorectal tumor (14) and etc. The transformer is a novel DL architecture which had first attracted much attention due to its ability in natural language processing (NLP), and has recently demonstrated promising results on tasks involving computer vision. Owing to its capability in modeling data with long-range dependencies, transformer exhibits superior performance than CNN on tasks like object detection (15), image classification (16), synthesis (17) and segmentation (18). All these successes have inspired us to utilize DL in EOC segmentation.
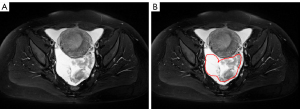
Despite the anticipated benefits that it may bring, automatic EOC segmentation on MRI images is facing the following challenges: (I) indistinct boundaries between ascites and cyst fluid: as Figure 1 shows, patients with EOC are often accompanied by ascites, which result in the blur boundary between ascites and cyst fluid since cyst walls are extremely thin and smooth; (II) morphological diversity of tumors: EOC lesions tend to be irregular in shape and vary in size (2–20 cm), which put forward higher requirements for the robustness of the automatic segmentation algorithm on different scales; (III) differences in signal of different tumor tissues: EOC lesions can be roughly divided into cystic, solid and complex (contain both solid and cystic components) tumor, with the cystic and solid components exhibiting different signal intensities on MRI; the solid components show low signal intensity on T2-weighted imaging with fat saturation (T2WI FS) whereas the cystic components show high signal intensity. For complex tumor which contains both components, this difference increases the difficulty of automatic EOC segmentation. Henceforth, doubts were cast on whether CNNs and transformers are applicable in the automatic segmentation of EOC.
In this study, we aimed to explore the feasibility of CNNs as well as transformers in the automatic EOC segmentation on MR images. 4 CNNs [U-Net (19), DeepLabv3 (20), U-Net++ (21), and PSPNet (22)] and 2 transformers [TransUnet (23) and Swin-Unet (24)] were applied and compared in our in-house EOC dataset which contains MRI images of 339 patients from 8 clinical centers. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-494/rc).
Methods
Dataset
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was reviewed and approved by the Ethics Committee of Jinshan Hospital of Fudan University, and informed consent was waived due to its retrospective nature. To evaluate the performance of the neural networks, an EOC dataset was constructed using T2WI FS of 339 patients from 8 clinical centers. The centers involved are listed as follows: (A) Obstetrics and Gynecology Hospital of Fudan University, (B) Jinshan Hospital of Fudan University, (C) Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medical, (D) Affiliated Hospital of Nantong University, (E) Guangdong Women and Children Hospital, (F) Affiliated Hospital of Xuzhou Medical University, (G) Huadong Hospital Affiliated to Fudan University and (H) Nantong Tumor Hospital. Details of MRI equipment are presented in Table 1 and examples of MR images acquired from different centers are presented in Figure 2. Considering that centers A and B share the same MR vendor and scanning protocol, a total of 229 patients from these 2 centers were grouped into 1 cohort. Since there are multiple subtypes of EOC, 70% of the patients of each subtype from centers A and B were randomly assigned for training, 10% were used for validation, while the rest were kept for internal testing. The patients from centers C–H were assigned as the external testing cohort. Hence, the training, validation, internal test and external test sets contain 154, 25, 50 and 110 patients, respectively. The International Federation of Gynecology and Obstetrics (FIGO) stages distribution of EOC in internal set is shown in Table 2 and histological types distribution of EOC in each center is shown in Table 3. The distribution of the FIGO stages and the histological types matches the general circumstances.
Table 1
| Centers | Specifications of MRI equipment | Protocol parameters | ||
|---|---|---|---|---|
| TR/TE (ms) | ST/IG (mm) | FOV (mm) | ||
| A | 1.5T, Avanto, Siemens | 8,000/83 | 4.0/1.0 | 256×256 |
| B | 1.5T, Symphony, Siemens | 8,000/83 | 4.0/1.0 | 256×256 |
| C | 3.0T, Twinspeed, GE | 3,840/105 | 6.0/0.5 | 350×350 |
| D | 1.5T/3.0T, Signa, GE | (6,600–8,000)/85 | 7.0/1.0 | (360–460)×(360–460) |
| E | 1.5T, Brivo MR 355, GE | 3,000/120 | 3.0/1.0 | 330×330 |
| F | 3.0T, 750W, GE; 3.0T, Ingenia, Philip | 4,000/142 | 6.0/1.0 | (256–512)×(256–512) |
| G | 3.0T, Trio/Verio/Skyra, Siemens | 5,020/114 | 4.0/0.8 | 260×260 |
| H | 1.5T, Espree/3.0T, Verio, Siemens | 4,120/108 | 7.0/1.0 | 320×320 |
MRI, magnetic resonance imaging; TR, repetition time; TE, echo time; ST, slice thickness; IG, intersection gap; FOV, field of view.
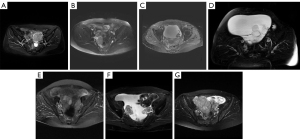
Table 2
| I | II | III | IV |
|---|---|---|---|
| 16 | 5 | 24 | 5 |
FIGO, International Federation of Gynecology and Obstetrics; EOC, epithelial ovarian cancer.
Table 3
| Centers | Total number of patients | |||||
|---|---|---|---|---|---|---|
| Total | Serous | Endometrioid | Mucinous | Clear cell | Undifferentiated | |
| A and B | 229 | 145 | 21 | 18 | 45 | |
| C | 45 | 23 | 5 | 7 | 9 | 1 |
| D | 24 | 15 | 2 | 1 | 6 | |
| E | 2 | 1 | 1 | |||
| F | 17 | 16 | 1 | |||
| G | 9 | 8 | 1 | |||
| H | 13 | 6 | 1 | 3 | 3 | |
EOC, epithelial ovarian cancer.
For each patient, the tumor region was delineated manually in a slice-by-slice manner by a board-certified radiologist with 5 years of experience specializing in gynecologic imaging using the Medical Imaging Interaction Toolkit (MITK) (version 2016.11.0; http://www.mitk.org/). The delineation was checked by another radiologist (reader 2) with 8 years of experience specializing in gynecologic imaging. If there raised a disagreement, the final delineation was settled by a radiologist (reader 3) with 20 years of experience specializing in gynecologic imaging.
Data preprocessing and augmentation
For each patient, slices that did not contain any tumor were discarded. A final count of 5,682 slices was obtained from 339 patients. The preprocessing flowchart is depicted in Figure 3. All slices were then resampled with the voxel size of 1 mm × 1 mm × 5 mm. Unnecessary regions that correspond to non-tumor areas were cropped. For most patients, slices were cropped to 224×288 pixels at the center; for patients whose tumor region exceeds the edge of the cropping box, their slices were treated as special cases and were cropped to 288×288 pixels at the center. Finally, all slices were resized to 256×256 pixels. The intensity range of each slice was normalized to 0–1 through min-max normalization. The same resampling, cropping, and resizing operations were also performed for the corresponding tumor region images.
Data augmentation was applied during the training phase, including a random horizontal flip with a probability of 0.5, a random rotation of ±20° and a random scaling from 80% to 120%.
Segmentation models
Six common-used algorithms, including CNNs (U-Net, DeepLabv3, U-Net++ and PSPNet) and transformers (TransUnet and Swin-Unet) were used for automatic EOC segmentation.
U-Net
U-Net is a symmetrical U-shaped model consisting of an encoder-decoder architecture. The encoder involves the use of down-sampling to obtain the feature map, whereas the decoder involves the use of up-sampling to restore the encoded features to the original image size, and to generate the final segmentation results. Skip-connection is added to the encoder-decoder networks to concatenate the high- and low-level features while retaining the detailed feature information required for the up-sampled output.
DeepLabv3
DeepLabv3 employs deep convolutional neural network (DCNN) to generate a low-level feature map and introduces it to the Atrous Spatial Pyramid Pooling (ASPP) module. ASPP probes multiscale convolutional features without increasing the number of convolution weights by applying atrous convolution at different rates. Additionally, DeepLabv3 augments the ASPP with image-level features and batch normalization. The dense feature maps extracted by ASPP are bilinearly up-sampled to generate full-resolution segmentation results, which could be considered as a naive decoder module.
U-Net++
U-Net++ is a modified U-Net model based on nested and dense skip connections. A series of nested, dense convolutional blocks are applied to connect the encoder and decoder, bringing the semantic level of the encoder feature maps closer to that of the feature maps awaiting in the decoder. Owing to the skip connections that are re-designed, U-Net++ is able to generate full resolution prediction maps at multiple semantic levels. The maps can then be averaged or selected to generate the best segmentation results.
PSPNet
PSPNet adapts ResNet with the dilated network strategy as the encoder to generate semantic feature maps. On top of the maps, a pyramid pooling module is utilized to simultaneously gather context information. Using a 4-level pyramid, the pooling kernels cover the whole, half of and small portions of the image. They are fused as the global prior. The prior is then concatenated with the original feature map in the final part of the pyramid pooling module, followed by a convolution layer to generate the final segmentation results.
TransUnet
TransUnet is the first transformer-based medical image segmentation framework. It employs a hybrid CNN-Transformer architecture to leverage both detailed high-resolution spatial information from CNN features and the global context encoded by transformers. TransUnet utilizes a three-layer CNN as the feature extractor to generate feature maps. The feature maps are then sent to the transformer after patch embedding to extract global contexts. The self-attentive features encoded by the transformer is then unsampled by a cascaded up-sampler. The up-sampler and the hybrid decoder are then brought together to form a u-shaped architecture that generates the final segmentation results.
Swin-Unet
Swin-Unet is the first pure transformer-based U-shaped architecture. The Encoder, bottleneck and decoder are all built based on the Swin Transformer block. The input medical images are split into individual non-overlapping image patches, that are treated as a token and fed into the transformer-based encoder to learn deep feature representations. The extracted context features are then up-sampled by the decoder with patch expanding layer, and fused with the multi-scale features from the encoder via skip connections, so as to restore the spatial resolution of the feature maps and further generate segmentation results.
Evaluation indicators
The segmentation performance was evaluated using five metrics, including dice similarity coefficient (DSC), Hausdorff distance (HD), average symmetric surface distance (ASSD), precision and recall. DSC, precision and recall are calculated based on the region overlap between manually and automatically segmented results. HD refers to the maximum value of all the distances from the closest point in automatically segmented results to manually segmented results. ASSD refers to the average value of all the distances from points on the boundary of automatically segmented results to the boundary of the manually segmented results. The equations are described in Eqs. [1-5], in which P represents the segmentation results predicted by the models; G represents the accurate ground truth; TP, FP, TN and FN represents the true positive, false positive, true negative and false negative; represents the Euclidean distance between pixel p and g; and represents the number of pixels.
Among the metrics, DSC, precision and recall return values in the range of (0, 1), where values toward unity indicate an adequate segmentation performance; HD and ASSD return values in the range of (0, ∞), where values toward null indicate an adequate segmentation performance.
Implement details
Models were implemented in Python 3.6 with the PyTorch 1.8.1 platform on a server equipped with an NVIDIA GeForce GTX 2080 Ti GPU. To enhance the training efficiency, pre-trained encoders and transformer modules (trained on the ImageNet dataset) were used to initialize the corresponding parameters of each layer. Adaptive moment estimation (Adam) with default parameters (β1=0.9, β2=0.999) was used as the optimizer. All the networks were trained using the optimal learning rate. The optimal hyperparameters were determined by the validation set. The maximum iteration was set to 200, and the learning rate was halved every ten iterations. The batch size was set to 4.
Results
Table 4 presents comparisons of the six models in terms of GPU RAM consumption, training time and prediction time. Among six models, DeepLabv3 requires the largest GPU RAM (7.20 GB) and the longest time (17.7 h) for training. PSPNet requires the smallest GPU RAM (2.42 GB) and the shortest time for training (9.7 h). For all models, the prediction time reaches within 1 s per patient, which is fast enough compared with manual segmentation (around 2–3 min and for some large-size tumor, such as mucinous tumor, the segmentation time is even longer).
Table 4
| Model | GPU RAM (GB) | Training time (h) | Prediction time (s/case) |
|---|---|---|---|
| U-Net | 2.75 | 10.1 | 0.34 |
| DeepLabv3 | 7.20 | 17.7 | 0.31 |
| U-Net++ | 3.45 | 12.7 | 0.74 |
| PSPNet | 2.42 | 9.7 | 0.32 |
| TransUnet | 6.20 | 13.5 | 0.56 |
| Swin-Unet | 3.56 | 12.9 | 0.50 |
GPU, graphics processing unit; RAM, random access memory.
The five metrics used in evaluating the performances of the model, along with 95% confidence interval on the internal and external tests of each model are summarized in Table 5 and Table 6, respectively. In the internal test set, U-Net++ performed best in DSC (0.851) and ASSD (1.75); TransUnet performed best in HD (19.8) and precision (0.847); while DeepLabv3 achieved the best recall (0.887). In the external test set, overall, metrics were lower than those in the internal test set for all models. This may be attributed to the significant variety of image appearance caused by different scanners and protocols. U-Net++ performed best in DSC (0.740), precision (0.825) and recall (0.725) whereas TransUnet performed best in HD (29.8) and ASSD (3.34). In addition, Swin-Unet was identified to have performed the poorest in both the internal and external test sets.
Table 5
| Model | DSC | HD (mm) | ASSD (mm) | Precision | Recall |
|---|---|---|---|---|---|
| U-Net | 0.845 (0.803–0.882) | 27.6 (21.6–33.6) | 1.93 (1.29–2.58) | 0.831 (0.788–0.873) | 0.876 (0.841–0.911) |
| DeepLabv3 | 0.847 (0.809–0.885) | 24.6 (19.0–30.1) | 1.94 (1.22–2.67) | 0.829 (0.782–0.876) | 0.887 (0.855–0.919) |
| U-Net++ | 0.851 (0.816–0.886) | 25.3 (20.0–30.7) | 1.75 (1.21–2.30) | 0.838 (0.795–0.881) | 0.882 (0.850–0.915) |
| PSPNet | 0.837 (0.799–0.874) | 29.5 (23.4–35.6) | 1.79 (1.28–2.31) | 0.834 (0.791–0.878) | 0.857 (0.818–0.895) |
| TransUnet | 0.848 (0.803–0.892) | 19.8 (14.7–24.8) | 1.79 (0.71–2.87) | 0.847 (0.799–0.895) | 0.860 (0.813–0.907) |
| Swin-Unet | 0.753 (0.695–0.812) | 32.4 (26.6–38.2) | 3.36 (2.14–4.58) | 0.707 (0.641–0.773) | 0.851 (0.804–0.898) |
DSC, dice similarity coefficient; HD, Hausdorff distance; ASSD, average symmetric surface distance.
Table 6
| Model | DSC | HD (mm) | ASSD (mm) | Precision | Recall |
|---|---|---|---|---|---|
| U-Net | 0.719 (0.666–0.772) | 58.4 (50.6–66.2) | 6.26 (4.72–7.80) | 0.778 (0.727–0.830) | 0.718 (0.663–0.773) |
| DeepLabv3 | 0.716 (0.664–0.768) | 39.9 (34.7–45.0) | 3.90 (3.01–4.78) | 0.782 (0.735–0.828) | 0.719 (0.666–0.772) |
| U-Net++ | 0.740 (0.690–0.789) | 42.5 (35.7–49.3) | 4.21 (2.96–5.46) | 0.825 (0.779–0.871) | 0.725 (0.673–0.776) |
| PSPNet | 0.637 (0.577–0.696) | 57.3 (49.6–65.0) | 7.92 (5.37–10.46) | 0.760 (0.705–0.814) | 0.618 (0.555–0.681) |
| TransUnet | 0.719 (0.651–0.787) | 29.8 (24.8–34.7) | 3.34 (2.18–4.50) | 0.805 (0.740–0.871) | 0.685 (0.613–0.757) |
| Swin-Unet | 0.359 (0.289–0.429) | 66.5 (57.5–75.5) | 11.76 (8.77–14.76) | 0.570 (0.492–0.649) | 0.348 (0.277–0.420) |
DSC, dice similarity coefficient; HD, Hausdorff distance; ASSD, average symmetric surface distance.
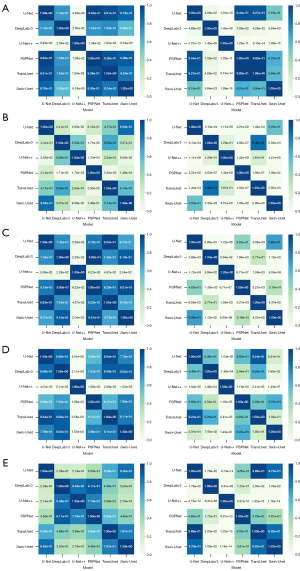
Statistical differences between the DL algorithms were determined by t-test. We used Python SciPy to test whether each metric followed normal distribution before t-test. The results turned out that all the metrics followed normal distribution (P>0.05). The significance levels are shown in Figure 4. The differences between two compared groups are considered to be statistically significant when P<0.05. Most models do not present statistical differences in the internal test set, indicating similar segmentation performance. Nevertheless, U-Net++ presents statistically significant differences from other algorithms in DSC, ASSD and precision, as observed in Figure 4A,4C,4D. In the external test set, some models present statistically significant differences. U-Net++ presents statistically significant differences from other algorithms in DSC, precision and recall, as observed in Figure 4A,4D,4E.
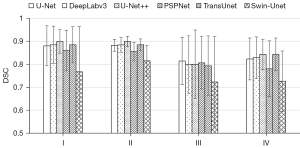
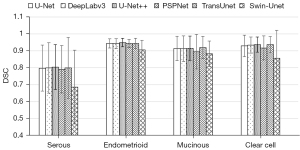
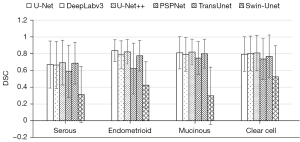
In addition, we compare the segmentation performance for different histological types and FIGO stages. We summarize the segmentation performance of different FIGO stages in the internal test set and the segmentation performance of different histological types in both internal and external test set, just as Figures 5-7 show. As shown in Figure 5, the mean DSCs of stage I and stage II tumor are relatively high (0.863 and 0.870, respectively) and the mean DSCs of stage III and stage IV tumor are relatively low (0.789 and 0.807, respectively). As shown in Figure 6, the mean DSC of serous tumor (0.780) is at least 14.0% lower than the mean DSC of other 3 types of tumors (0.938, 0.907 and 0.918, respectively). As shown in Figure 7, compared with the internal test set, the DSC of all types of tumors in the external test set decreased. The mean DSC of serous tumor is still the lowest (0.603), which is at least 15.3% lower than other types of tumors (0.711, 0.711 and 0.739, respectively).
To intuitively illustrate the performance of the segmentation models, four typical results are shown in Figure 8. Figure 8A shows a case where a solid tumor was surrounded by ascites. This tumor can be well segmented by all algorithms. The manual contour is best matched by U-Net++, while U-Net, TransUnet and Swin-Unet yielded various degrees of overestimation. Figure 8B shows a case where the patient has cystic tumor with ascites. U-Net, PSPNet, TransUnet and Swin-Unet misinterprets the ascites as tumor; DeepLabv3 and U-Net++ recognize the basic contour of the tumor; whereas U-Net++ yields segmentation result that fits the ground truth best. Figure 8C shows a representative case in which the solid tumor is attached to the uterus and the boundary is hard to identify. Consequently, all algorithms misinterpret the uterus as EOC, leading to mis-segmentation. Figure 8D shows another typical situation where the patient has severe ascites. The tumors contain both cystic and solid components, possessing sharp contrast in signal intensity. Moreover, the cystic components are adjacent to the ascites and share similar signal intensity, so most algorithms only manage to partially segment the solid components but fail to recognize the cystic components altogether.

Discussion
This study aims to explore the feasibility of CNNs as well as transformers in the automatic EOC segmentation on MR images. Overall, the performance of U-Net++ surpasses all other models in both test sets. We speculate that there might be two possible reasons. First, U-Net++ has a built-in ensemble of U-Nets, which enables image segmentation based on multi-receptive field networks, endowing it with the capability to segment tumors of varying sizes. Second, the redesigned skip connections enable the fusion of semantically rich decoder feature maps with feature maps of varying semantic scales from the intermediate layers of the architecture, assisting the former layers with more semantic information and refining segmentation results.
TransUnet can be considered as a combination of transformer and the original U-Net. When compared with U-Net, the results of TransUnet showed that it achieved remarkable improvement in HD, ASSD and precision. However, the improvement in DSC is limited and TransUnet performed worse than U-Net in terms of recall. We infer that there could be two reasons to this: Firstly, though transformer provides the capability to encode distant dependency, EOC lesions are usually clustered and located in the center of the slice, and long-range semantic information interactions provide little improvement on segmentation accuracy under this circumstance; Secondly, TransUnet computes self-attention on feature maps that have gone through four times of down-sampling, so the low-resolution attention maps acquired might be too rough for accurate identification of boundary regions. Building multi-resolution feature maps on transformers might be a better solution to this problem. In addition, Swin-Unet is identified to have performed the poorest in both the internal and external test sets. The possible explanation for this would be that Swin-Unet pays attention to the global semantic information interactions but lacks inner interactions for pixels in each patch, which is vital for pixel-dense work like segmentation.
We also aim to explore how FIGO stages and histological types impact the segmentation results. The segmentation accuracy of stage I and stage II tumor is higher than that of stage III and IV. We believe it is because early-stage tumors tend to be simple in shape and smooth in boundary, while advanced-stage tumors are infiltrative and usually tightly connect with surrounding tissues, making their boundaries irregular, which are hard to accurately outline. Moreover, the segmentation accuracy of serous tumor is poorer than that of endometrioid, mucinous and clear cell tumors. One possible explanation is that most serous tumors are at stages III and IV, as we analyze the FIGO stage distribution of serous tumors.
It is worth mentioning that model performance in external test set is poorer than that in the internal set. Most fully supervised methods heavily rely on large-scale training data with labels, when transferring the pre-trained model to unknown domain for testing, model performance usually decreases significantly due to domain shift between training and testing data (25,26). Research on how to deal with domain shift has been extensively conducted in the literature, such as meta learning (27), transfer learning (28), domain generalization (29), domain adaption (30) and etc. In further research, we will consider these methods as a solution for increasing the generalization and robustness of EOC segmentation in real-world clinical applications.
Few studies can be found on the automatic segmentation of EOC on MRI. Before the development of CNNs and transformers, Dastidar et al. (11) utilized the IARD algorithm for ovarian tumor segmentation on MRI images, which can be considered as an attempt of the traditional region-based method. To evaluate the segmentation performance, they compared the volumes estimated by the algorithm with the volumes measured and calculated at laparotomy. The study found that the algorithm was useful for measuring ovarian tumors in patients without significant ascites but failed to achieve the same results in patients with significant ascites. Compared with this previous study, the numbers of patients (6 in Dastidar’s study vs. 339 in this study) and clinical centers (1 in Dastidar’s study vs. 8 in this study) were substantially increased in this study, which could increase the versatility and robustness of the segmentation model. The accuracy of our models was also relatively higher, indicating the feasibility of DL methods as the solution for automatic EOC segmentation.
Nonetheless, this study poses several limitations. First, the performance of DL models has a strong dependence on the amount of data (31). Thus, larger MRI dataset is likely to improve the performance of DL models in automatic EOC segmentation. Second, some studies have pointed out that the use of MR images of various sequences (e.g., T1-weighted and diffusion-weighted images) might be helpful in improving the accuracy of automatic segmentation (32). Thus, more MRI modalities should be included in future work. Third, 3D network can capture the 3D spatial context contained in volumetric medical images (33). We will also try to implement 3D networks into EOC segmentation in our further study. Lastly, the algorithms that were evaluated are all generic segmentation methods. In the future, we will utilize some techniques, such as cascaded networks and boundary-aware networks, and use adjacent slice information to better accommodate the characteristics of EOC and design a network focus on EOC segmentation.
Conclusions
Segmentation is indispensable in determining the stage of EOC, which will subsequently impact the treatment strategy for a patient. In this study, the feasibility of DL methods in automatic segmentation was assessed. The results show promising outcome for DL to be applied in the automatic segmentation of EOC on T2-weighted MR images. Among all the models, U-Net++ reaches a relatively better segmentation performance with a reasonable computing consumption. As a result, it is considered the best model among the six models that we apply and compare.
Acknowledgments
The authors thank Dr. Fenghua Ma and Dr. Guofu Zhang from Department of Radiology, Obstetrics and Gynecology Hospital of Fudan University, Dr. Shuhui Zhao from Department of Radiology, Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medical, Dr. Jingxuan Jiang from Department of Radiology, Affiliated Hospital of Nantong University, Dr. Yan Zhang from Department of Radiology, Guangdong Women and Children Hospital, Dr. Keying Wang from Department of Radiology, Affiliated Hospital of Xuzhou Medical University, Dr. Guangwu Lin from Department of Radiology, Huadong Hospital Affiliated to Fudan University, and Dr. Feng Feng from Department of Radiology, Nantong Tumor Hospital for providing the data.
Funding: This work was supported by the Key Research and Development Program of Shandong Province (grant number: 2021SFGC0104), the Key Research and Development Program of Jiangsu Province (grant number: BE2021663), the National Natural Science Foundation of China (grant number: 81871439), Suzhou Science and Technology Plan Project (grant number: SJC2021014), the China Postdoctoral Science Foundation (grant number: 2022M712332), and the Jiangsu Funding Program for Excellent Postdoctoral Talent (grant number: 2022ZB879).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-494/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-494/coif). Jinan Guoke Medical Engineering and Technology Development Co., Ltd. is an affiliate of Suzhou Institute of Biomedical Engineering and Technology. XG is the leading researcher of Jinan Guoke Medical Engineering and Technology Development Co., Ltd. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Jinshan Hospital of Fudan University, and informed consent was waived due to its retrospective nature.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Ushijima K. Treatment for recurrent ovarian cancer-at first relapse. J Oncol 2010;2010:497429. [Crossref] [PubMed]
- Javadi S, Ganeshan DM, Qayyum A, Iyer RB, Bhosale P. Ovarian Cancer, the Revised FIGO Staging System, and the Role of Imaging. AJR Am J Roentgenol 2016;206:1351-60. [Crossref] [PubMed]
- Elsherif SB, Bhosale PR, Lall C, Menias CO, Itani M, Butler KA, Ganeshan D. Current update on malignant epithelial ovarian tumors. Abdom Radiol (NY) 2021;46:2264-80. [Crossref] [PubMed]
- Hennessy BT, Coleman RL, Markman M. Ovarian cancer. Lancet 2009;374:1371-82. [Crossref] [PubMed]
- Lheureux S, Braunstein M, Oza AM. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J Clin 2019;69:280-304. [Crossref] [PubMed]
- Forstner R, Hricak H, White S CT. Abdom Imaging 1995;20:2-8. [Crossref] [PubMed]
- Li Y, Jian J, Pickhardt PJ, Ma F, Xia W, Li H, et al. MRI-Based Machine Learning for Differentiating Borderline From Malignant Epithelial Ovarian Tumors: A Multicenter Study. J Magn Reson Imaging 2020;52:897-904. [Crossref] [PubMed]
- Araki T. Editorial for "MRI-Based Machine Learning for Differentiating Borderline From Malignant Epithelial Ovarian Tumors". J Magn Reson Imaging 2020;52:905. [Crossref] [PubMed]
- Jian J, Li Y, Pickhardt PJ, Xia W, He Z, Zhang R, Zhao S, Zhao X, Cai S, Zhang J, Zhang G, Jiang J, Zhang Y, Wang K, Lin G, Feng F, Wu X, Gao X, Qiang J. MR image-based radiomics to differentiate type I and type II epithelial ovarian cancers. Eur Radiol 2021;31:403-10. [Crossref] [PubMed]
- Derlatka P, Grabowska-Derlatka L, Halaburda-Rola M, Szeszkowski W, Czajkowski K. The Value of Magnetic Resonance Diffusion-Weighted Imaging and Dynamic Contrast Enhancement in the Diagnosis and Prognosis of Treatment Response in Patients with Epithelial Serous Ovarian Cancer. Cancers (Basel) 2022.
- Dastidar P, Mäenpää J, Heinonen T, Kuoppala T, Van Meer M, Punnonen R, Laasonen E. Magnetic resonance imaging based volume estimation of ovarian tumours: use of a segmentation and 3D reformation software. Comput Biol Med 2000;30:329-40. [Crossref] [PubMed]
- Liang J, Yang C, Zeng M, Wang X. TransConver: transformer and convolution parallel network for developing automatic brain tumor segmentation in MRI images. Quant Imaging Med Surg 2022;12:2397-415. [Crossref] [PubMed]
- Zhang J, Saha A, Zhu Z, Mazurowski MA. Hierarchical Convolutional Neural Networks for Segmentation of Breast Tumors in MRI With Application to Radiogenomics. IEEE Trans Med Imaging 2019;38:435-47. [Crossref] [PubMed]
- Zheng S, Lin X, Zhang W, He B, Jia S, Wang P, Jiang H, Shi J, Jia F. MDCC-Net: Multiscale double-channel convolution U-Net framework for colorectal tumor segmentation. Comput Biol Med 2021;130:104183. [Crossref] [PubMed]
- Carion N, Massa F, Synnaeve G, Usunier N, Kirillov A, Zagoruyko S. End-to-end object detection with transformers. European conference on computer vision. Springer; 2020:213-29.
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, Uszkoreit J, Houlsby N. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 2020.
- Jiang Y, Chang S, Wang Z. Transgan: Two pure transformers can make one strong gan, and that can scale up. Adv Neural Inf Process Syst 2021;34:14745-58.
- Guo R, Niu D, Qu L, Li Z. SOTR: Segmenting Objects with Transformers. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021:7157-66.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer; 2015:234-41.
- Chen LC, Papandreou G, Schroff F, Adam H. Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587 2017.
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 2018;11045:3-11. [Crossref] [PubMed]
- Zhao H, Shi J, Qi X, Wang X, Jia J. Pyramid Scene Parsing Network. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017:2881-90.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y. TransUNet: Transformers Make Strong Encoders for Medical Image Segmentation. arXiv preprint arXiv:2102.04306 2021.
- Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M. Swin-unet: Unet-like pure transformer for medical image segmentation. arXiv preprint arXiv:2105.05537 2021.
- Pan X, Luo P, Shi J, Tang X. Two at Once: Enhancing Learning and Generalization Capacities via IBN-Net. Proceedings of the European Conference on Computer Vision. 2018:464-79.
- Zhou K, Liu Z, Qiao Y, Xiang T, Loy CC. Domain Generalization: A Survey. IEEE Trans Pattern Anal Mach Intell 2022; Epub ahead of print. [Crossref] [PubMed]
- Luo S, Li Y, Gao P, Wang Y, Serikawa S. Meta-seg: A survey of meta-learning for image segmentation. Pattern Recognit 2022;126:108586. [Crossref]
- Karimi D, Warfield SK, Gholipour A. Transfer learning in medical image segmentation: New insights from analysis of the dynamics of model parameters and learned representations. Artif Intell Med 2021;116:102078. [Crossref] [PubMed]
- Peng D, Lei Y, Hayat M, Guo Y, Li W. Semantic-aware domain generalized segmentation. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2022:2594-605.
- Bian C, Yuan C, Ma K, Yu S, Wei D, Zheng Y. Domain Adaptation Meets Zero-Shot Learning: An Annotation-Efficient Approach to Multi-Modality Medical Image Segmentation. IEEE Trans Med Imaging 2022;41:1043-56. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Zhou T, Ruan S, Canu S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019;3-4:100004. [Crossref]
- Valanarasu JMJ, Sindagi VA, Hacihaliloglu I, Patel VM. KiU-Net: Overcomplete Convolutional Architectures for Biomedical Image and Volumetric Segmentation. IEEE Trans Med Imaging 2022;41:965-76. [Crossref] [PubMed]



