
A deep learning model based on the attention mechanism for automatic segmentation of abdominal muscle and fat for body composition assessment
Introduction
Lung cancer is the second most prevalent malignancy worldwide based on 2020 World Health Organization (WHO) global lung cancer statistics, accounting for 11.4% of all cancers and 19.3 million new cases, which is only slightly lower than the global proportion of breast cancer at 11.8%. In addition, lung cancer is the leading cause of death among all cancers, with more than 1.8 million/10 million people dying from lung cancer each year at a mortality rate of 18% (1). Approximately 70% of lung cancer patients are diagnosed at an advanced stage and have a dismal prognosis (2). Consequently, for patients with advanced lung cancer, identifying biomarkers that enable early accurate prediction of treatment-related toxicities, treatment response, and prognosis so that personalized treatment plans can be adjusted in a timely manner is the key to further improving patients’ prognosis and quality of life.
Muscle and fat quantification data obtained from body composition analysis are expected to be new stable biomarkers. Body composition analysis is an analytical method for measuring the proportion of body components (mainly muscle and fat); it is commonly used to assess nutritional status and manage obesity and its related problems, including metabolic diseases and cardiovascular diseases, such as hypertension (3). Currently, there is growing evidence that changes in body composition, such as sarcopenia (defined as decreased skeletal muscle mass) with intramuscular fat infiltration, are associated with a higher risk of cancer development, a poorer prognosis of cancer patients, and higher chemotherapy toxicity (4). Sarcopenia has a high prevalence among various cancer patients, especially for non-small cell lung cancer, with a prevalence as high as 43%. In particular, sarcopenia has been confirmed as an independent prognostic factor in patients with lung cancer (5). Therefore, how to accurately quantify muscle and fat has become a pressing need.
Computed tomography (CT) has emerged the gold standard for assessing the quantification of muscle and fat mass due to its ability to provide clear anatomical detail that allows for the direct assessment of muscle and fat mass volume (6). Surveys of CT are the standard of care for staging and monitoring a range of cancers and are essential in guiding diagnostic efforts (7-10). In patients with lung cancer, CT is one of the most widely used imaging modalities and is already included in the clinical workflow acquired for tumor staging, surgical planning, and treatment monitoring because CT scans are readily available without the need for additional ionizing radiation (11,12). CT is a tomographic image obtained by passing X-rays through body material with different attenuation coefficients, which clearly reveals human muscle and adipose tissue, allowing for the comprehensive monitoring and quantification of muscle loss associated with attenuation and sarcopenia throughout the entire course of lung cancer treatment. Thus, the ability to adequately use CT to track dynamic muscle changes can greatly improve the prognosis of those suffering from sarcopenia, making it of great value in tailoring individualized treatment plans. However, the development of CT-based body composition is still currently at the stage of manual, layer-by-layer segmentation and semiautomated segmentation by experts (13-16). How to achieve fast, accurate, and objective segmentation of muscles and fats on CT images is an urgent problem to be solved.
The expeditious development of deep learning techniques has shown the potential to overcome these limitations (17-22). Neural networks enable the automatic extraction of features (23), which dramatically overcomes the drawbacks and outperforms traditional medical image segmentation algorithms that rely excessively on the a priori knowledge of medical experts. Unlike existing methods, neural networks are more objective, and their generalization capability and portability can be quickly extended to different task scenarios through transfer learning (24). Traditional methods of measuring the abdominal region are time-consuming and cumbersome to perform manually, so medical experts will only measure the volume of the abdominal region by randomly selecting a layer of slices, which can only measure the area of the region, and the method of predicting the overall volume from the local area is imprecise. Several studies have reported high accuracy of automatic quantification of abdominal muscle and/or fat area through deep learning (25-36).
Similar to traditional methods, these deep learning methods are based on a single slice for measurement, while there are approximately 20 slices at the L3 level, and the area of the region is not consistent or even varies greatly for each slice. The selection of the level in existing clinical trials is randomized, and this approach may lead to relatively poor reproducibility between studies and can result in large random errors. Therefore, to reduce randomness, we aimed to segment each slice for each patient at the L3 level to calculate the regional volume more accurately and objectively. By segmenting all slices at the L3 level for each patient, we obtained 3-dimensional segmentation results by stacking slices and obtaining volumetric quantification results.
In this study, we used U-Net as the framework to target subcutaneous adipose tissue (SAT), skeletal muscle (SM), and visceral adipose tissue (VAT) to develop an automated abdominal CT image segmentation system in each patient’s abdominal CT slice sequence. We further incorporated an attention mechanism that assigns weights at multiple scales to allow the model to adaptively focus on more informative features and evaluated its performance. We present the following article in accordance with the MDAR reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-330/rc).
Methods
Data acquisition and preprocessing
We retrospectively derived data derived from scans of 43 patients with lung cancer (24 males; 19 females; age of 55.9±10.2 years) who received standard contrast-enhanced CT scans from a GE Medical Systems Revolution CT scanner (GE Healthcare, Chicago, IL, USA) between October 2018 and March 2021. The patients were randomly grouped into a training set and a testing set (545:301). Two patients only received noncontrast CT scans, which were used for independent testing. Based on each patient’s body weight, contrast CT scans were performed with a bolus of the contrast medium (Loversol, 350 mg/mL; Jiangsu Hengrui Medicine, Lianyungang, China) injected at a standard dosage of 1.5 mL/kg and a flow rate of 2.0–3.0 mL/s. The venous phase of the contrast-enhanced CT scans was acquired 50 s after injection of the contrast medium.
All the noncontrast and contrast-enhanced CT scans were performed under the following scanning parameter settings: tube voltage =120 kV, tube current selection from 150 to 600 mA, and matrix =512×512. The field of view (40–50 cm) was adapted to each patient’s size during scanning. The CT images were reconstructed using the standard algorithm with a reconstructed slice thickness of 1.25 mm and an adaptive statistical iterative reconstruction-V (ASIR-V) of 50%. All the obtained images were reviewed by 2 board-certified radiologists (PH and YR), and unsatisfactory images with a low signal-to-noise ratio or overt motion artifacts were removed. As a result, a total of 846 CT contrast-enhanced images in the venous phase of 43 patients (with the number of CT slices per patient ranging from 16 to 24) were obtained from the Chinese Academy of Medical Sciences Cancer Hospital, Shenzhen Hospital.
In our dataset, all CT scans collected covered the thorax, abdomen, and pelvis. However, it has been shown that whole-body adipose tissue and SM volumes show a correlation with those obtained from cross-sectional (axial) CT images of the third lumbar vertebra (L3) (14,25,26,37). Therefore, only abdominal CT images at the level of the third lumbar vertebra were selected for analysis. Two board-certified radiologists (PH and YR) manually segmented SAT, SM, and VAT on around 16–24 consecutive abdominal slices as ground truth labels, which were then reviewed by a senior radiologist (ZL) with any discrepancy dissolved through discussion. Subsequently, 28 patients with a total of 545 enhanced CT slices were used as the training and validation sets (Table 1; cohort 1), 15 patients with a total of 301 enhanced CT slices were used as independent test set 1 (Table 1; cohort 2) to validate the generalization of the model, and 2 patients with a total of 42 plain CT slices were used as independent test set 2 (Table 1; cohort 3) to validate the robustness of the model. The specific information related to the dataset is shown in Table 1.
Table 1
| Characteristics | Cohort 1 (training and validation set) (n=28) | Cohort 2 (test set 1) (n=15) | Cohort 3 (test set 2) (n=2) |
|---|---|---|---|
| Number of scans | 545 | 301 | 42 |
| Age (years) | 52.6±11.5 [32–72] | 59.2±8.9 [39–79] | 60.0±3.0 [57–63] |
| Male (%), (male:female) | 42.9 (12:16) | 80 (12:3) | 100 (2:0) |
| Slice thickness (mm) | 1.25 | 1.25 | 1.25 |
| Height (cm) | 162.5±8.8 [147–178] | 166.3±5.9 [158–178] | 176.0±2.0 [174–178] |
| Weight (kg) | 58.1±9.5 [40–78] | 60.7±7.9 [46–71] | 68.3±4.3 [64–72.5] |
| Enhanced/plain | Enhanced | Enhanced | Plain |
Data are presented as mean ± standard deviation (min – max).
Some research shows that abdominal muscles could be calibrated with a predefined threshold of –29 to +150 Hounsfield units (HU) and that abdominal adipose tissue could be demarcated with a predetermined threshold of –190 to –30 HU (38). Another study stated that setting the HU value to –128 for CT images above 127 and below –128 could improve the performance of convolutional neural network (CNN) models for soft tissue segmentation in CT images (39). Hence, in this study, we preprocessed the data accordingly before feeding them into the model. The HU values below –128 and above 150 in the CT images were set to –128 and 150, respectively, to reduce the amount of information given to the CNN model, and the feature matrix was z score–normalized as follows:
where , and ε is a small constant used to smooth the formula to prevent division by 0. The feature matrix had a mean of 0 and a standard deviation of 1 after processing.
Additionally, data augmentation was performed to the available training set, primarily random rotation, horizontal flipping, and vertical flipping, to reduce the possibility of network overfitting and enhance generalization (probability =0.5).
Training process
Figure 1 depicts an overview of the segmentation. The training dataset was labeled with the ground truth by medical experts, and the results were enhanced and normalized with augmentation for training images and labels. The ensemble technique was used to tackle the problem of potential overfitting in a small sample size. Due to the relatively small sample size, we performed a 5-fold cross-validation of the model to ensemble it into a more robust model in order to effectively use the limited data and obtain a more reasonable and accurate evaluation of the model. The training data with ground truth labels were used as input to the network to learn the parameters. The trained model was evaluated using the validation set for each epoch in terms of the Dice similarity coefficient (DSC) and network loss. The model with the highest average DSC in the validation set was retained for independent testing in cohort 2 after training. Subsequently, we normalized the test data and fed it into the best network model we saved (the model is an integration of 5 models generated through five-fold cross-validation) to obtain the deep learning model’s predicted output. We computed the prediction with the corresponding ground truth labels for relevant comparisons to analyze the performance of the model.
Module description
The model was an advancement in the framework of supervised U-Net (40). The overall framework is shown in Figure 2A. Specifically, the encoder and decoder subnet consisted of modules with a depth of 5, each of which contained a residual convolution submodule (41) and an amended efficient channel attention (ECA) (42) submodule. In the residual convolution submodule, the number of channels of the feature map was changed by adjusting the number of convolution kernels, which were summed with the original feature map after the convolution layer, batch normalization, and rectified linear unit (ReLU) activation function. The amended ECA submodule was named the selective efficient channel attention (SECA) block, as shown in Figure 2B.
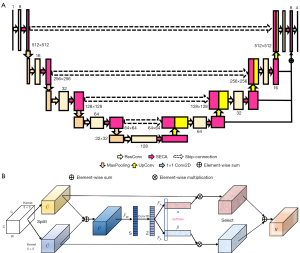
As illustrated in Figure 2B, the given feature map U=(u1,u2,…,uc) was split into 2 branches. Each branch was implemented with a convolution operation to obtain the information of different receptive fields. We implemented 2 conventional convolutions with kernel sizes of 3×3 and 5×5 and with the activation function of ReLU in this paper. We fused the different information from 2 branches via an element-wise summation:
Then, we implemented global average pooling (Fgp) and dimensionality reduction in each channel to obtain channel attention statistics , and the i-th element of S was calculated by shrinking as follows:
We avoided dimensionality reduction using 1-dimensional convolution and effective learning channel attention to obtain , and then Z was subjected to a softmax transformation to obtain the channel attention vectors and . We multiplied the attention vectors α and β with the result of the 2 convolutions and then added the results to obtain the final feature map V.
Our proposed SECANet channel-wise guides for the model on how to assign representations focused on the value of convolutional kernels. This was done by using information aggregated from multiscale feature maps and by effectively capturing the cross-channel interactions at multiple scales by avoiding dimensionality reduction through 1-dimensional convolution.
Loss function and optimization methods
The loss function used Lovasz-Softmax loss (43), a direct optimization method for joint loss-averaged intersection in neural networks proposed in the context of semantic segmentation. Lovasz-Sotfmax loss outperformed the conventional cross-entropy loss function for the multiclass segmentation task. The optimal hyperparameters were experimentally determined. A batch size of 16 sections was used. The learning rate was initially set to 0.001 with Adam optimization (44), and a cosine annealing algorithm was used to reduce the learning rate gradually to a minimum of 0.000001 and by training a total of 200 epochs, setting the 25th epoch as the iteration position for the first restart of the learning rate. The network was trained on a Nvidia GeForce RTX 3080 (Nvidia, Santa Clara, CA, USA) and written in Python 3.9.6 (Python Software Foundation, Beaverton, OR, USA) using PyTorch 1.9.0 (open source) backend. We performed 5-fold cross-validation and retained the best model for each fold. The entire cross-validation model training process took approximately 35 hours.
Assessment indicators
DSC is an ensemble similarity measure, which is ordinarily used as the primary evaluation metric in the field of medical image segmentation. DSC illustrates the degree of overlap between the ground truth labels and the number of pixels of the neural network output as calculated by the following equation:
where true positive (TP) is the number of pixels correctly detected (i.e., pixels included in both the ground truth label and the segmentation result), false positive (FP) is the number of pixels judged as positive samples but that are actually negative samples (i.e., pixels included in the segmentation result but not in ground truth labels), and false negative (FN) is the number of pixels that are judged as negative samples but are truly positive samples (i.e., pixels that are included in the ground truth label but not in the segmentation result).
Dice coefficients focus on the interior of the segmentation and have the defect of being insensitive to the boundary inscription. Hausdorff distance (HD) emphasizes the edges of the segmentation and is capable of complementing Dice. HD is the maximum value of the shortest distance from an element in one set to another set. Given 2 sets A=(a1,a2,…,ap) and B=(b1,b2,…,bq), the HD is the following:
where represents the distance paradigm of 2 sets, generally the Euclidean distance.
The Jaccard coefficient is mainly applied to the similarity between individuals of Boolean attributes as the ratio of the size of the intersection of A and B to the size of the concatenation of A and B, and is calculated as follows:
where p denotes the number of pixels where both samples A and B have a Boolean value of 1, q represents the number of pixels where sample A is 1 and B is 0, and r shows the number of pixels for which at A is 0 and B is 1.
In particular, we also considered precision and recall as auxiliary evaluation metrics, which are determined as follows:
Output of the model
Considering that the task objective of this study was to segment SM, SAT, and VAT, the model output was a 4-channel probability map with dimensions of 4×512×512. Each dimension corresponded to a probability matrix of background, SAT, SM, and VAT. The probability gave the confidence level that the model predicted each pixel to be in the region of interest (ROI). For each channel, pixels with a probability above 0.5 were classified as the result of the segmentation. For each slice in the test set, 5 different probability maps were predicted using the 5 models from 5-fold cross-validation. The final probability map for a test slice was calculated by combining the probabilities of the 5 probability maps.
Since the output was a probability map with 4 channels, the ground truth label needed to be one-hot coded accordingly when various evaluation metrics of output and label were being calculated. DSC, Jaccard coefficient, HD95, precision, and recall were used to measure the similarity between the segmentation output and the ground truth.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the ethics committee of Cancer Hospital Chinese Medical Sciences, Shenzhen Hospital and individual consent for this retrospective analysis was waived.
Results
Ensemble learning performance
The segmentation results were successfully generated by our model for the independent test dataset, and the evaluation of the model performance was accomplished with a variety of metrics. The prediction accuracies are shown in Table 2 for the ensemble of ResUNet, the ensemble of ResECAUNet models, the single SECAUNet, model, and the ensemble of SECAUNet models. The results showed that the model performed satisfactorily on the test set. The ensemble of SECAUNet models outperformed the ensemble of ResUNet models in terms of segmentation results for SAT, SM, and VAT from a quantitative perspective. In addition, the performance of the ensemble model using cross-validation was significantly better than that of the single model, while the performance of the single SECAUNet-based model was comparable to that of the ensemble model based on ResUNet. This result suggests the preeminence of SECAUNet and correspondingly illustrates that ensemble learning outperforms individual learning.
Table 2
| Measure | Tissue | DSC | Jaccard | Precision | Recall | HD95 |
|---|---|---|---|---|---|---|
| Ensemble-SECAUNet | SAT | 0.93±0.06* | 0.87±0.10* | 0.98±0.02 | 0.88±0.11* | 3.78±2.18* |
| SM | 0.96±0.02* | 0.92±0.03* | 0.95±0.02* | 0.96±0.02 | 4.47±0.95* | |
| VAT | 0.87±0.11* | 0.79±0.17 | 0.82±0.16 | 0.94±0.05* | 20.68±26.91 | |
| Single-SECAUNet | SAT | 0.91±0.06 | 0.85±0.10 | 0.98±0.02 | 0.87±0.10 | 4.19±2.11 |
| SM | 0.95±0.02 | 0.90±0.03 | 0.94±0.02 | 0.95±0.02 | 5.55±1.56 | |
| VAT | 0.86±0.12 | 0.77±0.17 | 0.81±0.17 | 0.93±0.06 | 21.80±26.46 | |
| Ensemble-ResUNet | SAT | 0.91±0.05 | 0.84±0.08 | 0.98±0.01 | 0.85±0.09 | 4.71±1.51 |
| SM | 0.95±0.02 | 0.91±0.03 | 0.94±0.03 | 0.97±0.02* | 4.70±1.36 | |
| VAT | 0.87±0.11* | 0.79±0.16* | 0.83±0.16* | 0.93±0.06 | 20.78±26.78 | |
| Ensemble-ResECAUNet | SAT | 0.92±0.06 | 0.86±0.10 | 0.99±0.01* | 0.87±0.10 | 3.81±2.16 |
| SM | 0.95±0.02 | 0.91±0.03 | 0.94±0.02 | 0.96±0.02 | 4.64±1.05 | |
| VAT | 0.87±0.12 | 0.78±0.17 | 0.82±0.16 | 0.94±0.06 | 20.26±26.28* |
Data are presented as mean ± standard. *, the numbers indicate the best values in the same group of attributes. SAT, subcutaneous adipose tissue; SM, skeletal muscle; VAT, visceral adipose tissue; DSC, Dice similarity coefficient; HD95, Hausdorff distance at 95th percentile.
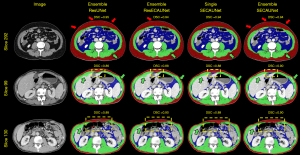
We randomly selected 3 original slices for the result presentation. In Figure 3, the ensemble of SECAUNet models had better continuous and accurate segmentation in the SAT and SM area compared to each of the remaining models. The other models were more prone to under- and oversegmentation in the SAT and SM areas. By comparing the yellow boxed areas of slice 99 and slice 130, it can be observed that, in the complex area, SECAUNet shows less miss-segmentation and exhibits superior performance.
We independently tested the segmentation performance of the model on plain CT slices with training exclusively using the enhanced CT slices in cohort 1. Table 3 illustrates the segmentation results of the ensemble ResUNet, ResECAUNet, and SECAUNet models on 42 plain CT slices. The quantitative results revealed that the model trained using only enhanced CT slices could also obtain superior segmentation results on plain CT slices during testing, which substantiated the robustness of the model. The qualitative results are shown in Figure 4A, which presents the slices of high DSC, median DSC, and low DSC in the plain CT dataset. In addition, Figure 4B illustrates the zoomed-in details of the ROIs for SM and VAT in Figure 4A. The qualitative results showed that our model outperformed the others in details compared to the ground truth.
Table 3
| Measure | Tissue | DSC | Jaccard | Precision | Recall | HD95 |
|---|---|---|---|---|---|---|
| Ensemble-SECAUNet | SAT | 0.90±0.09 | 0.82±0.01 | 0.99±0.01* | 0.83±0.02 | 5.26±0.36* |
| SM | 0.95±0.01* | 0.90±0.02* | 0.95±0.02* | 0.94±0.02 | 5.61±1.64* | |
| VAT | 0.92±0.03* | 0.86±0.05* | 0.93±0.03* | 0.92±0.04* | 7.40±3.38* | |
| Ensemble-ResUNet | SAT | 0.91±0.01* | 0.84±0.01* | 0.99±0.01 | 0.85±0.01* | 5.27±0.38 |
| SM | 0.93±0.01 | 0.88±0.02 | 0.93±0.03 | 0.95±0.02* | 7.13±2.53 | |
| VAT | 0.92±0.03 | 0.85±0.05 | 0.93±0.03 | 0.91±0.04 | 7.73±3.44 | |
| Ensemble-ResECAUNet | SAT | 0.90±0.01 | 0.82±0.01 | 0.99±0.01 | 0.83±0.02 | 5.28±0.38 |
| SM | 0.92±0.02 | 0.86±0.03 | 0.91±0.03 | 0.94±0.02 | 14.88±9.33 | |
| VAT | 0.92±0.03 | 0.86±0.05 | 0.93±0.04 | 0.92±0.04 | 7.81±3.59 |
Data are presented as mean ± standard. *, the numbers indicate the best values in the same group of attributes. SAT, subcutaneous adipose tissue; SM, skeletal muscle; VAT, visceral adipose tissue; DSC, Dice similarity coefficient; HD95, Hausdorff distance at 95th percentile.
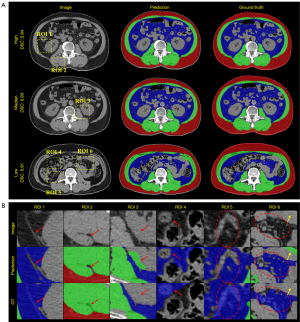
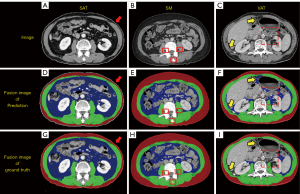
Difference between output and ground truth
In the low metric slices, we found that the model predictions differed qualitatively from the ground truth. Representative examples of SECAUNet-based segmentation are demonstrated in Figure 5. In Figure 5A, the model excludes the skin when segmenting the SAT area, but the ground truth classifies it as part of the SAT (Figure 5D,5G). In the SM area, the ground truth includes part of the cone, but prediction screened it as a region of no interest. The VAT area was at the lowest level of each of the metrics evaluated in our model output compared to ground truth. We found that slices with a more segmented area of VAT (Figure 3, Slice 292; overall DSC =0.94; DSC of VAT =0.96) had a correspondingly high metric, while slices with a less or extremely undersized segmented area of VAT (Figure 5C; overall DSC =0.92; DSC of VAT =0.49) had a lower or exceptionally low metric.
The Bland-Altman plots for all test datasets indicated good agreement between ground truth measurements and those of the SECAUNet-based segmentations for SAT, SM, and VAT, with mean differences of –14.42, 1.8, and 6.7 cm2, respectively (Figure 6). the consistencyof muscle was the best, in contrast to the SAT, which was slightly worse, probably due to the difference between ground truth with the segmentation results at the skin level. Overall, our results showed that the majority of test cases (n=299, 99.34%) were within the 95% consistency limits.

Discussion and conclusions
Our proposed ensemble model of SECAUNet based on the attention mechanism accurately segmented subcutaneous fat, SM, and visceral fat on abdominal CT images. The model was well-trained to perform on all evaluation metrics, which was in accordance with the previous deep learning studies on abdominal muscle and/or fat (32-35). Additionally, fully automated segmentation using a deep learning model only took a few seconds, while manual segmentation required more than 3 min, indicating comparable measurement performance of the model.
The SAT areas segmented by our model did not differ significantly from the ground truth for the slices of patients with thin skin areas that made up the majority of the test set. For the slices from patients with thick skin, our model excluded these when generating segmentation results. The SAT area of ground truth contained skin, which resulted in low values of DSC, especially in slices with few SAT areas. The inclusion or exclusion of skin in the SAT area greatly affected the DSC of SAT, resulting in the output of the segmentation model being actually more accurate than ground truth but showing a low DSC. In the SM area, the output of the segmentation model in the small target area was discordant with the ground truth.
We observed that, in the VAT area, our model did not perform well enough; hence we focused on analyzing the causes of this issue. In our test dataset, slices with less VAT area accounted for a large ratio, which led to the problem of unbalanced pixel proportions and seriously affected the segmentation of the VAT area. Additionally, comparing Figure 5F and Figure 5I showed that the VAT area derived from our model segmentation was more objective than the VAT area of ground truth, while in this case, the DSC of VAT was only 0.49, which greatly and wrongly affected the performance of evaluating our model.
In several previous studies that used deep learning to segment abdominal muscle and/or fat, the models were trained, validated, and tested using enhanced CT and plain CT (32-36). Our model was trained and validated on enhanced CT images only and independently tested on enhanced CT images and plain CT images, both of which had high accuracy results, which demonstrated the robustness of our model. In clinical practice, deep learning models may need to assess medical data generated by different scanners and acquisition protocols, and differences in machine vendors and data acquisition may interfere with the deep learning model, leading to inconsistent performance (45). But the performance and generalization of our model were further improved by the ensemble technique. Our model was trained, validated, and tested on a dataset containing all slices at the L3 level for each patient. Considering the results, our model can predict segmentation in the future if there is a new dataset of patients with CT slices at any position at the L3 level.
Our study had a number of limitations. Since the area of ROI is very small (e.g., SAT and skin), the model is prone to incorrect segmentation in some cases. There were large differences between individuals, especially in the VAT area, which suggested that the model had not been fully trained with diverse and heterogeneous images, which is a limitation of the model in the absence of extensive training. The proposed model can overcome such shortcomings by increasing the variability and heterogeneity of the training images, as well as by including images from a wider range of institutions. We used CT imaging to develop a deep learning model instead of using magnetic resonance imaging (MRI) or a combination of both, which might be more effective (46). The segmentation of small targets also remains a class of challenges to be solved. Images with little VAT were prone to the problem of category imbalance. One possible solution is to adjust the weight of the foreground and background in the loss function to amplify the weight of the foreground and suppress the weight of the background. Another possible solution is to minimize the amount of downsampling for the model to avoid excessive information loss in the target region after multiple instances of downsampling.
In this study, we proposed a method for the automatic outlining of subcutaneous fat, SM, and visceral fat areas on L3 cross-sectional CT images. We obtained better segmentation results compared to U-Net by proposing an improvement for the attention mechanism, which was accurate in terms of DSC and other assessment metrics. Our model was completely trained on enhanced CT images, and it obtained good results on plain CT images, demonstrating high robustness. Our model can be used for body composition analysis with minimal manual effort. It may be valuable for identifying new biomarkers of health and disease and in developing personalized treatment plans.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 62101540), the Shenzhen Excellent Technological Innovation Talent Training Project of China (No. RCJC20200714114436080), National High Level Hospital Clinical Research Funding (No. 2022-PUMCH-B-070), and the Shenzhen High-level Hospital Construction Fund.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-330/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-330/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol 2017;7:170070. [Crossref] [PubMed]
- Machann J, Horstmann A, Born M, Hesse S, Hirsch FW. Diagnostic imaging in obesity. Best Pract Res Clin Endocrinol Metab 2013;27:261-77. [Crossref] [PubMed]
- Shachar SS, Deal AM, Weinberg M, Williams GR, Nyrop KA, Popuri K, Choi SK, Muss HB. Body Composition as a Predictor of Toxicity in Patients Receiving Anthracycline and Taxane-Based Chemotherapy for Early-Stage Breast Cancer. Clin Cancer Res 2017;23:3537-43. [Crossref] [PubMed]
- Yang M, Shen Y, Tan L, Li W. Prognostic Value of Sarcopenia in Lung Cancer: A Systematic Review and Meta-analysis. Chest 2019;156:101-11. [Crossref] [PubMed]
- Guerri S, Mercatelli D, Aparisi Gómez MP, Napoli A, Battista G, Guglielmi G, Bazzocchi A. Quantitative imaging techniques for the assessment of osteoporosis and sarcopenia. Quant Imaging Med Surg 2018;8:60-85. [Crossref] [PubMed]
- Xia Q, Liu J, Wu C, Song S, Tong L, Huang G, Feng Y, Jiang Y, Liu Y, Yin T, Ni Y. Prognostic significance of (18)FDG PET/CT in colorectal cancer patients with liver metastases: a meta-analysis. Cancer Imaging 2015;15:19. [Crossref] [PubMed]
- Podoloff DA, Advani RH, Allred C, Benson AB 3rd, Brown E, Burstein HJ, et al. NCCN task force report: positron emission tomography (PET)/computed tomography (CT) scanning in cancer. J Natl Compr Canc Netw 2007;5:S1-22; quiz S23-2. [Crossref] [PubMed]
- Pastorino U, Bellomi M, Landoni C, De Fiori E, Arnaldi P, Picchio M, Pelosi G, Boyle P, Fazio F. Early lung-cancer detection with spiral CT and positron emission tomography in heavy smokers: 2-year results. Lancet 2003;362:593-7. [Crossref] [PubMed]
- Diederich S, Wormanns D, Semik M, Thomas M, Lenzen H, Roos N, Heindel W. Screening for early lung cancer with low-dose spiral CT: prevalence in 817 asymptomatic smokers. Radiology 2002;222:773-81. [Crossref] [PubMed]
- Imai K, Minamiya Y, Ishiyama K, Hashimoto M, Saito H, Motoyama S, Sato Y, Ogawa J. Use of CT to evaluate pleural invasion in non-small cell lung cancer: measurement of the ratio of the interface between tumor and neighboring structures to maximum tumor diameter. Radiology 2013;267:619-26. [Crossref] [PubMed]
- Wu MT, Chang JM, Chiang AA, Lu JY, Hsu HK, Hsu WH, Yang CF. Use of quantitative CT to predict postoperative lung function in patients with lung cancer. Radiology 1994;191:257-62. [Crossref] [PubMed]
- McDonald AM, Swain TA, Mayhew DL, Cardan RA, Baker CB, Harris DM, Yang ES, Fiveash JB. CT Measures of Bone Mineral Density and Muscle Mass Can Be Used to Predict Noncancer Death in Men with Prostate Cancer. Radiology 2017;282:475-83. [Crossref] [PubMed]
- Shen W, Punyanitya M, Wang Z, Gallagher D, St-Onge MP, Albu J, Heymsfield SB, Heshka S. Total body skeletal muscle and adipose tissue volumes: estimation from a single abdominal cross-sectional image. J Appl Physiol (1985) 2004;97:2333-8. [PubMed]
- Polan DF, Brady SL, Kaufman RA. Tissue segmentation of computed tomography images using a Random Forest algorithm: a feasibility study. Phys Med Biol 2016;61:6553-69. [Crossref] [PubMed]
- Kamiya N, Zhou X, Chen H, Muramatsu C, Hara T, Yokoyama R, Kanematsu M, Hoshi H, Fujita H. Automated segmentation of psoas major muscle in X-ray CT images by use of a shape model: preliminary study. Radiol Phys Technol 2012;5:5-14. [Crossref] [PubMed]
- Liu X, Song L, Liu S, Zhang Y. A Review of Deep-Learning-Based Medical Image Segmentation Methods. Sustainability 2021;13. [Crossref]
- Jones KI, Doleman B, Scott S, Lund JN, Williams JP. Simple psoas cross-sectional area measurement is a quick and easy method to assess sarcopenia and predicts major surgical complications. Colorectal Dis 2015;17:O20-6. [Crossref] [PubMed]
- Greco F, Mallio CA. Artificial intelligence and abdominal adipose tissue analysis: a literature review. Quant Imaging Med Surg 2021;11:4461-74. [Crossref] [PubMed]
- Attanasio S, Forte SM, Restante G, Gabelloni M, Guglielmi G, Neri E. Artificial intelligence, radiomics and other horizons in body composition assessment. Quant Imaging Med Surg 2020;10:1650-60. [Crossref] [PubMed]
- Huang Z, Zou S, Wang G, Chen Z, Shen H, Wang H, Zhang N, Zhang L, Yang F, Wang H, Liang D, Niu T, Zhu X, Hu Z. ISA-Net: Improved spatial attention network for PET-CT tumor segmentation. Comput Methods Programs Biomed 2022;226:107129. [Crossref] [PubMed]
- Huang Z, Tang S, Chen Z, Wang G, Shen H, Zhou Y, Wang H, Fan W, Liang D, Hu Y, Hu Z. TG-Net: Combining transformer and GAN for nasopharyngeal carcinoma tumor segmentation based on total-body uEXPLORER PET/CT scanner. Comput Biol Med 2022;148:105869. [Crossref] [PubMed]
- Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep Learning: A Primer for Radiologists. Radiographics 2017;37:2113-31. [Crossref] [PubMed]
- Pan SJ, Tsang IW, Kwok JT, Yang Q. Domain adaptation via transfer component analysis. IEEE Trans Neural Netw 2011;22:199-210. [Crossref] [PubMed]
- Lee H, Troschel FM, Tajmir S, Fuchs G, Mario J, Fintelmann FJ, Do S. Pixel-Level Deep Segmentation: Artificial Intelligence Quantifies Muscle on Computed Tomography for Body Morphometric Analysis. J Digit Imaging 2017;30:487-98. [Crossref] [PubMed]
- Burns JE, Yao J, Chalhoub D, Chen JJ, Summers RM. A Machine Learning Algorithm to Estimate Sarcopenia on Abdominal CT. Acad Radiol 2020;27:311-20. [Crossref] [PubMed]
- Decazes P, Tonnelet D, Vera P, Gardin I. Anthropometer3D: Automatic Multi-Slice Segmentation Software for the Measurement of Anthropometric Parameters from CT of PET/CT. J Digit Imaging 2019;32:241-50. [Crossref] [PubMed]
- Weston AD, Korfiatis P, Kline TL, Philbrick KA, Kostandy P, Sakinis T, Sugimoto M, Takahashi N, Erickson BJ. Automated Abdominal Segmentation of CT Scans for Body Composition Analysis Using Deep Learning. Radiology 2019;290:669-79. [Crossref] [PubMed]
- Hu P, Huo Y, Kong D, Carr JJ, Abramson RG, Hartley KG, Landman BA. Automated Characterization of Body Composition and Frailty with Clinically Acquired CT. Comput Methods Clin Appl Musculoskelet Imaging (2017) 2018;10734:25-35. [PubMed]
- Dabiri S, Popuri K, Cespedes Feliciano EM, Caan BJ, Baracos VE, Beg MF. Muscle segmentation in axial computed tomography (CT) images at the lumbar (L3) and thoracic (T4) levels for body composition analysis. Comput Med Imaging Graph 2019;75:47-55. [Crossref] [PubMed]
- Ackermans LLGC, Volmer L, Wee L, Brecheisen R, Sánchez-González P, Seiffert AP, Gómez EJ, Dekker A, Ten Bosch JA, Olde Damink SMW, Blokhuis TJ. Deep Learning Automated Segmentation for Muscle and Adipose Tissue from Abdominal Computed Tomography in Polytrauma Patients. Sensors (Basel) 2021;21:2083. [Crossref] [PubMed]
- Park HJ, Shin Y, Park J, Kim H, Lee IS, Seo DW, Huh J, Lee TY, Park T, Lee J, Kim KW. Development and Validation of a Deep Learning System for Segmentation of Abdominal Muscle and Fat on Computed Tomography. Korean J Radiol 2020;21:88-100. [Crossref] [PubMed]
- Dabiri S, Popuri K, Ma C, Chow V, Feliciano EMC, Caan BJ, Baracos VE, Beg MF. Deep learning method for localization and segmentation of abdominal CT. Comput Med Imaging Graph 2020;85:101776. [Crossref] [PubMed]
- Hemke R, Buckless CG, Tsao A, Wang B, Torriani M. Deep learning for automated segmentation of pelvic muscles, fat, and bone from CT studies for body composition assessment. Skeletal Radiol 2020;49:387-95. [Crossref] [PubMed]
- Edwards K, Chhabra A, Dormer J, Jones P, Boutin RD, Lenchik L, Fei B. Abdominal muscle segmentation from CT using a convolutional neural network. Proc SPIE Int Soc Opt Eng 2020;11317:113170L.
- Amarasinghe KC, Lopes J, Beraldo J, Kiss N, Bucknell N, Everitt S, Jackson P, Litchfield C, Denehy L, Blyth BJ, Siva S, MacManus M, Ball D, Li J, Hardcastle N. A Deep Learning Model to Automate Skeletal Muscle Area Measurement on Computed Tomography Images. Front Oncol 2021;11:580806. [Crossref] [PubMed]
- Mourtzakis M, Prado CM, Lieffers JR, Reiman T, McCargar LJ, Baracos VE. A practical and precise approach to quantification of body composition in cancer patients using computed tomography images acquired during routine care. Appl Physiol Nutr Metab 2008;33:997-1006. [Crossref] [PubMed]
- Prado CM, Lieffers JR, McCargar LJ, Reiman T, Sawyer MB, Martin L, Baracos VE. Prevalence and clinical implications of sarcopenic obesity in patients with solid tumours of the respiratory and gastrointestinal tracts: a population-based study. Lancet Oncol 2008;9:629-35. [Crossref] [PubMed]
- Shahedi M, Ma L, Halicek M, Guo R, Zhang G, Schuster DM, Nieh P, Master V, Fei B. A semiautomatic algorithm for three-dimensional segmentation of the prostate on CT images using shape and local texture characteristics. Proc SPIE Int Soc Opt Eng 2018;10576:1057616.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Springer International Publishing, 2015.
- Wu B, Waschneck B, Mayr CG. Convolutional Neural Networks Quantization with Double-Stage Squeeze-and-Threshold. Int J Neural Syst 2022;32:2250051. [Crossref] [PubMed]
-
Wang Q Wu B Zhu P Li P Zuo W Hu Q. ECA-Net: Efficient Channel Attention for Deep Convolutional Neural Networks. 2019 . Available online: https://arxiv.org/abs/1910.03151 - Berman M, Triki AR, Blaschko MB, editors. The Lovasz-Softmax Loss: A Tractable Surrogate for the Optimization of the Intersection-Over-Union Measure in Neural Networks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2018.
-
Kingma D Ba J. Adam: A Method for Stochastic Optimization. Computer Science 2014 . Available online: https://arxiv.org/abs/1412.6980 - Castro DC, Walker I, Glocker B. Causality matters in medical imaging. Nat Commun 2020;11:3673. [Crossref] [PubMed]
- Amjad A, Xu J, Thill D, O'Connell N, Li A. Deep Learning-based Auto-segmentation on CT and MRI for Abdominal Structures. International Journal of Radiation OncologyBiologyPhysics 2020;108:S100-S101. [Crossref]



