
A machine learning approach for preoperatively assessing pulmonary function with computed tomography in patients with lung cancer
Introduction
Among all cancer types, the prevalence and fatality rate of lung cancer rank first (1). Surgical removal of the affected lobe is one of the main therapies for lung cancer. However, as many patients with lung cancer are of advanced ages, heavy smokers, or have chronic obstructive pulmonary disease (COPD), they bear a high risk for postsurgical complications (2). Hence, preoperative pulmonary function test (PFT) is of great significance to pass patients through the perioperative period safely (3). However, due to variations in patient compliance and underlying diseases, some patients have low tolerance and compliance to routine PFT. Studies on preoperative evaluation of pulmonary function of patients with lung cancer are lacking. A few researchers have evaluated the pulmonary function of patients with lung cancer from the aspect of quantitative computed tomography (CT) parameters (4). However, there are no reports focusing on preoperative evaluation of pulmonary function in patients with lung cancer through lung CT imaging using machine learning approaches. Based on CT scans of patients with lung cancer, we have developed and verified a method which automatically extracts the radiomics features and uses a machine learning model to evaluate pulmonary function using the CT images. The method can supplement current pulmonary function assessment, which has several limitations, including long detection time, difficult patient cooperation, a high rate of false negatives, and frequent contraindications (5,6). Our long-term goal is to maximize the value of CT imaging of patients with lung cancer by developing a rapid and simple method for assessing pulmonary function.
Radiomics is an accurate and noninvasive tool that can be used to attain quantitative features based on images from regions of interest (ROIs) and analyzing them to develop decision-support tools (7). Being widely used in the assessment of lung nodules, radiomics can be performed to extract image features from each nodule and predict secondary cancers (8). Radiomics has shown great promise in extracting information from clinical images (9,10). An important field of radiomics is in cancer imaging, in which radiomics has been used for staging cancer, assessing treatment efficacy, and predicting patient survival (11). In lung cancer, radiomics has been used for differentiating between benign and malignant lesions (12), predicting histological subtypes of lung cancer (13), assessing the effect of cancer treatment such as radiation-induced lung injury (14), and estimating patient prognosis (15,16). As a newly developed methodology, radiomics in its development is closely associated with machine learning (17,18). We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-70/rc).
Methods
Data sets
We enrolled 188 patients from our hospital, including 24 with COPD, in this study. The patients were admitted to our hospital between 2016 and 2019. The demographic and clinical characteristics of the cohort are displayed in Table 1. The inclusion criteria were as follows: (I) having undergone preoperative PFT with good coordination, (II) having obtained a preoperative thoracic CT scan without obvious artifacts, and (III) an interval time between PFT and CT scanning not exceeding 3 days. The exclusion criteria were as follows: patients who met the above inclusion criteria but failed to be segmented by LK software (19). We collected preoperative pulmonary function examination data of patients, including forced vital capacity (FVC), the percentage of measured forced expiratory volume in the predicted value [FEV1 (% of predicted)], forced expiratory volume in the first second/forced vital capacity (FEV1/FVC), maximum vital capacity (VCmax), and CT data. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethics board of The First Affiliated Hospital of Guangzhou Medical University (No. 2022-70), and informed consent was provided by all the patients.
Table 1
| Characteristics | Median (range) or n (%) |
|---|---|
| Male: female | 64:124 (34%:66%) |
| Age (years) | 59.0 (50.0, 65.0) |
| Height (cm) | 161.00 (155.5, 167.0) |
| Weight (kg) | 60.20 (54.59, 67.91) |
| BMI (kg/m2) | 23.15 (21.56, 25.12) |
| FEV1 (% of predicted) | 100.65 (89.74, 110.44) |
| FVC (L) | 3.04 (2.57, 3.60) |
| FEV1/FVC | 79.65 (74.22, 82.90) |
| VCmax (L) | 3.11 (2.61, 3.61) |
BMI, body mass index; FEV1 (% of predicted), the percentage of measured forced expiratory volume in the predicted value; FVC, forced vital capacity; FEV1/FVC, forced expiratory volume in the first second/forced vital capacity; VCmax, maximum vital capacity.
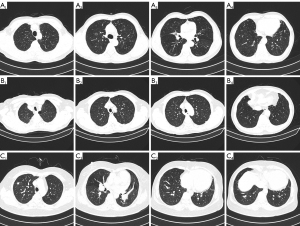
Figure 1 shows some typical cases included in our cohort. Figure 1A is a 71-year-old male with invasive adenocarcinoma (T1N0M0) of the upper right lung. Figure 1B is a 79-year-old male with invasive adenocarcinoma (T1N0M0) of the upper left lobe. Figure 1C is a 59-year-old female with invasive adenocarcinoma (T1aN0M0) of the upper right lung.
Clinical PFT
We carried out PFT by using the Cosmed Quark PFT series (Cosmed, Rome, Italy) pulmonary function instrument. All PFTs met the instrument quality control standards recommended by the American Thoracic Society (ATS)/European Respiratory Association (ERS). During the examination, patients adopted the sitting position while the main parameters, including FVC, FEV1 (% of predicted), FEV1/FVC (%), and VCmax, were measured.
CT imaging protocol and parameters
We used CT scanners from two manufacturers: Siemens SOMATOM Definition AS+ 128-slice CT or 64-slice CT (Siemens, Erlangen, Germany) and GE Revolution 256-slice CT (GE Healthcare, Chicago, IL, USA). Spiral CT was performed from head to foot under deep inspirations and breath-holding state, ranging from the upper edge of the lung apex to the lower edge of the costal diaphragmatic angle. The Siemens SOMATOM scanning parameters were as follows: tube voltage 120 kV, tube current automatic tube current modulation technology, field of view (FOV) 400 mm × 400 mm, matrix 512×512, slice thickness 1 mm, and slice spacing 1 mm. The GE Revolution scanning parameters were as follows: tube voltage 120 kV, tube current automatic tube current modulation technology, FOV 350 mm × 350 mm, matrix 512×512, slice thickness 0.625 mm, and slice spacing 0.625 mm. All patients only received one preoperative CT scan in our study. The quality of all the CT images obtained in our study was verified by two independent chest radiologists.
Experimental procedure
First, for the CT images of the 188 patients, the whole lung lobe and airway were segmented by the automatic segmentation module of LK software and 3 ROIs were obtained. Second, 107 radiomics features were extracted from the 3 ROIs using the PyRadiomics module of AK software (Artificial Intelligence Kit v.3.3.0; GE Healthcare). For the extracted features, we used the median method to replace missing values, the cap method to process outliers, and Z score to standardize. The 188 cases were randomly grouped in a ratio of 7:3, 70% of which were used to train the machine learning model and 30% to test the model. Third, we adopted Spearman correlation and stepwise regression to reduce the dimension of features and used logistic regression machine learning models to construct the prediction models evaluated by correlation coefficient and root-mean-square error (RMSE). All statistical analyses were performed using the R language, version 3.6.3 (R Foundation for Statistical Computing, Vienna, Austria). A P value <0.05 was considered statistically significant. The flowchart of our image processing and validation is shown in Figure 2. The major steps included extracting the CT images of the 188 patients, separating them into a training set and test set, preprocessing all the CT images, and training a machine learning model. The ground truth was the results of the PFT measurements of the patients. The machine learning model was evaluated on the test set using Spearman r correlation coefficient and RMSE.
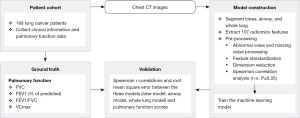
Image preprocessing
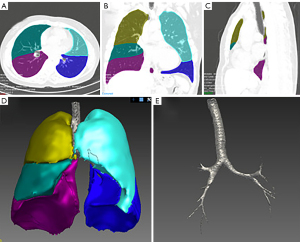
Our image pre-processing included several steps. The first step was to segment the lung CT images. We used the automatic segmentation module of the LK software to segment the lobes, airway, and the whole lung region. Then, three types of ROIs were marked: the lobes, airway, and the whole lungs. All segmentation results were reviewed by two experienced radiologists, and images with poor segmentation were excluded. There were 75 cases with poor extraction of airway in grades 3–4 in the airway segmentation results, so they were excluded from the airway model. The whole lung segmentation is a combination of airway and lobe segmentation regions. All 188 included cases were accurately segmented. Figure 3 shows the segmentation results of a typical case.
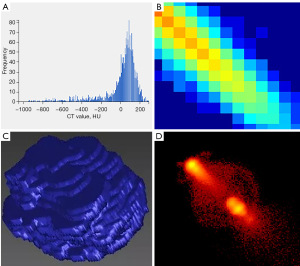
We then used the PyRadiomics module in the AK software to extract 107 radiomics features, including 18 first-order features, 14 shape characteristics, 16 gray-level size zone matrix (GLSZM) features (20), 24 gray-level cooccurrence matrix (GLCM) features (21), 5 neighborhood gray-tone difference matrix (NGTDM) features (22), 14 gray-level dependence matrix (GLDM) features (23), and 16 gray-level run-length matrix (GLRLM) features (24). For the extracted features, we used the median method to replace missing values and used the cap method to process outliers. All the features were standardized by the Z score. Figure 4 shows some radiomics features.
Feature reduction
Spearman correlation analysis and stepwise regression analysis were used to select the valuable features. In Spearman correlation analysis, the correlation coefficient and corresponding P value of each feature and the ground truth were calculated. When the correlation coefficient was greater than the specific threshold and the corresponding P value was less than 0.1, this feature was retained. In order to keep the number of features eventually included in the model at the same level to maintain comparability between models, the correlation coefficient thresholds were different under different models and prediction tasks. The thresholds for different models and prediction tasks are shown in Table 1. Then, both backward and forward stepwise selection was performed by using the likelihood ratio test with Akaike information criterion (AIC) as the stopping rule.
The remaining features of each model after stepwise regression analysis are shown in Table 2. There were 5, 6, 6, and 6 features remaining in the lobe model predicting FEV1 (% of predicted), FVC, FEV1/FVC, and VCmax, respectively. There were 4, 7, 8, and 5 features remaining in the airway model predicting FEV1 (% of predicted), FVC, FEV1/FVC, and VCmax, respectively. There were 5, 5, 6, and 5 features remaining in whole-lung model predicting FEV1 (% of predicted), FVC, FEV1/FVC, and VCmax, respectively.
Table 2
| Pulmonary function indicators | Lobe model | Airway model | Whole-lung model |
|---|---|---|---|
| FEV1 (% of predicted) | (I) glrlm_GLNUN | (I) shape_Sphericity | (I) glcm_JointEnergy |
| (II) glszm_SZNUN | (II) glszm_SZNUN | (II) glcm_Idn | |
| (III) glszm_GLV | (III) glcm_DA | (III) glrlm_LRE | |
| (IV) glszm_SAE | (IV) glcm_Contrast | (IV) glrlm_RunVariance | |
| (V) firstorder_Kurtosis | (V) glszm_GLNUN | ||
| FVC | (I) shape_LAL | (I) shape_VoxelVolume | (I) shape_LAL |
| (II) shape_SurfaceArea | (II) shape_MeshVolume | (II) shape_MiAL | |
| (III) shape_M2DDS | (III) glszm_SALGLE | (III) shape_SurfaceArea | |
| (IV) shape_MiAL | (IV) glszm_SZNU | (IV) glszm_GLNU | |
| (V) shape_MV | (V) firstorder_TotalEnergy | (V) gldm_DNU | |
| (VI) firstorder_TotalEnergy | (VI) firstorder_Energy | ||
| (VII) glcm_InverseVariance | |||
| FEV1/FVC | (I) shape_SVR | (I) firstorder_Mean | (I) shape_SVR |
| (II) glrlm_LRHGLE | (II) glrlm_HGLRE | (II) firstorder_10Percentile | |
| (III) glszm_HGLZE | (III) glrlm_RunEntropy | (III) firstorder_Mean | |
| (IV) glszm_SAHGLE | (IV) glcm_SumAverage | (IV) firstorder_RMS | |
| (V) firstorder_10Percentile | (V) glcm_DifferenceEntropy | (V) glrlm_LRHGLE | |
| (VI) firstorder_Median | (VI) glcm_SumEntropy | (VI) gldm_LDHGLE | |
| (VII) gldm_HGLE | |||
| (VIII) gldm_SDE | |||
| VCmax | (I) shape_LAL | (I) shape_SurfaceArea | (I) shape_MaAL |
| (II) shape_SurfaceArea | (II) shape_MiAL | (II) shape_MiAL | |
| (III) shape_M2DDS | (III) shape_MaAL | (III) shape_SurfaceArea | |
| (IV) shape_MiAL | (IV) shape_VoxelVolume | (IV) glszm_GLNU | |
| (V) shape_MeshVolume | (V) firstorder_TotalEnergy | (V) gldm_DNU | |
| (VI) firstorder_TotalEnergy |
PFT, pulmonary function test; FEV1 (% of predicted), the percentage of measured forced expiratory volume in the predicted value; glrlm, gray-level run length matrix; GLNUN, GrayLevelNonUniformityNormalized; glcm, gray-level co-occurrence matrix; glszm, gray-level size zone matrix; SZNUN, SizeZoneNonUniformityNormalized; GLV, GrayLevelVariance; DA, DifferenceAverage; LRE, LongRunEmphasis; SAE, SmallAreaEmphasis; FVC, forced vital capacity; LAL, LeastAxisLength; MiAL, MinorAxisLength; M2DDS, Maximum2DDiameterSlice; SALGLE, SmallAreaLowGrayLevelEmphasis; SZNU, SizeZoneNonUniformity; GLNU, GrayLevelNonUniformity; MV, MeshVolume; gldm, gray-level dependence matrix; DNU, DependenceNonUniformity; FEV1/FVC, forced expiratory volume in the first second/forced vital capacity; SVR, SurfaceVolumeRatio; LRHGLE, LongRunHighGrayLevelEmphasis; HGLRE, HighGrayLevelRunEmphasis; HGLZE, HighGrayLevelZoneEmphasis; SAHGLE, SmallAreaHighGrayLevelEmphasis; RMS, RootMeanSquared; LDHGLE, LargeDependenceHighGrayLevelEmphasis; HGLE, HighGrayLevelEmphasis; SDE, SmallDependenceEmphasis; VCmax, maximum vital capacity; MaAL, MajorAxisLength.
Training and testing the machine learning model
We used linear regression (LR) (25) as the machine learning technique for predicting pulmonary functions from lung CT. We used FVC, FEV1 (% of predicted), FEV1/FVC (%), and VCmax of PFTs as the ground truth. We randomly selected 70% of the cases for training and the remaining 30% for testing. We applied the trained LR on the test set. Our criteria for evaluation were Spearman correlation coefficient r and RMSE.
Statistical analysis
All statistical analyses were performed in R language version 3.6.3 software. A P value of <0.05 was considered statistically significant. The nominal variables are represented by frequency and percentage, and the continuous variables are represented by the median and interquartile range (IQR). The correlation between the actual lung function index and the predicted results was calculated using Spearman correlation coefficient. The RMSE was calculated as the difference between the actual lung function index and the predicted results as follows:
where yi the actual value and is the predicted results.
Results
The baseline characteristics of 188 patients included are summarized in Table 1. Patients had a median in age of 59.00 (IQR, 50.00–65.00) years, height of 161.00 (IQR, 155.50–167.00) cm, weight of 60.20 (IQR, 54.59–67.91) kg, and body mass index (BMI) of 23.15 (IQR, 21.56–25.12) kg/m2, and there were 64 males (34%). In clinical lung function, the results showed that the median FEV1 (% of predicted) was 100.65 (IQR, 89.74–110.44), the FVC was 3.04 (IQR, 2.57–3.60), the FEV1/FVC was 79.65 (IQR, 74.22–82.90), and the VCmax was 3.11 (IQR, 2.61–3.61).
In the lobe model, 113 of the 188 cases were included to train and test the model, and the remaining 75 cases were excluded due to poor segmentation results. There were 76 cases in the training set and 37 cases in the test set. We used the FEV1 (% of predicted), FVC, FEV1/FVC, and VCmax given by the PFTs as the ground truth and calculated the Spearman correlation coefficient r and RMSE. The results are shown in Table 3. Both r and RMSE indicated that the lobe model performed better in predicting FVC and VCmax than in predicting FEV1 (% of predicted) and FEV1/FVC.
Table 3
| Pulmonary function indicators | Train | Test | |||||
|---|---|---|---|---|---|---|---|
| r | P | RMSE | r | P | RMSE | ||
| FEV1 (% of predicted) | 0.413 | <0.001 | 17.627 | 0.018 | 0.915 | 18.133 | |
| FVC | 0.794 | <0.001 | 0.499 | 0.714 | <0.001 | 0.582 | |
| FEV1/FVC | 0.611 | <0.001 | 7.933 | −0.08 | 0.613 | 109.878 | |
| VCmax | 0.800 | <0.001 | 0.464 | 0.687 | <0.001 | 0.573 | |
RMSE, root-mean-square error; PFT, pulmonary function test; FEV1 (% of predicted), the percentage of measured forced expiratory volume in the predicted value; FVC, forced vital capacity; FEV1/FVC, forced expiratory volume in the first second/forced vital capacity; VCmax, maximum vital capacity.
In the airway model, 113 of the 188 cases were included to train and test the model, and the remaining 75 cases were excluded due to poor segmentation results. There were 76 cases in the training set and 37 cases in the test set. The results are shown in Table 4, from which we can see that the model performed better in predicting FEV1/FVC and VCmax.
Table 4
| Pulmonary function indicators | Train | Test | |||||
|---|---|---|---|---|---|---|---|
| r | P | RMSE | r | P | RMSE | ||
| FEV1 (% of predicted) | 0.450 | <0.001 | 17.268 | 0.370 | 0.024 | 14.713 | |
| FVC | 0.720 | <0.001 | 0.569 | 0.509 | 0.001 | 0.695 | |
| FEV1/FVC | 0.520 | <0.001 | 8.564 | 0.603 | <0.001 | 6.268 | |
| VCmax | 0.650 | <0.001 | 0.588 | 0.642 | <0.001 | 0.538 | |
RMSE, root-mean-square error; PFT, pulmonary function test; FEV1 (% of predicted), the percentage of measured forced expiratory volume in the predicted value; FVC, forced vital capacity; FEV1/FVC, forced expiratory volume in the first second/forced vital capacity; VCmax, maximum vital capacity.
In the whole-lung model, all 188 cases were included to train and test the model. There were 132 cases in the training set and 56 cases in the test set. The result is shown in Table 5. From the table, we can see that the whole-lung model had better performance in predicting FVC and VCmax.
Table 5
| Pulmonary function indicators | Train | Test | |||||
|---|---|---|---|---|---|---|---|
| r | P | RMSE | r | P | RMSE | ||
| FEV1 (% of predicted) | 0.344 | <0.001 | 16.499 | 0.315 | 0.018 | 14.863 | |
| FVC | 0.777 | <0.001 | 0.494 | 0.704 | <0.001 | 0.584 | |
| FEV1/FVC | 0.393 | <0.001 | 8.010 | −0.028 | 0.839 | 8.109 | |
| VCmax | 0.746 | <0.001 | 0.494 | 0.693 | <0.001 | 0.594 | |
RMSE, root-mean-square error; PFT, pulmonary function test; FEV1 (% of predicted), the percentage of measured forced expiratory volume in the predicted value; FVC, forced vital capacity; FEV1/FVC, forced expiratory volume in the first second/forced vital capacity; VCmax, maximum vital capacity.
The scatter diagram of Figure 5 shows the results of LR analysis of the whole-lung model for predicting FVC. From this figure, we can see that the FVC results obtained by the model are linearly positively correlated with the clinical results in both the training set and the test set.
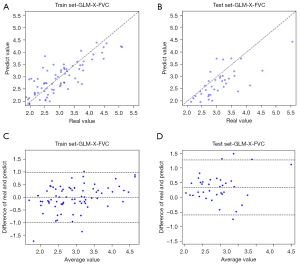
In Figure 6, we present the scatter plots of the correlation between the predicted and true values of lung function indices with good correlation between the results obtained in different models and clinical outcomes.

Discussion
In this study, we have developed a machine learning-based method to predict pulmonary functions from CT scans of patients with lung cancer. It is only in recent years that quantitative analysis of CT for assessing pulmonary function has been recognized (26). We hypothesized that, with the careful extraction of radiomics features from lung CT and the appropriate design of a machine learning model, we could use lung CT to predict the pulmonary functions of patients with lung cancer. In this study, we employed the LR as the machine learning technique for predicting pulmonary functions from CT images. We trained the LR based on three types of structural delineations of the lungs: the lobes, the airway, and the whole lungs. Our test results showed that the three types of delineations had different performance in predicting the PFT results.
- When using the lobe model, we found that the machine learning model had higher accuracy in predicting FVC and VCmax than in predicting FEV1/FVC and FEV1 (% of predicted).
- When using the airway model, we found that the machine learning model had higher accuracy in predicting FEV1/FVC and VCmax than in predicting FVC and FEV1 (% of predicted). The results of Gawlitza et al. (27) showed that the accuracy of predicting FEV1/FVC was higher than that of the other three metrics, which is slightly different from our results.
- When using the whole-lung model, we found that the machine learning model had higher accuracy in predicting FVC and VCmax than in predicting FEV1/FVC and FEV1 (% of predicted). We believe the reason behind the high prediction accuracy of FVC and VCmax is that the CT images were obtained at maximum inhalation, which can best reflect the maximum air content of lung tissues, whereas FVC and VCmax emphasize the activities initiated after maximum inhalation.
- The prediction accuracy of the whole-lung model was similar to that of the lobe model. We believe this is because the pathological process of our case mostly manifested as changes at the level of lung parenchyma, making the changes in lung parenchyma the dominant changes of the whole lung lesions. Therefore, these two models had similar performance. Overall, except for the better prediction accuracy of FEV1/FVC in the airway model, the prediction accuracies of FVC and VCmax were better in the lobe model and the whole-lung model. Across the three models, it appeared that FEV1 (% of predicted) had the lowest accuracy of being predicted by the machine learning model. FEV1 (% of predicted) is an important basis for the preoperative evaluation of lung cancer surgery and is also a parameter that clinicians should pay more attention to in PFT. The poor correlation of our machine evaluation with FEV1 (% of predicted) is unfortunate and perhaps occurred because of the difference in the number of predicted positions, such as recumbent position on CT and a seated position on PFT.
The rich information contained in images offers us a new opportunity for us to explore to disease behaviors and prognosis (28,29). Radiomics has shown its potential in the extraction of useful evidence from CT and other modalities for diagnosis, classification, prediction, and assessment of diseases. Due to the large amount of information present in radiomics, the analysis of the information is closely integrated with machine learning (10). Machine learning, including deep learning, can apply highly nonlinear analysis to the input, from which knowledge can be learned and applied to predict the research object (30).
In this work, we used PFT results as the ground truth to evaluate the accuracy of the radiomics approach. However, it is fair to note that, in practice, there are many factors affecting the PFT readings, including a patient’s demographic characteristics (e.g., age, gender, height, and weigh) and clinical presentation (e.g., the size and location of the tumor and the presence of comorbidities like diabetes), which can affect lung function (31). Therefore, it is recommended that characteristics about an individual should be taken into consideration when interpreting the PFT results (32-34). The chest anatomical structures of patients also display certain group-wise physiology characteristics, so care must be taken when establishing reference values for normal or abnormal PFT readings (35).
Our study had some limitations that should be noted. First, the sample size was rather small, and further studies with larger sample sizes are required to confirm our findings. It should be noted that, although sophisticated machine learning models like deep learning models can achieve better performance, we chose a linear model because of the small sample size, which limited the learning ability of sophisticated models. Future research could focus on sophisticated machine learning models with large sample sizes to obtain higher accuracy. Second, only 60% of patients had their airways accurately segmented by the LK software. One of the reasons is that our inclusion criteria were relatively strict, and only the segmentation results of the grade 3–4 airway that had been successfully extracted were included in the tracheal model. Another reason may be that the accuracy of LK airway segmentation is affected by many factors, such as image quality and the segmentation algorithm. The application of the airway model and the lobe model to the general population requires the development of imaging techniques with high spatial and temporal resolution and robust airway segmentation techniques. Fortunately, LK has good accuracy and generalization in the whole-lung segmentation. Third, owing to the limitation of a single site, the outcomes of this study might have been affected and may not be widely applicable to other hospitals. This study also has a few clinical limitations. Usually, physicians use preoperative FEV1 and then estimate the predicted postoperative FEV1 based on preoperative CT scans (quantitative CT or tomographic densitometry), but this study was limited to only comparing preoperative lung function. In addition, the lack of evaluation for diffusing capacity for carbon monoxide (DLCO) is also a limitation of this study. In the European Respiratory Society/European Society of Thoracic Surgery (ERS/ESTS) guidelines (36), DLCO should be a routine preoperative examination for patients undergoing pneumonectomy. However, whether DLCO should be a routine preoperative examination for all patients or only for patients with a low preoperative FEV1 is still controversial.
Conclusions
At present, most applications of machine learning on the lungs focus on the detection of abnormalities and the classification of the status of the abnormalities, such as pulmonary nodules, pulmonary embolism assessment, and COPD (37-39). Recently, researchers have been working to use machine learning in other aspects of lung disease diagnosis (40). For example, Walsh et al. developed an algorithm with the ability to classify pulmonary fibrotic lesions on CT images (41). González et al. developed a convolutional neural network that can distinguish patients with COPD and predict the risk of adverse events (42).
At present, biphasic respiratory CT images provide more abundant information for COPD, which provides better evaluation and identification of COPD. In future study, we aim to use the CT image information of biphasic respiration to model and predict lung function and determine whether this effect can better reflect the real clinical situation.
Acknowledgments
Funding: This study was supported by the Natural Science Foundation of Guangdong Province, China (No. 2019A1515011382 to H Chen).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-70/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-70/coif). Y Liao is an employee of GE Healthcare, Guangzhou, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of The First Affiliated Hospital of Guangzhou Medical University (No. 2022-70), and informed consent was provided by all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Zhang BW, Zhang Y, Ye JD, Qiang JW. Use of relative CT values to evaluate the invasiveness of pulmonary subsolid nodules in patients with emphysema. Quant Imaging Med Surg 2021;11:204-14. [Crossref] [PubMed]
- Arenberg D. Searching for red shirts. Emphysema as a lung cancer screening criterion? Am J Respir Crit Care Med 2015;191:868-9. [Crossref] [PubMed]
- Lee SJ, Yoo JW, Ju S, Cho YJ, Kim JD, Kim SH, Jang IS, Jeong BK, Lee GW, Jeong YY, Kim HC, Bae K, Jeon KN, Lee JD. Quantitative severity of pulmonary emphysema as a prognostic factor for recurrence in patients with surgically resected non-small cell lung cancer. Thorac Cancer 2019;10:421-7. [Crossref] [PubMed]
- Suliman YA, Dobrota R, Huscher D, Nguyen-Kim TD, Maurer B, Jordan S, Speich R, Frauenfelder T, Distler O. Brief Report: Pulmonary Function Tests: High Rate of False-Negative Results in the Early Detection and Screening of Scleroderma-Related Interstitial Lung Disease. Arthritis Rheumatol 2015;67:3256-61. [Crossref] [PubMed]
- Berry MF, Villamizar-Ortiz NR, Tong BC, Burfeind WR Jr, Harpole DH, D'Amico TA, Onaitis MW. Pulmonary function tests do not predict pulmonary complications after thoracoscopic lobectomy. Ann Thorac Surg 2010;89:1044-51; discussion 1051-2. [Crossref] [PubMed]
- Alahmari SS, Cherezov D, Goldgof D, Hall L, Gillies RJ, Schabath MB. Delta Radiomics Improves Pulmonary Nodule Malignancy Prediction in Lung Cancer Screening. IEEE Access 2018;6:77796-806.
- Hawkins S, Wang H, Liu Y, Garcia A, Stringfield O, Krewer H, Li Q, Cherezov D, Gatenby RA, Balagurunathan Y, Goldgof D, Schabath MB, Hall L, Gillies RJ. Predicting Malignant Nodules from Screening CT Scans. J Thorac Oncol 2016;11:2120-8. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Ma J, Wang Q, Ren Y, Hu H, Jun Z. Automatic lung nodule classification with radiomics approach. In: Medical Imaging 2016: PACS and Imaging Informatics: Next Generation and Innovations. SPIE, 2016;9789:26-31.
- Wu W, Parmar C, Grossmann P, Quackenbush J, Lambin P, Bussink J, Mak R, Aerts HJ. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front Oncol 2016;6:71. [Crossref] [PubMed]
- Moran A, Daly ME, Yip SSF, Yamamoto T. Radiomics-based Assessment of Radiation-induced Lung Injury After Stereotactic Body Radiotherapy. Clin Lung Cancer 2017;18:e425-31. [Crossref] [PubMed]
- Parmar C, Leijenaar RT, Grossmann P, Rios Velazquez E, Bussink J, Rietveld D, Rietbergen MM, Haibe-Kains B, Lambin P, Aerts HJ. Radiomic feature clusters and prognostic signatures specific for Lung and Head & Neck cancer. Sci Rep 2015;5:11044. [Crossref] [PubMed]
- Zhang Y, Oikonomou A, Wong A, Haider MA, Khalvati F. Radiomics-based Prognosis Analysis for Non-Small Cell Lung Cancer. Sci Rep 2017;7:46349. [Crossref] [PubMed]
- Parmar C, Grossmann P, Bussink J, Lambin P, Aerts HJWL. Machine Learning methods for Quantitative Radiomic Biomarkers. Sci Rep 2015;5:13087. [Crossref] [PubMed]
- Thawani R, McLane M, Beig N, Ghose S, Prasanna P, Velcheti V, Madabhushi A. Radiomics and radiogenomics in lung cancer: A review for the clinician. Lung Cancer 2018;115:34-41. [Crossref] [PubMed]
- Wei W, Hu XW, Cheng Q, Zhao YM, Ge YQ. Identification of common and severe COVID-19: the value of CT texture analysis and correlation with clinical characteristics. Eur Radiol 2020;30:6788-96. [Crossref] [PubMed]
- Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 2014;61:630-7. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics 1973;SMC-3:610-21. [Crossref]
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics 1989;19:1264-74. [Crossref]
- Van Gool L, Dewaele P, Oosterlinck A. Texture analysis anno 1983. Computer Vision, Graphics, and Image Processing 1985;29:336-57. [Crossref]
- Galloway MM. Texture analysis using gray level run lengths. Computer Graphics and Image Processing 1975;4:172-9. [Crossref]
- Wilkinson GN, Rogers CE. Symbolic description of factorial models for analysis of variance. J R Stat Soc Ser C Appl Stat 1973;22:392-9.
- Occhipinti M, Paoletti M, Bartholmai BJ, Rajagopalan S, Karwoski RA, Nardi C, Inchingolo R, Larici AR, Camiciottoli G, Lavorini F, Colagrande S, Brusasco V, Pistolesi M. Spirometric assessment of emphysema presence and severity as measured by quantitative CT and CT-based radiomics in COPD. Respir Res 2019;20:101. [Crossref] [PubMed]
- Gawlitza J, Sturm T, Spohrer K, Henzler T, Akin I, Schönberg S, Borggrefe M, Haubenreisser H, Trinkmann F. Predicting Pulmonary Function Testing from Quantified Computed Tomography Using Machine Learning Algorithms in Patients with COPD. Diagnostics (Basel) 2019;9:33. [Crossref] [PubMed]
- Chassagnon G, Vakalopolou M, Paragios N, Revel MP. Deep learning: definition and perspectives for thoracic imaging. Eur Radiol 2020;30:2021-30. [Crossref] [PubMed]
- Vallières M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471-96. [Crossref] [PubMed]
- Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics 2017;37:505-15. [Crossref] [PubMed]
- Litonjua AA, Lazarus R, Sparrow D, Demolles D, Weiss ST. Lung function in type 2 diabetes: the Normative Aging Study. Respir Med 2005;99:1583-90. [Crossref] [PubMed]
- Liang BM, Lam DC, Feng YL. Clinical applications of lung function tests: a revisit. Respirology 2012;17:611-9. [Crossref] [PubMed]
- Ma YN, Wang J, Dong GH, Liu MM, Wang D, Liu YQ, Zhao Y, Ren WH, Lee YL, Zhao YD, He QC. Predictive equations using regression analysis of pulmonary function for healthy children in Northeast China. PLoS One 2013;8:e63875. [Crossref] [PubMed]
- Nysom K, Ulrik CS, Hesse B, Dirksen A. Published models and local data can bridge the gap between reference values of lung function for children and adults. Eur Respir J 1997;10:1591-8. [Crossref] [PubMed]
- Celli BR, Halbert RJ, Isonaka S, Schau B. Population impact of different definitions of airway obstruction. Eur Respir J 2003;22:268-73. [Crossref] [PubMed]
- Brunelli A, Charloux A, Bolliger CT, Rocco G, Sculier JP, Varela G, Licker M, Ferguson MK, Faivre-Finn C, Huber RM, Clini EM, Win T, De Ruysscher D, Goldman LEuropean Respiratory Society. European Society of Thoracic Surgeons Joint Task Force on Fitness for Radical Therapy. The European Respiratory Society and European Society of Thoracic Surgeons clinical guidelines for evaluating fitness for radical treatment (surgery and chemoradiotherapy) in patients with lung cancer. Eur J Cardiothorac Surg 2009;36:181-4. [Crossref] [PubMed]
- Humphries SM, Notary AM, Centeno JP, Strand MJ, Crapo JD, Silverman EK, Lynch DA. Genetic Epidemiology of COPD (COPDGene) Investigators. Deep Learning Enables Automatic Classification of Emphysema Pattern at CT. Radiology 2020;294:434-44. [Crossref] [PubMed]
- Ma J, Song Y, Tian X, Hua Y, Zhang R, Wu J. Survey on deep learning for pulmonary medical imaging. Front Med 2020;14:450-69. [Crossref] [PubMed]
- Han F, Wang H, Zhang G, Han H, Song B, Li L, Moore W, Lu H, Zhao H, Liang Z. Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging 2015;28:99-115. [Crossref] [PubMed]
- Bianconi F, Fravolini ML, Pizzoli S, Palumbo I, Minestrini M, Rondini M, Nuvoli S, Spanu A, Palumbo B. Comparative evaluation of conventional and deep learning methods for semi-automated segmentation of pulmonary nodules on CT. Quant Imaging Med Surg 2021;11:3286-305. [Crossref] [PubMed]
- Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med 2018;6:837-45. [Crossref] [PubMed]
- González G, Ash SY, Vegas-Sánchez-Ferrero G, Onieva Onieva J, Rahaghi FN, Ross JC, Díaz A, San José Estépar R, Washko GRCOPDGene and ECLIPSE Investigators. Disease Staging and Prognosis in Smokers Using Deep Learning in Chest Computed Tomography. Am J Respir Crit Care Med 2018;197:193-203. [Crossref] [PubMed]


