
New 3D phase-unwrapping method based on voxel clustering and local polynomial modeling: application to quantitative susceptibility mapping
Introduction
The magnetic susceptibility in magnetic resonance imaging (MRI) is used to measure the degree of tissue magnetization after an external magnetic field is applied. It is defined as the ratio of the tissue magnetization per unit volume with an external magnetic field to the strength of the external magnetic field, and part per million (ppm) is the commonly used unit (1,2). The inverse solution of the underlying magnetic susceptibility distribution in quantitative susceptibility mapping (QSM) from the collected magnetic resonance signals involves multichannel coil combination, field map estimation, phase unwrapping, background field removal, and dipole inversion (2-5). Error at any step is likely to be introduced, even propagated and accumulated in the following process. Researchers have proposed many algorithms for addressing the challenges in these steps over the past years, and these have been successfully applied to measure iron in brain tissue, distinguish between hemorrhage and calcification, measure the cerebral metabolic rate for oxygen and liver iron deposition, and reveal the cortical development in children with drug-resistant epilepsy (3,6-8).
Phase unwrapping is a key step in QSM reconstruction, and its reliability directly influences the accuracy of QSM results. The collected magnetic resonance signals are complex-valued, and the phase, usually obtained using the arctangent function, is generally wrapped into the fixed range of (−π, π] (9-11). Therefore, a phase-unwrapping algorithm is needed to remove phase discontinuities in order to obtain the underlying smooth true phase. However, phase unwrapping will fail in the presence of severe noise, disconnected regions, rapid phase changes, and open-ended lines in the phase data (9-13). Many 3-dimensional (3D) phase-unwrapping algorithms have been proposed and used to deal with these problems, and applied to QSM, including path-following methods (14-17) and Laplacian-based unwrapping methods (18-20). In the path-following methods, if the phase transformation of adjacent voxels is greater than π along a certain path, a wrap can be determined to have occurred. The value of +2π or –2π is added to unwrapped voxels along this path, and the sign of the added value depends on whether the phase jump is positive or negative. Unless an error occurs, this processing will accurately restore the underlying true phase. Due to the influence of noise, rapid phase variations, and other factors, it is easy to generate mistakes in the process of unwrapping, which leads to propagation and accumulation in the unwrapping pipeline. The Laplacian-based unwrapping methods try to identify the position of the wrapping phase, because the local derivatives of the unwrapping phase and the wrapping phase are mostly the same. However, these phase-unwrapping methods will modify the underlying true phase values of all voxels in the original data so that the phase change of neighboring voxels is less than π. The higher the number of dimensions, the less the phase-unwrapping method is sensitive to noise but the greater the computational complexity. Therefore, a robust 3D phase-unwrapping method is needed prior to the generation of QSM.
Recently, we proposed a 2-dimensional (2D) phase-unwrapping method based on pixel clustering and local surface fitting (CLOSE) (10). The phasor is defined as the complex MR signal divided by the magnitude (21). A local difference of phasors represents the local variations of the underlying true phase (LDTP), and in theory it is less than or equal to the corresponding local difference of the wrapped phase (LDWP). Firstly, the CLOSE method clusters pixels into 2 classes: smooth blocks that are easy-to-unwrap, and residual pixels that are difficult-to-unwrap. Next, a local surface fitting method based on region growing is used to unwrap the intrablock, interblock, and residual-pixel phases. The proposed 2D CLOSE method can obtain an accurate and reliable underlying true phase image even when there is serious noise, rapid phase changes, or disconnected regions in the phase image. To do this, we extend CLOSE from 2 dimensions to 3 dimensions and apply it to QSM. The extended CLOSE uses a 3D 26-connected neighborhood centered at each voxel to calculate the local variations of phase and phasor, divides the phase data into 3D blocks or sub-blocks, and finally takes advantage of a 3D local polynomial model to recover the local true phase. Simulation data and in vivo QSM data were collected to evaluate the performance of the proposed 3D CLOSE (CLOSE3D) method, which was compared with the common region growing (RG) method (22) and the region-expanding labeling for unwrapping estimates (PRELUDE) method (23).
Methods
Problem formulation
In MRI, the acquired complex signal S can be written as , where M represents the magnitude information and ϕ is the true phase. The phase φ calculated using the arctangent function is defined as:
where (·) is an arctangent function. The calculated phase φ will be wrapped into the range of (-π, π] radians, thereby φ is also known as the wrapped phase. The relationship between the wrapped phase and the underlying true phase can be written as follows:
where k is an integer. The process of phase unwrapping is performed to find the correct phase compensation 2kπ for the wrapped phase of each voxel to recover the underlying true phase, based on the assumption that the true phase is spatially smooth (10,23).
Proposed CLOSE3D method for phase unwrapping
In the MR image, the variation of wrapped phases is rapid in the region far from the tissue boundary but near the wrapped position, and the change in the corresponding phasors is slow. Motivated by these observations, our original CLOSE method first classified the pixels into smooth blocks that are easy-to-unwrap, and residual pixels that are difficult-to-unwrap. Next, a local surface fitting method is used to unwrap intrablock, interblock, and residual-pixel phases with a region growing strategy. CLOSE can be extended from 2 dimensions to 3 dimensions, in which the key step is to calculate LDTP and LDWP using the 3D neighborhood and then cluster the phase data into 3D blocks or sub-blocks. Finally, a 3D local polynomial model is used to recover the underlying true phase in the local window.
Local polynomial modeling
In the original CLOSE method, a local surface function model is used to model the smooth true phase in the local window. In the proposed CLOSE3D method, we use a local polynomial model to approximate the true phase in the local window, which can be represented as follows:
where Cl,m,n denotes the polynomial fitting coefficients. L, M and N are the orders of the polynomial function in the x, y, and z directions, respectively. represents the fitting error. The phase-unwrapping problem is effectively transformed into a parameter estimation problem using the 3D local polynomial model. The coefficients Cl,m,n can be determined through the fitting Eq. [3] utilizing the information of the unwrapped voxels in the local window. If there are P unwrapped voxels in a local window contented at the growing point , then Eq. [3] can be reformulated:
here Φ is a column vector containing the phase of P unwrapped voxels; X is a matrix of , and each row of X contains a polynomial basis of unwrapped voxels. is a column vector consisting of a fitting coefficient; R is a column vector consisting of a residual error. Through the fitting Eq. [4] to these P unwrapped voxels, the fitting coefficients are estimated by the least square method. The integer compensation of the growing voxel is calculated as follows:
where is the polynomial basis of the growing voxel , and round(z) is a function that computes the closest integer to z.
Computing 3D LDTP and LDWP
We use the 26-connected neighborhood to compute the local variations of phasors and phases, namely:
where N(x0, y0, z0) represents the set of voxels in a local window centered on voxel . and are the wrapped phases at the voxel (x, y, z) and in space, respectively; is a function to calculate absolute value.
Proposed CLOSE3D method
The CLOSE3D method presented herein first applies the threshold to the LDTP map and classifies the voxels in the phase data into 2 classes: easy-to-unwrap 3D blocks and difficult-to-unwrap residual voxels. The phase unwrapping of intrablock, interblock, and residual-voxels is then performed in turn. For intrablock phase unwrapping, CLOSE3D first applies the threshold to the LDWP map and divides each block into 3D sub-blocks without phase unwrapping and residual voxels, and then the phase unwrapping of the inter-sub-blocks and residual voxels is performed by exploring the 3D local polynomial fitting method. Intrablock unwrapping begins with the largest sub-block and processes the closest sub-blocks and residual voxels. The implementation of the interblock phase unwrapping is similar to the process of inter-sub-block unwrapping. Phase unwrapping of residual voxels in the region of interest (ROI) is performed similarly to the phase unwrapping of residual voxels in a block.
Experiments
Simulations
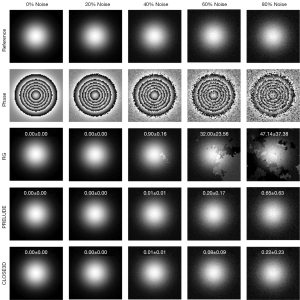
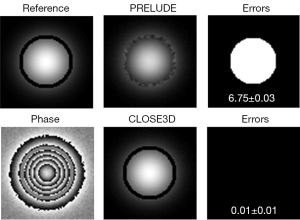
Simulation datasets were used to quantitatively and qualitatively evaluate the performance of CLOSE3D. A Gaussian function with a standard deviation (SD) of 20 voxels was used to generate a 3D spherical phase map with a size of 80×80×80. The magnitude of the simulation data was 100, and the true phase height was set to 40 radians. To test the performance of CLOSE3D at different noise levels, Gaussian noises with mean of 0 and SDs of 0, 20, 40, 60, and 80 were independently added to the real and imaginary parts of the generated complex data. Therefore, the noise levels of the simulated data were 0%, 20%, 40%, 60%, and 80%, respectively, as shown in Figure 1. To test the performance of CLOSE3D with phase data with disconnected regions, the value of the voxels in a preset region was set to 0. Finally, Gaussian noise with a mean of 0 and SD of 20 was added to the real part and imaginary part of the generated complex data, as shown in Figure 2.
In addition, a Gaussian phase cylinder was generated to test the performance of the CLOSE3D method under different signal-to-noise ratios (SNRs) and varying phase levels. The synthesized true phase was defined as follows:
For each xOy plane, the width of each section of data was π/6 radians, and the magnitude of the signal increased from 10 to 120 with an interval of 10 in the anticlockwise direction. Random noise with a mean of 0 and SD of 20 was independently added to the real part and imaginary part of the generated complex data. Therefore, the SNRs increased from 0.5 to 6 at a rate of 0.5 in the anticlockwise direction. The different z values had different phase change levels, so this simulated dataset could be used to verify the performance of the proposed method under different SNRs and phase levels, as shown in Figure 3 and Table 1.
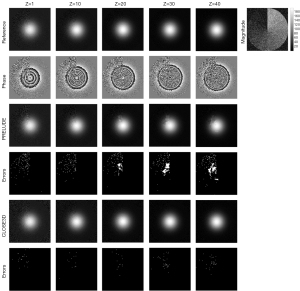
Table 1
| Z | PRELUDE | CLOSE3D |
|---|---|---|
| 1 | 0.50±0.06 | 0.21±0.08 |
| 5 | 0.49±0.13 | 0.20±0.07 |
| 10 | 0.51±0.10 | 0.21±0.03 |
| 15 | 0.50±0.13 | 0.25±0.03 |
| 20 | 0.53±0.12 | 0.22±0.05 |
| 25 | 0.72±0.23 | 0.21±0.02 |
| 30 | 1.13±0.13 | 0.23±0.09 |
| 35 | 1.99±0.37 | 0.23±0.04 |
| 40 | 3.19±0.73 | 0.39±0.03 |
Values are given as mean ± standard deviation. Z, the Nth slice; MCR, number of incorrect unwrapping voxels divided by the total voxels in the region of interest.
In vivo brain QSM data
To verify the validity of the proposed CLOSE3D method, we used brain QSM data collected on a 3.0T MR machine (Siemens Skyra; Siemens, Erlangen, Germany) from 3 adult volunteers with cerebral hemorrhage. The 3D multi-echo gradient-echo sequence was used to obtain the required images for testing and comparing of the phase-unwrapping methods. The scanning parameters of the QSM data were: repetition time (TR) =2,300 ms, echo time (TE)1 =2.74 ms, echo space =5 ms, echo number =8, FA =15°, resolution =0.9×0.9×2 mm3, and field of view (FOV) =24 cm. In addition, new wrapped phase data were obtained by multiplying the original vector data of the brain QSM data by 5 to evaluate the performance of CLOSE3D when the open-ended cutline was present.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of the First Affiliated Hospital of Zhengzhou University (No. 2018-KY-88) and informed consent was provided by all the participants.
Implementation and parameters
The CLOSE3D algorithm was implemented using MATLAB (R2010a; MathWorks, Natick, MA, USA) on a desktop (Dell, Intel Core 2 Duo, 3.5 GB RAM). The classical 3D RG method implemented in the QSM toolkit (22) and PRELUDE with default parameters implemented in the fMRI software library (FSL; Oxford Center for Functional Magnetic Resonance Imaging of the brain, UK) (24) were compared with our proposed method. All 3 methods used a mask to remove voxels from the background regions. The simulated data obtained the mask in a similar way to that of our original CLOSE method. For the application of the in vivo QSM data, the BET function in the FSL (24) was used to extract the brain mask.
The parameters of CLOSE3D included a 26-connected neighborhood, which is more robust to noise and can reflect phase variation in every direction, and was used to calculate LDTP and LDWP. An empirical value of π/4 as the clustering threshold could appropriate cluster the voxels into (sub)blocks and residual voxels in the simulated experiments, and was applied to the in vivo data. To eliminate the effect of small (sub)blocks, a block or sub-block with <100 voxels was classified as residual voxels. The polynomial orders L, M, and N were all set to 2, consistent with the report by Friedlander and Francos (25). In the inter-sub-block unwrapping, the fitting points were selected as the 100 voxels closest to the unwrapped regions in the growing region and the 100 voxels closest to the growing (sub)block in the unwrapped regions. The fitting size of 100 voxels was experientially determined to be equal to the minimum block size, consistent with the report by Cheng et al. (10). The fitting window size was set to 11, and applied to all the following experiments based on observing the simulation results under different noise levels.
In the simulation experiments, the reference phase image was defined as the sum of the synthesized original phase and the phase changes caused by noise (23). This reference phase was subtracted from the unwrapped phase image, and if the absolute value of the phase difference of voxels was >π/10 radian, the voxels were not considered to be unwrapped correctly. The misclassification ratio (MCR) was used to quantitatively and qualitatively evaluate the performance of CLOSE3D. The MCR was defined as the number of incorrect unwrapping voxels divided by the total voxels in the ROI. Each simulation experiment was repeated 50 times to reduce the variability and compute the corresponding means and SDs of MCRs.
In order to evaluate the performance of CLOSE3D with the vivo QSM data, the acquired in vivo 3D multi-echo GRE data were used to reconstruct the QSM. First, the multi-echo magnitude and phase data were used to generate the wrapped total field map by a non-linear least square fitting method. Subsequently, the RG, PRELUDE, and CLOSED methods were used to obtain the unwrapped total field map. After that, the projection onto dipole fields (PDF) (26,27) method was used to exclude the background fields to obtain the tissue field. Finally, the morphology-enabled dipole inversion (MEDI) algorithm (28) was used to inverted the tissue field to the QSM maps. PDF with default parameters, and MEDI with λ=1,000 in the MEDI_toolbox (22) were used. If the phase-unwrapping method cannot obtain an accurate underlying true phase image, there will be serious artifacts in the susceptibility image.
Results
Evaluation of simulation data
Figure 1 shows the phase-unwrapping results using the RG, PRELUDE, and CLOSE3D methods under different noise levels. The first to the fifth rows report the reference phase, the wrapped phase, and the phase-unwrapping results of RG, PRELUDE, and CLOSE3D, respectively. The values in the last 3 rows are the mean and SDs of the MCRs (mean ± SD, %) for 50 independent repetitions. When the noise level was ≤20%, all 3 methods obtained perfect unwrapped results, and the corresponding MCR was 0.00%±0.00%. When the noise level ≥40%, the phase-unwrapping image generated by the RG method had obvious residual wraps, whereas there were no significant residual wraps in the phase-unwrapping results of PRELUDE and CLOSE3D. The MCRs of the 3 methods all increased with added noise levels, but the MCR results for PRELUDE and CLOSE3D were consistently <0.65%±0.63%, and the MCR value of CLOSE3D was lower than that of PRELUDE. The statistical comparisons of the error ratios of RG and PRELUDE with those of CLOSE3D over the simulated data with different noise levels are shown in Table S1. When the noise levels were equal to 60% and 80%, the error ratios of RG (P=0.000 and P=0.000) and PRELUDE (P=0.007, P=0.014) were significantly higher than those of CLOSE3D.
Figure 2 shows the 1-slice phase-unwrapping results using PRELUDE and CLOSE3D with datasets with disconnected regions. PRELUDE generated serious unwrapped errors (mean and SD of MCR, 6.75%±0.03%). In contrast, CLOSE3D generated a promising underlying true phase image with a small number of errors (MCR 0.01%±0.01%).
In Figure 3, we show the phase-unwrapping results for PRELUDE and CLOSE3D with different phase variations levels (z-direction) and SNRs (xOy plane). When Z is ≤10, both PRELUDE and CLOSE3D obtained accurate true phase images without significant error residuals in the results. However, when Z increased to 20, the phase-unwrapping result of PRELUDE showed significant residual wraps. In contrast, when Z changed from 1 to 40, CLOSE3D continued to produce accurate phase-unwrapping results. The number of error unwrapped voxels with both the PRELUDE and CLOSE3D methods increased with the decrease in SNRs, but the number of incorrectly unwrapped voxels in PRELUDE was higher than that in CLOSE3D. The result for CLOSE3D was consistently better than that for PRELUDE under different SNRs and different phase variations levels.
In Table 1, we show the means and SDs of the MCRs of PRELUDE and CLOSE3D with 50 independent repetitions. When Z was ≤20, the mean MCRs of CLOSE3D was equal to about half of that of PRELUDE, all of which generated accurate phase-unwrapping results, and the corresponding mean MCR was not more than 0.53%. When Z was ≥25, CLOSE3D continued to generate a lower mean and SD of the MCRs than PRELUDE. When Z =40, the mean MCR of CLOSE3D was about 1/8 of the mean MCR of PRELUDE.
Performance with in vivo data
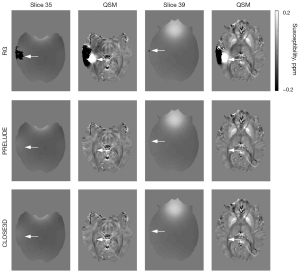
In Figure 4, we show the phase-unwrapping results using the RG, PRELUDE, and CLOSE3D methods using clinical MR brain data acquired at 3.0T. In contrast to the RG method, which shows significant residual wraps (as indicated by the arrow), the PRELUDE method shows slight residual wraps (as indicated by the arrow) in the limbic region of the brain, but there are no obvious residual wraps in the results of the CLOSE3D method.
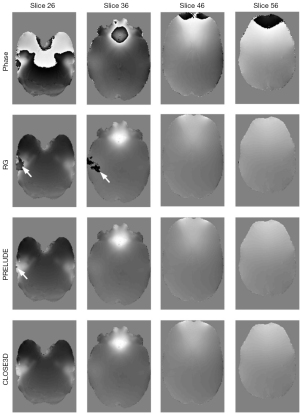
Figure 5 shows the QSM results based on the phase-unwrapping results of RG, PRELUDE, and CLOSE3D methods. As shown by the arrows, there are residual wraps in the unwrapping results of the RG method, which results in serious artifacts in the QSM results. In contrast, no significant residual wraps are found in the unwrapping results by the PRELUDE and CLOSE3D methods, and no significant artifacts are found in the corresponding QSM results. The susceptibility of the deep grey matter nuclei (substantia nigra, red nucleus, caudate nucleus, putamen, globus pallidus, and substantia nigra) were measured and reported in the Appendix 1.
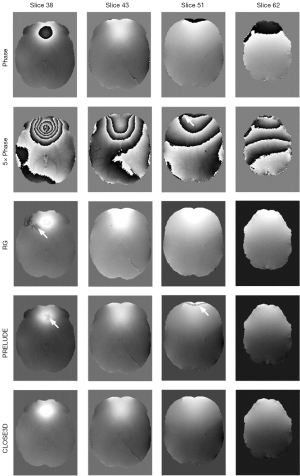
In Figure 6, we show the comparison of RG, PRELUDE, and CLOSE3D with the simulated phase data with open-ended cutline. The arrow in the second row points to the location of an open-ended cutline. The arrows in the third and fourth rows point to obvious unwrapping errors. The results for the RG and PRELUDE methods both have serious residual wraps (as indicated by the arrows), but the CLOSE3D method obtained accurate phase-unwrapping results. When there is an open-ended cutline in the image, CLOSE3D can still achieve an accurate phase-unwrapping result, whereas PRELUDE cannot achieve a smooth underlying true phase image (as indicated by the arrow).
Discussion
We have proposed a new 3D phase-unwrapping method to be applied to QSM to obtain brain tissue susceptibility results. The CLOSE3D method firstly applies the threshold to the LDTP to cluster voxels into easy-to-unwrap 3D smooth blocks and difficult-to-unwrap residual voxels. Next, a local polynomial fitting method based on region growing is performed to unwrap the intrablock, interblock, and residual-voxels in turn. The results of simulation and clinical MR data experiments showed that even when there was severe noise, disconnected regions, rapid phase changes, or open-ended cutlines in the phase data, the CLOSE3D method still obtained accurate 3D phase-unwrapping results. The CLOSE3D method will help phase-related 3D MRI applications such as susceptibility weighted imaging (SWI) and QSM.
There are significant differences among the RG, PRELUDE, and CLOSE3D methods for data with different SNRs and rapid phase changes. With the RG method, errors that can occur at any point along the unwrapping path are passed to the phase unwrapping of subsequent voxels and accumulate, producing serious errors in the resulting image, as shown in Figure 1. PRELUDE assumes that the underlying true phase change of neighboring voxels is <π, and if that condition is not met, PRELUDE will fail, as shown in Figure 6. The CLOSE3D method first unwraps 3D smooth blocks that are easy-to-unwrap, and then uses the phase information of the already unwrapped region to process the unwrapped residual voxels. This strategy can avoid voxels of low SNR appearing in the unwrapping path early, which can reduce error propagation and accumulation. In addition, the CLOSE3D uses a local polynomial model to model the true phase in the local window, and the local polynomial model is not sensitive to noise, which means the CLOSE3D method can obtain accurate phase-unwrapping results.
For in vivo QSM experiments, the phase-unwrapping results have an important effect on the accurate determination of magnetic susceptibility. Local error in the phase-unwrapping results of the RG algorithm leads to serious artifacts in the QSM results, making it difficult to estimate tissue susceptibility, as shown in Figure 5. Compared with the RG algorithm, the QSM results obtained by unwrapping using the PRELUDE and CLOSE3D methods were obviously improved, and artifacts were eliminated. In the simulation experiment with an open-ended cutline, due to the presence of voxels with true phase changes larger than π, there were serious residual wraps in the PRELUDE result. CLOSE3D classifies the voxels with phase changes greater than π and the voxels around them are residual voxels, and it utilizes a local polynomial fitting method to unwrap the residual voxels using the information of the already unwrapped regions around them, which helps to reduce error propagation and accumulation. In addition, the local polynomial model is robust to the disconnected regions and the cases in which the underlying true phase variation of neighboring voxels is greater than π.
The CLOSE3D method retains the shortcomings of our original CLOSE method, such as too many parameters needing to be selected and optimized, and the code running efficiency is relatively low. These need to be improved prior to CLOSE3D being applied to clinical Dixon water-fat separation and QSM. In similarity to the original CLOSE method, the CLOSE3D method is limited by the polynomial model used. The CLOSE3D method cannot obtain accurate phase-unwrapping results if the true phase changes do not match the polynomial model, because the local polynomial function cannot accurately model the underlying true phase in the local window. One solution to this problem is to use higher-order polynomial models such as B-spline interpolation and Chebyshev polynomials (29). However, a higher-order polynomial model would slightly improve fitting quality but increase the complexity of the algorithm and reduce the computational efficiency, which is consistent with other reports (25,30), and is demonstrated in the Appendix 1. Another solution is to use a hybrid model that uses local polynomial fitting to unwrap the phase and then the result is unwrapped with a nonparametric phase-unwrapping method such as a least squares algorithm. These shortcomings will be addressed in our future research.
Conclusions
A robust 3D phase imaging method was proposed and applied to QSM to obtain the susceptibility results. The CLOSE3D method firstly clusters voxels into easy-to-unwrap 3D smooth blocks and difficult-to-unwrap residual voxels by applying the threshold to the LDTP, and then the phase-unwrapping of intrablock, interblock, and residual-voxels is performed in sequence by the region growing local polynomial model. Compared with the classical RG and PRELUDE methods, the results with both simulated and in vivo MRI data showed that even with serious noise, disconnected regions, rapid phase changes, or open-ended cutlines in the phase data, the CLOSE3D method can accurately unwrap 3D phase data. The proposed CLOSE3D method will contribute to phase-related applications such as SWI and QSM in MRI.
Acknowledgments
Funding: This work was supported by the Union Project of Medical and Technology Research Program of Henan Province (No. LHGJ20190159), the Science and Technology Planning Program of Henan Province (Nos. 212102310088, 212102310886, and 2020GGJS123), Guangdong Basic and Applied Basic Research Foundation (No. 2019A1515111182), and National Natural Science Foundation of China (No. 62101144).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-525/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Board of the First Affiliated Hospital of Zhengzhou University (No. 2018-KY-88) and informed consent was provided by all the participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Haacke EM, Liu S, Buch S, Zheng W, Wu D, Ye Y. Quantitative susceptibility mapping: current status and future directions. Magn Reson Imaging 2015;33:1-25. [Crossref] [PubMed]
- Wang Y, Liu T. Quantitative susceptibility mapping (QSM): Decoding MRI data for a tissue magnetic biomarker. Magn Reson Med 2015;73:82-101. [Crossref] [PubMed]
- Langkammer C, Schweser F, Shmueli K, Kames C, Li X, Guo L, Milovic C, Kim J, Wei H, Bredies K, Buch S, Guo Y, Liu Z, Meineke J, Rauscher A, Marques JP, Bilgic B. Quantitative susceptibility mapping: Report from the 2016 reconstruction challenge. Magn Reson Med 2018;79:1661-73. [Crossref] [PubMed]
- Chen J, Zhang Z, Nie X, Xu Y, Liu C, Zhao X, Wang Y. Predictive value of thrombus susceptibility for cardioembolic stroke by quantitative susceptibility mapping. Quant Imaging Med Surg 2022;12:550-7. [Crossref] [PubMed]
- Zheng Q, Xu L, Xiong L, Cui X, Nan J, He T. Coil combination using linear deconvolution in k-space for phase imaging. Quant Imaging Med Surg 2019;9:1792-803. [Crossref] [PubMed]
- Zhang Y, Shi J, Wei H, Han V, Zhu WZ, Liu C. Neonate and infant brain development from birth to 2 years assessed using MRI-based quantitative susceptibility mapping. Neuroimage 2019;185:349-60. [Crossref] [PubMed]
- Lorio S, Sedlacik J, So PW, Parkes HG, Gunny R, Löbel U, Li YF, Ogunbiyi O, Mistry T, Dixon E, Adler S, Cross JH, Baldeweg T, Jacques TS, Shmueli K, Carmichael DW. Quantitative MRI susceptibility mapping reveals cortical signatures of changes in iron, calcium and zinc in malformations of cortical development in children with drug-resistant epilepsy. Neuroimage 2021;238:118102. [Crossref] [PubMed]
- Bachrata B, Trattnig S, Robinson SD. Quantitative susceptibility mapping of the head-and-neck using SMURF fat-water imaging with chemical shift and relaxation rate corrections. Magn Reson Med 2022;87:1461-79. [Crossref] [PubMed]
- Cheng J, Guan J, Mei Y, Xu L, Liu X, Feng Q, Chen W, Feng Y. A novel phase-unwrapping method by using phase-jump detection and local surface fitting: application to Dixon water-fat MRI. Magn Reson Med 2018;80:2630-40. [Crossref] [PubMed]
- Cheng J, Mei Y, Liu B, Guan J, Liu X, Wu EX, Feng Q, Chen W, Feng Y. A novel phase-unwrapping method based on pixel clustering and local surface fitting with application to Dixon water-fat MRI. Magn Reson Med 2018;79:515-28. [Crossref] [PubMed]
- Cheng J, Liu B, Mei Y, Guo Y, Chen M, Liu X, Chen W, Feng Y. A robust 3D phase unwrapping method with application to quantitative susceptibility mapping. In Proceedings of the 25th Annual Meeting of ISMRM, Honolulu, USA, 2017. p1956.
- Lindemeyer J, Oros-Peusquens AM, Shah NJ. Quality-based UnwRap of SUbdivided Large Arrays (URSULA) for high-resolution MRI data. Med Image Anal 2019;52:13-23. [Crossref] [PubMed]
- Chavez S, Xiang QS, An L. Understanding phase maps in MRI: a new cutline phase unwrapping method. IEEE Trans Med Imaging 2002;21:966-77. [Crossref] [PubMed]
- Lu Y, Wang X, He G. Phase unwrapping based on branch cut placing and reliability ordering. Optical Engineering 2005;44:055601. [Crossref]
- Cusack R, Papadakis N. New robust 3-D phase unwrapping algorithms: application to magnetic field mapping and undistorting echoplanar images. Neuroimage 2002;16:754-64. [Crossref] [PubMed]
- Abdul-Rahman HS, Gdeisat MA, Burton DR, Lalor MJ, Lilley F, Moore CJ. Fast and robust three-dimensional best path phase unwrapping algorithm. Appl Opt 2007;46:6623-35. [Crossref] [PubMed]
- Schofield MA, Zhu Y. Fast phase unwrapping algorithm for interferometric applications. Opt Lett 2003;28:1194-6. [Crossref] [PubMed]
- Makhoul J. A fast cosine transform in one and two dimensions. IEEE Transactions on Acoustics, Speech, and Signal Processing 1980;28:27-34. [Crossref]
- Haralick RM. A storage efficient way to implement the discrete cosine transform. IEEE Transactions on Computers 1976;25:764-5. [Crossref]
- Hockney RW. A fast direct solution of Poisson’s equation using Fourier analysis. JACM 1965;12:95-113. [Crossref]
- Xiang QS. Two-point water-fat imaging with partially-opposed-phase (POP) acquisition: an asymmetric Dixon method. Magn Reson Med 2006;56:572-84. [Crossref] [PubMed]
- MEDI. Accessed January 30, 2020. Available online: http://weill.cornell.edu/mri/QSM/Online.zip
- Jenkinson M. Fast, automated, N-dimensional phase-unwrapping algorithm. Magn Reson Med 2003;49:193-7. [Crossref] [PubMed]
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23:S208-19. [Crossref] [PubMed]
- Friedlander B, Francos JM. Model based phase unwrapping of 2-D signals. IEEE Transactions on Signal Processing 1996;44:2999-3007. [Crossref]
- Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR, Wang Y. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage 2012;59:2560-8. [Crossref] [PubMed]
- Liu T, Khalidov I, de Rochefort L, Spincemaille P, Liu J, Tsiouris AJ, Wang Y. A novel background field removal method for MRI using projection onto dipole fields (PDF). NMR Biomed 2011;24:1129-36. [Crossref] [PubMed]
- Liu T, Liu J, de Rochefort L, Spincemaille P, Khalidov I, Ledoux JR, Wang Y. Morphology enabled dipole inversion (MEDI) from a single-angle acquisition: comparison with COSMOS in human brain imaging. Magn Reson Med 2011;66:777-83. [Crossref] [PubMed]
- Langley J, Zhao Q. A model-based 3D phase unwrapping algorithm using Gegenbauer polynomials. Phys Med Biol 2009;54:5237-52. [Crossref] [PubMed]
- Liang ZP. A model-based method for phase unwrapping. IEEE Trans Med Imaging 1996;15:893-7. [Crossref] [PubMed]


