
Automatic deep learning method for detection and classification of breast lesions in dynamic contrast-enhanced magnetic resonance imaging
Introduction
Breast cancer is the most common cancer in women and the leading cause of cancer deaths in women worldwide (1). Accurate, early diagnosis has an important impact on treatment planning and improving survival rates for breast cancer patients (2). Magnetic resonance imaging (MRI) plays a significant role in the diagnosis of breast lesions, staging of known cancers, screening for high-risk women, and evaluation of response to neoadjuvant chemotherapy (3-7). Dynamic contrast-enhanced (DCE)-MRI is the backbone of breast MRI protocol and is the most sensitive imaging technique for the diagnosis of breast cancer, providing excellent morphological and semi-quantitative enhancement kinetic information, even for dense breast tissue (4,6).
The American College of Radiology (ACR) introduced the Breast Imaging Reporting and Data System (BI-RADS) for image diagnostic specifications, which standardizes the description, classification, and corresponding treatment recommendations of breast MRI (8). Breast lesions are classified as 0–6 based on their morphology and type of enhancement kinetics, which typically relies on visual evaluation and substantial expertise by the radiologist. As manual slice-by-slice analysis of breast MR images is both time-consuming and error-prone, various computer-aided detection (CAD) approaches have been proposed to assist radiologists in detecting and reporting the malignancy status of breast lesions (9,10). Deep learning (DL), which can obtain the best features directly from raw images through end-to-end learning (11,12), has been successfully used in many CAD systems such as photographic, pathological, and radiographic images, and is reported to be promising in diverse clinical tasks (13-15). Convolutional neural network (CNN) models have been widely used as a common DL approach for breast MRI, including segmentation (16), detection (17), and classification of lesions (13,18-20), as well as prediction of the molecular subtype (21), lymph node metastasis (22), and response to therapy (23). However, considering its independent clinical application, DL needs to simultaneously detect and classify lesions in MRI, which has not been well investigated.
The Faster region-based CNN (R-CNN) (24) is a well-known DL model that is based on a CNN with additional components for detecting, localizing, and classifying objects in images. Faster R-CNN is employed for the detection and classification of breast lesions in mammography, where it first learns the regional proposals from mammography, from which it then predicts the location and classification of the whole images (25). Based on the Faster R-CNN, feature pyramid networks (FPNs) with multi-scale and pyramidal hierarchy architectures (26) have also been used to detect breast masses by reducing the false positive rate of DL-based systems without significantly decreasing the detection sensitivity (27,28). In addition, to prevent the overfitting problems and further reduce false positives, Cai et al. (29) proposed a multi-stage detection architecture named the cascade model, which consists of a sequence of detectors trained with increasing intersection over union (IoU) thresholds. Furthermore, the DL-based system with cascade FPN (CFPN) has shown promising results in the detection of pulmonary nodules (30). We were interested to determine how the CFPN performed for the automatic detection and classification of breast lesions in DCE-MRI.
The detection of small breast lesions has always troubled radiologists and has been a challenge for CAD systems. Therefore, the main goal of this study was to evaluate the performance of the CFPN system in detecting and classifying breast lesions in DCE-MRI compared to Faster R-CNN and FPN systems and to evaluate their performance in detecting breast lesions of various sizes. We present the following article in accordance with the STARD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-323/rc).
Methods
Study population
This retrospective study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Review Board of the Second Affiliated Hospital of Xi’an Jiaotong University (No. 2018-080). The requirement for informed consent was waived due to the retrospective nature of the study. MRI data archives of our Picture Archiving and Communication Systems (PACS) between 1 January 2016 and 31 July 2020 were searched consecutively to identify patients meeting the following criteria: (I) underwent DCE-MRI examination within 7 days before biopsy or surgery; (II) had benign or malignant lesions confirmed by biopsy or pathology after surgery; (III) did not receive neoadjuvant chemoradiotherapy (NAC) before baseline biopsy or DCE-MRI examination. Finally, a total of 191 patients with 157 malignant and 34 benign lesions were enrolled for analysis, and their clinical and MRI features are shown in Table 1. There were no significant differences in fibroglandular tissue (FGT), background parenchymal enhancement (BPE), and lesion type between benign and malignant patients. We randomly divided the 191 patients into a training set comprising ~153 patients with 126 malignant and 27 benign lesions and a validation set comprising ~38 patients with 31 malignant and 7 benign lesions under 5-fold cross-validation (Table 2).
Table 1
| Characteristics | Benign (n=34) | Malignant (n=157) | P value |
|---|---|---|---|
| Age, years, mean (SD) | 40.09 (10.8) | 48.78 (10.3) | <0.001 |
| Lesion size on MRI, mm, mean (SD) | 15.03 (4.5) | 21.60 (7.8) | <0.001 |
| FGT classification, n (%) | 0.08 | ||
| Almost entirely fat | 1 (2.9) | 0 | |
| Scattered FGT | 4 (11.8) | 33 (21.0) | |
| Heterogeneous FGT | 14 (41.2) | 71 (45.2) | |
| Extreme FGT marked | 15 (44.1) | 53 (33.8) | |
| BPE classification, n (%) | 0.61 | ||
| Minimal | 7 (20.6) | 19 (12.1) | |
| Mild | 16 (47.1) | 83 (52.9) | |
| Moderate | 10 (29.4) | 48 (30.6) | |
| Marked | 1 (2.9) | 7 (4.4) | |
| BI-RADS classification, n (%) | <0.001 | ||
| I | 0 | 0 | |
| II | 3 (8.8) | 0 | |
| III | 10 (29.4) | 3 (1.9) | |
| IV | 10 (29.4) | 36 (22.9) | |
| V | 11 (32.4) | 118 (75.2) | |
| Type of lesions, n (%) | 0.80 | ||
| Mass | 33 (97.1) | 151 (96.2) | |
| Non-mass | 1 (2.9) | 6 (3.8) | |
BI-RADS, Breast Imaging Reporting and Data System; BPE, background parenchymal enhancement; FGT, fibroglandular tissue; MRI, magnetic resonance imaging; SD, standard deviation.
Table 2
| Fold | Training data | Testing data | |||
|---|---|---|---|---|---|
| Patients number | # Images before/after augmentation | Patients number | # Images | ||
| Fold-1 | 152 | 767/5,369 | 39 | 195 | |
| Fold-2 | 152 | 763/5,341 | 39 | 199 | |
| Fold-3 | 153 | 778/5,446 | 38 | 184 | |
| Fold-4 | 153 | 775/5,425 | 38 | 187 | |
| Fold-5 | 154 | 765/5,335 | 37 | 197 | |
MRI protocol
DCE-MRI examination was performed on a 3.0 T system (Signa HDxt; GE Medical Systems, Milwaukee, WI, USA) with a dedicated 8-channel breast coil. Pre- and post-contrast phases were acquired before and after the injection of 0.2 mmol/kg body weight of gadolinium-diethylenetriaminepentaacetic acid (DTPA) (Magnevist, Schering, Germany) for 64 s, 128 s, 192 s, 256 s, and 318 s at a rate of 2.0 mL/s with a power injector followed by 20 mL saline solution, where the dose of gadolinium-DTPA followed (31). We used T1-weighted 3-dimensional (3D) fast spoiled gradient-recalled echo sequence with parallel imaging (VIBRANT) sequence on the transverse plane with the following parameters: repetition time/echo time/inversion time (TR/TE/TI) =4.4/2.1/125 ms, flip angle 14°, matrix size =416×320, section thickness =1.6 mm, and field of view =350 mm × 350 mm. The contrast-enhanced MRI acquired at 128 s was used for the detection of breast lesions as it had the best signal intensity contrast.
Workflow of DL-based analysis
Figure 1 shows the framework for DL-based analysis. The proposed DL-based workflow included the annotation of breast lesions, the augmentation of training data, and the detection and classification by different systems, including the Faster R-CNN, FPN, and CFPN systems.
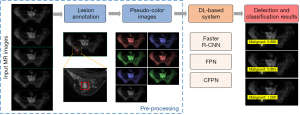
Pre-processing
Two radiologists (W Gao and J Zhong, with 6 and 3 years of experience in breast MRI, respectively) who were blinded to the clinical information and final pathology independently reviewed the MRI data of all patients and selected the approximately middle 5 consecutive slices for each breast lesion. On each slice, the bounding box was first annotated by drawing the minimum circumscribed rectangle of each lesion using LabelImg software (https://github.com/tzutalin/labelimg), then the lesion type was labeled as malignant or benign based on the pathological record.
Considering the limited sample size, the 3-channel artificial red, blue, green (RGB) image fusion method proposed by Yap et al. (32) was used to increase the training data. Specifically, each pseudo-color image was synthesized by concatenating the original MRI, the image sharpened by the Laplace operator and enhanced by histogram equalization. Changing the order of these 3 images with full permutation, together with the original MRI, produced a total of 7 (A33+1=7) pseudo-color images per slice. The training and testing images before and after augmentation are summarized in Table 2.
Detection and classification with Faster R-CNN, FPN, and CFPN system
The pseudo-color images were separately fed into the classical Faster R-CNN, FPN, or CFPN system for detection and classification of breast lesions. The classical Faster R-CNN developed by Ren et al. (24) was used as a comparison for detection performance with the other systems.
As shown in Figure 2A, the FPN and CFPN systems shared the same underlying modules, including a backbone network for feature extraction, an FPN for multi-scale feature extraction (Figure 2B), and a region proposal network (RPN) for generating proposal boxes (candidate detection boxes). However, the prediction networks for detecting and classifying breast lesions were different, with the basic prediction network (Figure 2C) and the cascaded prediction network (Figure 2D) used for the FPN and CFPN systems, respectively.
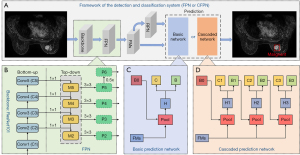
The shared backbone and FPN modules between the 2 systems are shown in Figure 2B. Specifically, ResNet101 (33) pre-trained on the ImageNet dataset was used as the backbone network to extract features from pseudo-color images with 5 convolution blocks, 4 of which had residual blocks. Feature maps (FMs) of different scales obtained from the last 4 convolution blocks (C2, C3, C4, C5) were fed into the FPN to generate the integrated multi-scale FMs (P2, P3, P4, P5, P6), which were then input into the RPN to generate proposal boxes. The anchor sizes in the RPN were set as 362, 722, 1442, 2882, 5762 with aspect ratios of 1:1, 1:2, and 2:1, according to the statistics of lesion dimensions. Other details are summarized in the Supplementary methods section (Appendix 1).
For the FPN system, the generated proposal boxes and multi-scale FMs were passed into the basic prediction network that had 2 output paths (Figure 2C): one for detecting breast lesions via bounding box regression, and the other for classifying the lesions as benign and malignant. To alleviate the class-imbalance issue, the focal loss (34) was used to construct the classification loss of the RPN as well as the final classification network, as follows:
where y and y' represent the ground-truth labels and predicted results of breast lesions. Factor α∈[0,1] was used to balance the uneven distribution between benign and malignant samples. Parameter γ (generally greater than zero) was used to tune the weight between the simple and hard samples. A larger γ indicates that more simple samples are suppressed and that more difficult samples are focused on. In this study, we empirically set the values of α and γ as 0.5 and 2, respectively.
FPN systems are often challenged when determining the IoU threshold, namely, the division of positives and negatives. Low IoU thresholds usually result in false positives because many areas without lesions are detected, whereas high thresholds also degrade the performance because of the increase of missed detections caused by model overfitting. To reduce missed detections, especially false detections in the FPN system, we further constructed a CFPN detection system with sequentially increasing IoU thresholds. Based on the FPN model with the optimal settings, we further improved the prediction module to obtain a system with better performance. This CFPN system used the same underlying modules and focal loss, but replaced the basic prediction network of the FPN system with a cascaded prediction network. As detailed in Figure 2D, the cascade prediction network extended the single-stage prediction of the basic network to 3 detection stages, each with IoU thresholds of 0.5, 0.6, and 0.7, respectively. Specifically, the proposal boxes from the previous stage and the FMs were fed into the region of interest (ROI) pooling (Pool) and the prediction module to generate the classification and proposal boxes used to train the next stage with an increased IoU threshold. At inference, the same 3-stage cascade procedure was applied to guarantee the match between training and inference distributions. Other details can be found in the Supplementary methods section (Appendix 1).
Detection of large and small breast lesions
According to the current eighth edition of the American Joint Committee Cancer Staging System for breast cancer, 2 cm is the diagnostic threshold for stage T1 and T2 lesions (35), and the majority of enhancing lesions larger than 2 cm are malignant (18). We divided all the patients in this study into large (>2 cm) and small (≤2 cm) breast lesion groups to test the detection and classification performance of different systems. Of the 191 patients (962 slices), there were 106 malignant (530 slices) and 12 benign (80 slices) in the large lesion group and 51 malignant (249 slices) and 22 benign (103 slices) in the small lesion group.
Implementation
All DL systems (Faster R-CNN, FPN, and CFPN) were built on the PyTorch framework (v1.3.1) with Python (v3.6) language (https://www.python.org). The experimental server used the Ubuntu operating system (v16.04; Canonical, London, UK) with an 8-core Intel processor (i7-9700 k, 32 GB RAM). To improve the training speed, a graphics processing unit (GPU) device (RTX 2080TI, 11 GB memory) was used for acceleration under the CUDA environment (v10.1).
Since the backbone for building FPN and CFPN was ResNet101, the same as that for building Faster R-CNN, the hyper-parameter setting in our study referred to Faster R-CNN in the open-source code on GitHub (https://github.com/jwyang/faster-rcnn.pytorch) and were adjusted according to the loss decline trend in the training set. Specifically, we used stochastic gradient descent (SGD) as the optimizer to update the parameters of each DL model. Following a stepwise decay strategy (StepLR), the initial learning rate of 0.001 was decayed by a factor of 0.1 every 5 epochs, and the batch size and the maximum epochs were set as 1 and 20, respectively.
Performance evaluation and statistical analysis
All experiments were performed under 5-fold cross-validation. Metrics including precision-recall (PR) curve, average precision (AP), mean AP (mAP), F1-score, free-response receiver operating characteristic (FROC) curve, sensitivity, and false positives were used to evaluate different systems. To evaluate the performance in real detection, we kept only the detected box with the highest confidence level on each slice as the final result, and calculated the precision, recall, F1-score, sensitivity, and FPs per slice and per patient, respectively. Metrics for each patient were obtained by majority voting on the results of all slices for that patient.
where TP, FP, and FN represent the true positive, false positive, and false negative detection, respectively. The AP is the average precision under every recall value by traversing the IoU threshold during detection, and the mAP denotes the mean average precision between benign and malignant lesions.
Cohen’s kappa scores for the detection and classification of breast lesions were provided to assess the agreement between the CFPN system and the other 2 systems (Faster R-CNN and FPN). Student’s t-test was adopted to compare the performance of different systems in terms of the aforementioned metrics, followed by the Holm-Bonferroni method for adjusted P values in the comparison among multiple groups. A P value <0.05 was considered statistically significant.
Results
Evaluation of FPN system with different settings
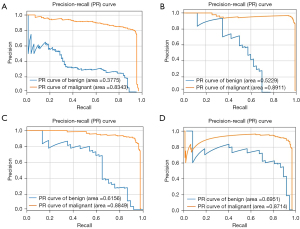
To evaluate the detection performance under different settings, we used the original FPN, the FPN augmented by the pseudo-color image fusion, the FPN with the focal loss, and the FPN with both augmentation and focal loss to detect different classifications of the breast lesions. As shown in Table S1, for both benign and malignant breast lesions, the area under the PR curve (AP value) was higher for FPN with augmentation or focal loss than the original FPN. Compared to FPN systems with other settings, the FPN with both augmentation and focal loss had the highest AP value and mAP value after 5-fold cross-validation. PR curves of the FPN system with different settings are shown in Figure 3.
Detection and classification results for Faster R-CNN, FPN, and CFPN systems
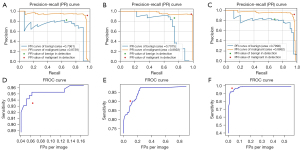
The CFPN and FPN systems with the optimal settings (including augmentation and focal loss), as well as the classical Faster R-CNN were used to detect and classify benign and malignant breast lesions. As shown in Table 3, the AP values of CFPN for benign and malignant lesions were 0.731±0.075 and 0.921±0.048, respectively, which were both better than or comparable to the other 2 systems. In particular, the mAP values of CFPN outperformed the FPN and Faster R-CNN with a value of 0.826±0.051. The results in Table 4 show the sensitivity of the CFPN was better than the other systems under the same number of FPs per image. The CFPN also had the highest sensitivity, and there was a significant difference between it and the Faster R-CNN systems in the highest sensitivity (P=0.008). The PR curves and FROC curves of median results in 5-fold cross-validation are shown in Figure 4.
Table 3
| System | Fold | AP for benign | AP for malignant | mAP |
|---|---|---|---|---|
| Faster R-CNN | Fold-1 | 0.667 | 0.873 | 0.770 |
| Fold-2 | 0.688 | 0.980 | 0.834 | |
| Fold-3 | 0.779 | 0.986 | 0.882 | |
| Fold-4 | 0.591 | 0.889 | 0.740 | |
| Fold-5 | 0.800 | 0.865 | 0.833 | |
| Mean ± SD | 0.705±0.076 | 0.919±0.053 | 0.812±0.051 | |
| FPN | Fold-1 | 0.695 | 0.871 | 0.783 |
| Fold-2 | 0.611 | 0.981 | 0.796 | |
| Fold-3 | 0.667 | 0.987 | 0.827 | |
| Fold-4 | 0.708 | 0.892 | 0.799 | |
| Fold-5 | 0.557 | 0.876 | 0.717 | |
| Mean ± SD | 0.648±0.056 | 0.921±0.052 | 0.784±0.037 | |
| CFPN | Fold-1 | 0.679 | 0.875 | 0.777 |
| Fold-2 | 0.654 | 0.965 | 0.809 | |
| Fold-3 | 0.840 | 0.994 | 0.917 | |
| Fold-4 | 0.681 | 0.887 | 0.784 | |
| Fold-5 | 0.800 | 0.886 | 0.843 | |
| Mean ± SD | 0.731±0.075 | 0.921±0.048 | 0.826±0.051 |
AP, average precision; DL, deep learning; CFPN, cascade feature pyramid network; FPN, feature pyramid network; Faster R-CNN, faster region-based convolutional neural network; mAP, mean average precision; SD, standard deviation.
Table 4
| Sensitivities | Fold | Faster R-CNN | FPN | CFPN |
|---|---|---|---|---|
| Sensitivity (FPs =0.125) | Fold-1 | 0.856 | 0.810 | 0.836 |
| Fold-2 | 0.905 | 0.905 | 0.900 | |
| Fold-3 | 0.951 | 0.902 | 0.984 | |
| Fold-4 | 0.808 | 0.893 | 0.861 | |
| Fold-5 | 0.873 | 0.777 | 0.888 | |
| Mean ± SD | 0.879±0.048 | 0.857±0.053 | 0.894±0.050 | |
| Sensitivity (FPs =0.250) | Fold-1 | 0.918 | 0.908 | 0.928 |
| Fold-2 | 0.935 | 0.930 | 0.940 | |
| Fold-3 | 0.962 | 0.978 | 0.989 | |
| Fold-4 | 0.909 | 0.925 | 0.931 | |
| Fold-5 | 0.924 | 0.878 | 0.949 | |
| Mean ± SD | 0.930±0.018 | 0.924±0.033 | 0.947±0.022 | |
| Highest sensitivity | Fold-1 | 0.918 (FPs =0.210) | 0.954 (FPs =0.446) | 0.954 (FPs =0.415) |
| Fold-2 | 0.935 (FPs =0.201) | 0.950 (FPs =0.553) | 0.985 (FPs =0.558) | |
| Fold-3 | 0.962 (FPs =0.169) | 0.984 (FPs =0.891) | 0.989 (FPs =0.130) | |
| Fold-4 | 0.909 (FPs =0.246) | 0.947 (FPs =0.610) | 0.963 (FPs =0.417) | |
| Fold-5 | 0.924 (FPs =0.218) | 0.944 (FPs =0.523) | 0.959 (FPs =0.355) | |
| Mean ± SD | 0.930±0.018 (FPs =0.209) | 0.956±0.014 (FPs =0.605) | 0.970±0.014 (FPs =0.375) |
DL, deep learning; Faster R-CNN, faster region-based convolutional neural network; FPN, feature pyramid network; CFPN, cascade feature pyramid network; FPs, false positives; SD, standard deviation.
At both the slice and patient level (Table S2), the F1-scores of the CFPN for both benign and malignant lesions were superior to those of the other systems. In particular, the CFPN achieved the highest sensitivities in detection at the lowest FPs at both the slice and patient level (Table 5). The Cohen’s kappa scores between the CFPN system and the Faster R-CNN and FPN systems were 0.961 and 0.878, respectively. This indicated the CFPN and Faster R-CNN systems were similar in detecting and classifying breast lesions, which was consistent with the F1-scores of the 3 systems.
Table 5
| Systems | Sensitivity | FPs |
|---|---|---|
| Slice level | ||
| Faster R-CNN | 0.784 | 0.102 |
| FPN | 0.902 | 0.105 |
| CFPN | 0.967 | 0.033 |
| Patient level | ||
| Faster R-CNN | 0.827 | 0.027 |
| FPN | 0.895 | 0.098 |
| CFPN | 0.974 | 0.026 |
FPs, false positives; Faster R-CNN, faster region-based convolutional neural network; FPN, feature pyramid; CFPN, cascade feature pyramid network.
Performance on large and small breast lesions
The detection and classification results for large and small breast lesions are shown in Figure 5 and Table 6. For both large and small lesions, the mAP values of CFPN were higher than those of FPN and Faster R-CNN systems. Notably, in the small lesion group, CFPN had the highest AP values in both benign and malignant lesions. Correspondingly, CFPN achieved the best performance across all metrics in the real detection of small lesions at the slice and patient level for both benign and malignant lesions, except for the recall of malignant lesions at the patient level, which was comparable to other systems (Table 7). The Cohen’s kappa scores between the CFPN system and the other 2 systems were 0.933 for Faster R-CNN and 0.835 for FPN, showing the same trend as for all breast lesions. For large lesions, CFPN had both higher recall and F1-score for benign lesions, and higher precision and F1-score for malignant lesions at the slice level than the other 2 systems (Table S3). The same Cohen’s kappa score of 0.866 between the CFPN and the other systems indicated that the performances of the Faster R-CNN and CFPN were comparable to each other.
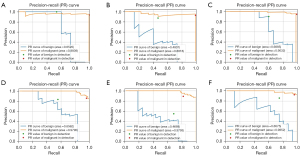
Table 6
| System | Fold | AP for benign (large/small) | AP for malignant (large/small) | mAP (large/small) |
|---|---|---|---|---|
| Faster R-CNN | Fold-1 | 0.288/0.527 | 0.843/0.774 | 0.565/0.651 |
| Fold-2 | 0.652/0.578 | 0.884/0.812 | 0.768/0.695 | |
| Fold-3 | 0.878/0.704 | 0.969/0.986 | 0.924/0.845 | |
| Fold-4 | 0.275/0.648 | 0.852/0.991 | 0.563/0.820 | |
| Fold-5 | 0.846/0.656 | 0.909/0.979 | 0.878/0.818 | |
| Mean ± SD | 0.588±0.262/0.623±0.062 | 0.891±0.045/0.908±0.095 | 0.740±0.152/0.766±0.078 | |
| FPN | Fold-1 | 0.373/0.552 | 0.844/0.742 | 0.609/0.647 |
| Fold-2 | 0.481/0.319 | 0.882/0.715 | 0.681/0.517 | |
| Fold-3 | 0.812/0.559 | 0.937/0.902 | 0.874/0.730 | |
| Fold-4 | 0.008/0.467 | 0.930/0.979 | 0.505/0.723 | |
| Fold-5 | 0.919/0.785 | 0.984/0.964 | 0.952/0.861 | |
| Mean ± SD | 0.519±0.325/0.531±0.143 | 0.915±0.048/0.860±0.111 | 0.717±0.166/0.696±0.113 | |
| CFPN | Fold-1 | 0.453/0.506 | 0.887/0.809 | 0.667/0.657 |
| Fold-2 | 0.720/0.660 | 0.903/0.852 | 0.811/0.756 | |
| Fold-3 | 0.992/0.718 | 0.995/0.920 | 0.994/0.819 | |
| Fold-4 | 0.235/0.658 | 0.882/0.985 | 0.559/0.821 | |
| Fold-5 | 0.831/0.801 | 0.896/0.992 | 0.864/0.897 | |
| Mean ± SD | 0.646±0.270/0.669±0.097 | 0.913±0.042/0.912±0.072 | 0.779±0.152/0.790±0.080 |
AP, average precision; mAP, mean average precision; Faster R-CNN, faster region-based convolutional neural network; FPN, feature pyramid network; CFPN, cascade feature pyramid network; SD, standard deviation.
Table 7
| System | Benign, mean ± SD | Malignant, mean ± SD | |||||
|---|---|---|---|---|---|---|---|
| Precision | Recall | F1-score | Precision | Recall | F1-score | ||
| Slice level | |||||||
| Faster R-CNN | 0.730±0.055 | 0.638±0.237 | 0.655±0.140 | 0.857±0.103 | 0.907±0.033 | 0.877±0.050 | |
| FPN | 0.682±0.174 | 0.570±0.147 | 0.600±0.107 | 0.830±0.078 | 0.878±0.072 | 0.850±0.052 | |
| CFPN | 0.757±0.137 | 0.700±0.176 | 0.718±0.139 | 0.888±0.084 | 0.913±0.056 | 0.899±0.058 | |
| Patient level | |||||||
| Faster R-CNN | 0.773±0.137 | 0.700±0.253 | 0.709±0.156 | 0.880±0.100 | 0.902±0.063 | 0.887±0.060 | |
| FPN | 0.693±0.155 | 0.650±0.138 | 0.658±0.117 | 0.851±0.056 | 0.862±0.081 | 0.854±0.052 | |
| CFPN | 0.775±0.160 | 0.850±0.258 | 0.806±0.199 | 0.932±0.106 | 0.900±0.069 | 0.914±0.082 | |
Faster R-CNN, faster region-based convolutional neural network; FPN, feature pyramid network; CFPN, cascade feature pyramid network; SD, standard deviation.
Discussion
In this study, we compared the DL-based CFPN system with the FPN and Faster R-CNN systems in the detection and classification of benign and malignant breast lesions, and further evaluated their performance for detecting large and small breast lesions. The results demonstrated that the mAP values and sensitivity of the CFPN outperformed the FPN and Faster R-CNN systems. CFPN achieved the highest sensitivities in detection at the lowest FPs at both the slice level and the patient level. This is crucial for the early follow-up treatment of patients with malignancy and for reducing overtreatment of patients without malignant lesions. In addition, the mAP values in overall evaluation and the F1-scores at the slice level and the patient level in real detection demonstrated the superiority of the CFPN system in detecting large and especially small breast lesions. Considering the difficulty of detecting small lesions, the developed detection system was expected to achieve accurate detection of breast lesions smaller than 2 cm, providing the possibility of detecting lesions of various size.
In DL of breast MRI, CNN models are currently a popular architecture for image analysis (3,36). However, other DL-based systems including Faster R-CNN, FPN, and CFPN, which have been successfully applied to mammography, ultrasound, and computed tomography (25,27,28,30,37), have not been systematically studied. Although automated methods can help reduce the inter-observer variation and improve reproducibility, automatic identification and object localization of breast cancers based on images is a challenge. Accordingly, an aim of our work was to perform and compare the results of using 3 different DL-based methods for the detection and classification of breast lesions. A previous study by Zhou et al. (17) showed that weakly supervised 3D DL could be used for breast cancer classification and localization in DCE-MRI. In contrast to their study, our study achieved simultaneous detecting, localizing, and classifying of the same lesion. On the other hand, the weakly supervised localization task could only detect lesions with a high malignancy probability, whereas our study also detected benign lesions.
Several other studies have evaluated the DL-based classification of breast lesions in MRI (13,18,19). However, as Ribli et al. (25) found, a lesion detector is clinically more useful than a simple image classifier, which can only give a single score per case or breast but cannot localize cancer that is essential for further diagnostic testing or treatment. The encouraging results in our study suggest that the DL-based CFPN system may be a candidate with promising development perspective for the simultaneous detection and classification of breast lesions in MRI. Regarding DL-based breast MRI classification, Dalmiş et al. (19) found that combining all non-contrast data yielded inferior results to DCE-MRI, which both confirms its importance and is why we followed this approach in this study. In contrast to the study by Zhou et al. (13), which included only mass-type lesions with limited size, weakening its general applicability in clinical applications, our study included both mass and non-mass type lesions and did not impose a limitation on lesions size. Although Truhn et al. (18) investigated the diagnostic performance of lesions smaller than 2 cm, lesion detection performance was not evaluated.
Our DL-based systems provide an automatic and accurate method for breast detection and classification with high sensitivity and low FPs. In particular, the CFPN achieved an optimal sensitivity of 0.970±0.014 at 0.375 FPs, which was consistent with the results of other studies (28,38). Regarding the low specificity and high sensitivity of the clinical MRI report, our proposed method could assist reporting systems to improve the specificity of cancer screening and potentially avoid unnecessary biopsy or overtreatment, and overfitting could be mitigated due to the adoption of data augmentation and focus loss strategies. Although the cascade model with increasing IoU thresholds played a significant role in the CFPN system, fewer samples still resulted in the inferior performance of benign lesions compared to malignant lesions in terms of overall evaluation (AP values) and real detection (F1-scores). This limitation can be appropriately addressed as we collect more samples in the future. Our results in real detection showed the F1-scores at patient level after majority voting were better than those at slice level in all DL-based systems for both benign and malignant breast lesions. The patient-level sensitivities were also improved at lower FPs compared to the corresponding results at the slice level. This suggested that the missed detection and FPs occurring on a single slice could be mitigated by integrating the detection results on all slices of each lesion. Nonetheless, the performance of our DL-based system will improve by increasing the number of training cases.
Our study had several limitations. First, the dataset was small for DL, with an imbalance of benign and malignant lesions, and a larger patient cohort is needed for further investigation to improve the performance of CFPN architecture. Second, the imaging data we used was collected from a single clinical center, which limits the robustness and generalizability of the model. Future studies should focus on collecting multi-institutional datasets to test and evaluate DL-based systems for lesion detection and classification. Finally, only DCE-MRI images were used for the DL-based systems, excluding non-enhanced MRI images, or a combination of both.
Conclusions
Our study shows that DL-based systems can automatically detect and classify breast lesions on DCE-MRI, and that CFPN has the highest sensitivity at the lowest FPs and accurately detects lesions smaller than 2 cm. These results illustrate the potential use of this technique in a clinically relevant setting.
Acknowledgments
Funding: This study was supported in part by grants from the Shaanxi Key R&D Plan (Nos. 2020SF042 and 2020SF036), Shaanxi Youth Innovation Team Project (No. 22JP087), and Shaanxi International Science and Technology Cooperation Program (No. 2021KW-55).
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-323/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-323/coif). XW is an employee of GE Healthcare, China. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Review Board of the Second Affiliated Hospital of Xi’an Jiaotong University (No. 2018-080). The requirement for informed consent was waived due to the retrospective nature of the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Byers T, Wender RC, Jemal A, Baskies AM, Ward EE, Brawley OW. The American Cancer Society challenge goal to reduce US cancer mortality by 50% between 1990 and 2015: Results and reflections. CA Cancer J Clin 2016;66:359-69. [Crossref] [PubMed]
- Ou WC, Polat D, Dogan BE. Deep learning in breast radiology: current progress and future directions. Eur Radiol 2021;31:4872-85. [Crossref] [PubMed]
- Mann RM, Cho N, Moy L. Breast MRI: State of the Art. Radiology 2019;292:520-36. [Crossref] [PubMed]
- Mann RM, Balleyguier C, Baltzer PA, Bick U, Colin C, Cornford E, et al. Breast MRI: EUSOBI recommendations for women's information. Eur Radiol 2015;25:3669-78. [Crossref] [PubMed]
- Leithner D, Wengert GJ, Helbich TH, Thakur S, Ochoa-Albiztegui RE, Morris EA, Pinker K. Clinical role of breast MRI now and going forward. Clin Radiol 2018;73:700-14. [Crossref] [PubMed]
- Meyer-Base A, Morra L, Tahmassebi A, Lobbes M, Meyer-Base U, Pinker K. AI-Enhanced Diagnosis of Challenging Lesions in Breast MRI: A Methodology and Application Primer. J Magn Reson Imaging 2021;54:686-702. [Crossref] [PubMed]
- D’Orsi CJ, Mendelson EB, Morris EASEA. ACR BI-RADS® Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology, 2013.
- Yamaguchi K, Schacht D, Newstead GM, Bradbury AR, Verp MS, Olopade OI, Abe H. Breast cancer detected on an incident (second or subsequent) round of screening MRI: MRI features of false-negative cases. AJR Am J Roentgenol 2013;201:1155-63. [Crossref] [PubMed]
- Gubern-Mérida A, Martí R, Melendez J, Hauth JL, Mann RM, Karssemeijer N, Platel B. Automated localization of breast cancer in DCE-MRI. Med Image Anal 2015;20:265-74. [Crossref] [PubMed]
- Mohammadi A, Afshar P, Asif A, Farahani K, Kirby J, Oikonomou A, Plataniotis KN. Lung Cancer Radiomics: Highlights from the IEEE Video and Image Processing Cup 2018 Student Competition. IEEE Signal Process Mag 2019;36:164-73. [Crossref] [PubMed]
- Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional Neural Networks for Radiologic Images: A Radiologist's Guide. Radiology 2019;290:590-606. [Crossref] [PubMed]
- Zhou J, Zhang Y, Chang KT, Lee KE, Wang O, Li J, Lin Y, Pan Z, Chang P, Chow D, Wang M, Su MY. Diagnosis of Benign and Malignant Breast Lesions on DCE-MRI by Using Radiomics and Deep Learning With Consideration of Peritumor Tissue. J Magn Reson Imaging 2020;51:798-809. [Crossref] [PubMed]
- Sitaula C, Aryal S. Fusion of whole and part features for the classification of histopathological image of breast tissue. Health Inf Sci Syst 2020;8:38. [Crossref] [PubMed]
- Lin H, Xiao H, Dong L, Teo KB, Zou W, Cai J, Li T. Deep learning for automatic target volume segmentation in radiation therapy: a review. Quant Imaging Med Surg 2021;11:4847-58. [Crossref] [PubMed]
- Zhang L, Mohamed AA, Chai R, Guo Y, Zheng B, Wu S. Automated deep learning method for whole-breast segmentation in diffusion-weighted breast MRI. J Magn Reson Imaging 2020;51:635-43. [Crossref] [PubMed]
- Zhou J, Luo LY, Dou Q, Chen H, Chen C, Li GJ, Jiang ZF, Heng PA. Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J Magn Reson Imaging 2019;50:1144-51. [Crossref] [PubMed]
- Truhn D, Schrading S, Haarburger C, Schneider H, Merhof D, Kuhl C. Radiomic versus Convolutional Neural Networks Analysis for Classification of Contrast-enhancing Lesions at Multiparametric Breast MRI. Radiology 2019;290:290-7. [Crossref] [PubMed]
- Dalmiş MU, Gubern-Mérida A, Vreemann S, Bult P, Karssemeijer N, Mann R, Teuwen J. Artificial Intelligence-Based Classification of Breast Lesions Imaged With a Multiparametric Breast MRI Protocol With Ultrafast DCE-MRI, T2, and DWI. Invest Radiol 2019;54:325-32. [Crossref] [PubMed]
- Wan KW, Wong CH, Ip HF, Fan D, Yuen PL, Fong HY, Ying M. Evaluation of the performance of traditional machine learning algorithms, convolutional neural network and AutoML Vision in ultrasound breast lesions classification: a comparative study. Quant Imaging Med Surg 2021;11:1381-93. [Crossref] [PubMed]
- Ha R, Mutasa S, Karcich J, Gupta N, Pascual Van Sant E, Nemer J, Sun M, Chang P, Liu MZ, Jambawalikar S. Predicting Breast Cancer Molecular Subtype with MRI Dataset Utilizing Convolutional Neural Network Algorithm. J Digit Imaging 2019;32:276-82. [Crossref] [PubMed]
- Ren T, Cattell R, Duanmu H, Huang P, Li H, Vanguri R, Liu MZ, Jambawalikar S, Ha R, Wang F, Cohen J, Bernstein C, Bangiyev L, Duong TQ. Convolutional Neural Network Detection of Axillary Lymph Node Metastasis Using Standard Clinical Breast MRI. Clin Breast Cancer 2020;20:e301-8. [Crossref] [PubMed]
- Ha R, Chin C, Karcich J, Liu MZ, Chang P, Mutasa S, Pascual Van Sant E, Wynn RT, Connolly E, Jambawalikar S. Prior to Initiation of Chemotherapy, Can We Predict Breast Tumor Response? Deep Learning Convolutional Neural Networks Approach Using a Breast MRI Tumor Dataset. J Digit Imaging 2019;32:693-701. [Crossref] [PubMed]
- Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Trans Pattern Anal Mach Intell 2017;39:1137-49. [Crossref] [PubMed]
- Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with Deep Learning. Sci Rep 2018;8:4165. [Crossref] [PubMed]
- Lin TY, Dollar P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. 30th IEEE/CVF conference on computer vision and pattern recognition (CVPR); 2017, Jul 21-26; Honolulu, HI. IEEE, 2017.
- Sajda P, Spence C, Pearson J. Learning contextual relationships in mammograms using a hierarchical pyramid neural network. IEEE Trans Med Imaging 2002;21:239-50. [Crossref] [PubMed]
- Peng J, Bao C, Hu C, Wang X, Jian W, Liu W. Automated mammographic mass detection using deformable convolution and multiscale features. Med Biol Eng Comput 2020;58:1405-17. [Crossref] [PubMed]
- Cai Z, Vasconcelos N. Cascade R-CNN: Delving into high Quality object detection. 31st IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2018 Jun 18-23; Salt Lake City, UT. IEEE, 2018.
- Mu G, Chen Y, Wu D, Zhan Y, Zhou X, Gao Y. Relu Cascade of feature pyramid networks for CT pulmonary nodule detection. 10th International Workshop on Machine Learning in Medical Imaging (MLMI)/22nd International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2019 Oct 13-17; Shenzhen, China. Springer, Cham, 2019.
- Sabel MS. Nonsurgical ablation of breast cancer: future options for small breast tumors. Surg Oncol Clin N Am 2014;23:593-608. [Crossref] [PubMed]
- Yap MH, Goyal M, Osman F, Martí R, Denton E, Juette A, Zwiggelaar R. Breast ultrasound region of interest detection and lesion localisation. Artif Intell Med 2020;107:101880. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016, Jun 27-30; Seattle, WA. IEEE, 2016.
- Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal Loss for Dense Object Detection. IEEE Trans Pattern Anal Mach Intell 2020;42:318-27. [Crossref] [PubMed]
- Kalli S, Semine A, Cohen S, Naber SP, Makim SS, Bahl M. American Joint Committee on Cancer's Staging System for Breast Cancer, Eighth Edition: What the Radiologist Needs to Know. Radiographics 2018;38:1921-33.
- Reig B, Heacock L, Geras KJ, Moy L. Machine learning in breast MRI. J Magn Reson Imaging 2020;52:998-1018. [Crossref] [PubMed]
- Zhou Y, Chen H, Li Y, Wang S, Cheng L, Li J. 3D multi-view tumor detection in automated whole breast ultrasound using deep convolutional neural network. Expert Systems with Applications 2021;168:114410. [Crossref]
- Agarwal R, Díaz O, Yap MH, Lladó X, Martí R. Deep learning for mass detection in Full Field Digital Mammograms. Comput Biol Med 2020;121:103774. [Crossref] [PubMed]


