
Application of deep learning to identify ductal carcinoma in situ and microinvasion of the breast using ultrasound imaging
Introduction
Ductal carcinoma in situ (DCIS) of the breast comprises abnormal proliferation of ductal endothelial cells without invasion of the basement membrane. Following a DCIS diagnosis, the 10-year breast cancer-specific mortality rate is 1.1% (1). However, some patients with breast DCIS also exhibit microinvasion (MIC) (2-5), where cancer cells within the duct break through the basement membrane and infiltrate the surrounding stroma with a diameter of no more than 1 mm (2-6). In the clinical and pathological anatomical stages, DCIS cases are given Tis (tumor in situ) classification (7). The invasive T1 tumors are subdivided into T1mi (invasive carcinoma of 1 mm or smaller), T1a (invasive carcinoma larger than 1 mm, up to and including 5 mm), T1b (invasive carcinoma larger than 5 mm, up to and including 10 mm), or T1c (invasive carcinoma larger than 10 mm, up to and including 20 mm) (7,8). The DCIS (with or without MIC) are divided into low, intermediate, and high grades. After breast-conserving surgery, high-risk DCIS patients (i.e., those with comedonecrosis or high nuclear grade) have a higher recurrence rate (9,10). High nuclear grade and comedonecrosis are more common in MIC patients (11-13). Overall, DCIS with MIC carries a nearly two-fold increase in mortality rate compared to DCIS without MIC (14). Patients with MIC are typically treated similarly to those with T1a breast cancer as recommended by current guidelines from the National Comprehensive Cancer Network (NCCN) (15).
The clinical diagnosis of breast tumors is based principally on imaging examination, including mammography, magnetic resonance imaging (MRI), and ultrasound. Mammography is an important method for breast cancer screening in developed countries. However, for dense breast tissue without calcification, mammography is only useful for identifying lesions (16-19). An auxiliary breast ultrasound screening can help detect mammographically-occult breast cancers, with typical detection rates of between 0.8 and 10 cancers detected in every 1,000 women screened (20). Breast volumes in Chinese women are typically smaller and denser than the global average, making ultrasound a more suitable option for Chinese DCIS patients due to its higher sensitivity and specificity when used in dense breast tissue (21,22). Ultrasound provides higher sensitivity to MIC than mammography (23).
Although MRI also provides high sensitivity, it is expensive, and there is conflicting evidence as to whether preoperative MRI is beneficial to DCIS patients (24-28). A recent systematic review by Canelo-Aybar et al. (29) showed that preoperative MRI did not reduce the rate of repeat surgery or mastectomy. With increasing access to imaging technology and screening, patients with DCIS can be identified earlier. The provision of ultrasound imaging findings of MIC inpatients with DCIS provides surgeons with more information from which to make clinical decisions. Techniques such as sentinel lymph node biopsy (SLNB) may also be considered to determine axillary lymph node status (9).
Determining the contour of a DCIS lesion using traditional manual data-driven methods is challenging because the heterogeneity of DCIS makes the boundary contour between the tumor and normal tissue unclear. Contour extraction is often based on a single image, while the diagnosis is typically based on multiple images of a lesion, which is more consistent with clinical practice. Deep neural networks (DNNs) are the most widely used models in medical image recognition and provide obvious advantages in the diagnosis of breast cancers. Impressive achievements have been made in automating the prediction of benign and malignant breast cancers and lymph node metastasis, and in simulating human decision-making (30-33).
The objectives of this study were to investigate the feasibility of using DNN models to identify MIC on ultrasound images of patients with DCIS and to evaluate the classification efficiency of deep learning using external verification. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-46/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee on Biomedical Research of West China Hospital, Sichuan University (No. 2020-1219). The committee waived individual consent for this retrospective analysis because it only involved the use of ultrasound images and did not involve patients’ personal information.
Patient population
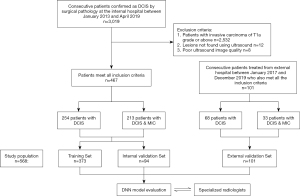
The medical records of consecutive patients with DCIS confirmed by surgical pathology were collected from January 2013 to April 2019 to form a training data set. The inclusion and exclusion criteria were based on the gold standard of pathology (Figure 1). The inclusion criteria were as follows: (I) the patient underwent an ultrasound examination in the 14 days before surgery; (II) the ultrasound images showed lesions that could be used for evaluation; (III) the patient underwent a simple mastectomy, and the lesions were completely removed for testing, after which the comprehensive pathological report confirmed either DCIS or DCIS MIC; (IV) if the patient had a multi-center or multi-focus DCIS, only the largest lesion was included in the analysis; and (V) the image background was uniform, with appropriate gain, and the lesion was clearly visible. The exclusion criteria were as follows: (I) any patient with invasive carcinoma of T1a or above; (II) any patient with a pathological diagnosis of DCIS or DCIS MIC where the tumor could not be located using an ultrasound; and (III) the background quality of the ultrasound image was poor, such that the tumor could not be recognized. Subsequent comprehensive pathological reports indicated DCIS with and without MIC in 213 and 254 patients, respectively. External verification data were collected using the same methodology from Sichuan Provincial People’s Hospital , comprising 101 patients with DCIS (68 without MIC; 33 with MIC).
Pathological assessment
The criteria for MIC were the presence of invasive foci on continuous sections of tumor tissue and a maximum diameter of the largest invasion less than or equal to 1 mm, according to the American Joint Committee on Cancer (AJCC) guidelines (6). The final surgical pathological report recorded the nuclear grade (low, medium, or high), the presence/absence of necrosis, and the expression statuses of estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), and the nuclear protein Ki-67. For the classification criteria of molecular markers, see Appendix 1.
Ultrasound image acquisition and visual inspection
Ultrasound images were acquired using machines equipped with a linear array high-frequency probe with a frequency range of 4 to 15 MHz, including the Philips (HD11, IU22, EPIQ5; Philips, Amsterdam, Netherlands), GE Logic E9 (GE Healthcare, Chicago, IL, USA), HI VISION Preirus (Hitachi Medical Corp., Tokyo, Japan), Esaote MyLab90 (Esaote, Maastricht, Holland), Supersonic Imagine (Aix-en-Province, France), and Siemens ACUSON OXANA 2 (Siemens Healthineers, Erlangen, Germany). Ultrasound examination used a portable probe for radial scanning of the whole breast. The standard acquisition procedure for ultrasound images is to obtain longitudinal and transverse sections at the maximum diameter of the lesion with the lesion located in the middle of the image. Supplementary images are obtained that show any malignant signs of the lesion, such as calcification or structural distortion.
For both internal and external verification of ultrasound images, the reading was conducted blind and the clinician reading the image was unaware of the clinicopathological status of the patient. Ultrasound images were assessed by three experienced breast radiologists (with 10, 12, and 30 years of experience), who reached a consensus through discussion. The clinicians read all acquired ultrasound images. Ultrasound characteristics were classified by gland background, phenotype, and calcification. The ultrasonic phenotype was summarized as either a mass or another type. A mass was defined as a lesion that could be recognized as a mass on the ultrasound images, including on the longitudinal and transverse sections. Depending on the mass composition, it was classified as either a solid texture mass or a cystic solid texture mass. Other types were characterized as having an indistinct hypoechoic area, dilated duct, architectural distortion, complicated cyst, or local thickening of the glands.
Image preprocessing
Only ultrasonographic images were input into the DNN. Data preprocessing included scaling, flipping, and rotating the original image to increase the datasets’ diversity and improve the model’s robustness (Figure 2). Images from the same patient showed similarities; therefore, lesions from the same patient were not divided into subsets. The DNN analysis was performed on the balanced data set. This comprised convolution feature extraction, nonlinear activation function mapping, pooling, and classification of full connection layers and the Softmax layer. The maximum characteristics of each image were recorded for each case. The results were obtained by combining four images.
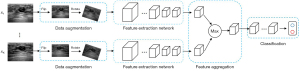
The data collected from West China Hospital of Sichuan University (the internal dataset) were randomly divided into training and validation sets at a ratio of 8:2 (Figure S1) to test five different DNN models. The data collected from Sichuan Provincial People’s Hospital formed the external validation dataset. There were 2,492 original images from both hospitals (1474 for DCIS; 1018 for MIC). During the training and validation phases, the number of images per case was set at four. For a case with fewer than four images, existing images were randomly chosen to be duplicated to make the total up to four. Otherwise, four images were randomly selected from each case. After the image preprocessing, only four images were included in each case, making the total number of images 2,272.
Deep learning
The model input comprised multiple breast ultrasonography images of the same lesion {Xn=x1, … xk, …, xK}. The output was the prediction Yn ∈ {MIC, non-MIC}, where Xn denotes the n-th set of the same lesion containing K images, and xk indicates the kth image in the set. Breast ultrasonography images were in RGB (red-green-blue) format with a size of 1,024×768×3 voxels. Original images were resized to 299×299×3 voxels using a bilinear interpolation algorithm from the Python Imaging Library (Secret Labs AB, Östergötland, Sweden) to save on computational resources without sacrificing model performance. Online data augmentation, including random horizontal flipping (probability 0.5) and random rotation by 0 to 180 degrees, was applied to resized images to augment the training data in real-time. The preprocessed image values were then divided by 255 to convert values into the range of 0.0 to 1.0.
Multiple ImageNet pre-trained networks were explored and fine-tuned on the dataset, namely Inception-v3, ResNet-50, ResNet-101, DenseNet-161, and DenseNet-169, all of which were implemented using PyTorch version 0.4.1 (Meta AI, New York, NY, USA). Each DNN model was split into two parts, one for extracting features from input images and one for classification The extraction process comprised aggregating high-level features using max aggregation operators (Figure 2). The classification process comprised transforming aggregated features into the classification space, Y. The DNN models were trained using Adadelta as the optimizer, with a learning rate of 0.1 and a weight decay rate of 10−4 for 200 epochs (34). The mini-batch size was fixed at 8. Dropout was applied to the last fully connected layer of the second sub-network to reduce overfitting, with a drop probability of 0.7. Finally, Inception-v3 was the highest performing network and was used in the final analysis (35).
DNN performance evaluation and statistical analysis
From each sample in the evaluation set, the probability score of the Softmax function and corresponding tag value were recorded. The scores were sorted from high to low, and a threshold was set. If the probability that the sample belonged to the positive sample was greater than the threshold, it was considered a positive sample; otherwise, it was a negative sample. Each time a different threshold was selected, a set of true-positive and false-positive rates were obtained corresponding to points on the receiver operating characteristic (ROC) curve. A cut-off value for the median score of 0.5 was used. The ROC curve was plotted, and the area under the ROC curve (AUC) was calculated. The accuracy was then calculated.
Statistical analysis was performed in SPSS version 22.0 (IBM Corp., Armonk, NY, USA). The characteristics of the training, internal, and external verification sets were compared using a t-test and the chi-square test/Fisher exact test. Continuous variables were described using mean ± standard deviation. Categorical variables were described using proportion (%). The clinical pathology and ultrasound characteristics were analyzed using univariate logistic regression to identify MIC. Statistical significance was considered at a threshold of P<0.05 (two-tailed).
Results
Clinical characteristics
Between January 2013 and April 2019, a total of 3,019 patients underwent surgery with a pathological diagnosis of DCIS, and 2,532 patients were excluded from the study due to the presence of an invasive carcinoma of T1a grade or above. A total of 12 patients were excluded due to negative ultrasound findings, and 8 cases were excluded due to poor ultrasound image quality. Between January 2017 and December 2019, 101 patients with DCIS with or without MIC were finally reported by pathology after surgery in Sichuan Provincial People’s Hospital. All 568 patients included in the study were women, 322 (56.7%) without MIC, and 246 (43.3%) with MIC. The training, internal evaluation, and external verification sets consisted of 373, 94, and 101 patients, respectively, with ages 48.79±11.12 (range, 21 to 83) years, 46.64±10.71 (range, 21 to 81) years, and 45.41±8.85 (range 25 to 70) years, respectively.
The characteristics of the training, internal verification, and external verification datasets are shown in Table 1. Age (P=0.086), menstrual status (P=0.158), histology (P=0.977), gland background (P=0.949), phenotype (P=0.751), and calcification (P=0.407) did not differ between the three datasets. Nuclear grade (P=0.115), necrosis (P=0.930), ER (P=0.233), PR (P=0.316), HER2 (P=0.184), Ki-67 (P=0.103), and lymphatic metastasis (P=0.663) did not differ between the training and internal validation sets.
Table 1
| Characteristics | Training (n=373), n (%) | Internal validation (n=94), n (%) | External validation (n=101), n (%) | P value* |
|---|---|---|---|---|
| Age, mean ± SD [range] | 48.79±11.12 [21–83] | 46.64±10.71 [21–81] | 45.41±8.85 [25–70] | 0.086 |
| Menstrual status | 0.158 | |||
| Premenopausal | 216 (57.91) | 62 (65.96) | 71 (70.30) | |
| Postmenopausal | 157 (42.09) | 32 (34.04) | 30 (29.70) | |
| Histology | 0.977 | |||
| DCIS | 203 (54.42) | 51 (54.26) | 68 (67.33) | |
| DCIS MIC | 170 (45.58) | 43 (45.74) | 33 (32.67) | |
| Nuclear grade | 0.115 | |||
| High | 220 (58.98) | 47 (50.00) | NA | |
| Low and medium | 148 (39.70) | 46 (48.94) | NA | |
| Null | 5 (1.32) | 1 (1.06) | NA | |
| Necrosis | 0.930 | |||
| Yes | 123 (32.96) | 32 (34.04) | NA | |
| No | 245 (65.68) | 61 (64.89) | NA | |
| Null | 5 (1.36) | 1 (1.07) | NA | |
| ER | 0.233 | |||
| Positive | 174 (46.65) | 45 (47.87) | NA | |
| Negative | 115 (30.83) | 33 (35.11) | NA | |
| Null | 84 (22.52) | 16 (17.02) | NA | |
| PR | 0.316 | |||
| Positive | 155 (41.55) | 42 (44.68) | NA | |
| Negative | 133 (35.66) | 36 (38.30) | NA | |
| Null | 85 (22.79) | 16 (17.02) | NA | |
| HER2 | 0.184 | |||
| Negative | 171 (45.84) | 49 (52.13) | NA | |
| Positive | 118 (31.64) | 29 (30.85) | NA | |
| Null | 84 (22.52) | 16 (17.02) | NA | |
| Ki-67 | 0.103 | |||
| Low expression | 184 (49.33) | 55 (58.51) | NA | |
| High expression | 105 (28.15) | 23 (24.47) | NA | |
| Null | 84 (22.52) | 16 (17.02) | NA | |
| Lymphatic metastasis | 0.663 | |||
| Yes | 6 (1.60) | 1 (1.11) | NA | |
| No | 367 (98.40) | 93 (98.89) | NA | |
| Gland background | 0.949 | |||
| Homogeneous | 201 (53.89) | 51 (54.26) | 46 (45.54) | |
| Heterogeneous | 172 (46.11) | 43 (45.74) | 55 (54.46) | |
| Phenotype | 0.751 | |||
| Mass | 148 (39.70) | 39 (41.49) | 51 (50.50) | |
| Other type** | 225 (60.30) | 55 (58.51) | 50 (49.50) | |
| Calcification | 0.407 | |||
| Present | 232 (62.20) | 54 (57.45) | 63 (62.38) | |
| Absent | 141 (37.80) | 40 (42.55) | 38 (37.62) |
*, T-test between training and internal validation cohort. **, Other type includes indistinct hypoechoic area; dilated duct; architectural distortion; complicated cyst; and local thickening of glands. SD, standard deviation; DCIS, ductal carcinoma in situ; MIC, microinvasion; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; NA, not available.
Univariate analysis of characteristics and ultrasound visual inspection
The clinicopathology of patients treated in the external hospital was not available; therefore, the univariate analysis was only performed on patients recruited through the internal hospital. High nuclear grade (P<0.001), necrosis (P=0.006), an ER negative result (P=0.003), a PR negative result (P=0.001), an HER2 positive result (P=0.034), lymphatic metastasis (P=0.008), and calcification (P<0.001) were found to be predictive of MIC (Table 2). Age (P=0.989), menstrual status (P=0.200), Ki-67 (P=0.091), gland background (P=0.096), and phenotype (P=0.050) were not found to have any predictive value.
Table 2
| Characteristics | Group | DCIS (n=254), n (%) | MIC (n=213), n (%) | P value |
|---|---|---|---|---|
| Age (years) | <40 | 44 (17.32) | 37 (17.37) | 0.989 |
| ≥40 | 210 (82.68) | 176 (82.63) | ||
| Menstrual status | Premenopausal | 158 (62.20) | 120 (56.34) | 0.200 |
| Postmenopausal | 96 (37.80) | 93 (43.66) | ||
| Nuclear grade | Low and medium | 145 (57.09) | 49 (23.00) | <0.001 |
| High | 103 (40.55) | 164 (77.00) | ||
| Null | 6 (2.36) | 0 (0.00) | ||
| Necrosis | No | 184 (72.44) | 122 (57.28) | 0.006 |
| Yes | 64 (25.20) | 91 (42.72) | ||
| Null | 6 (2.36) | 0 (0.00) | ||
| ER | Negative | 63 (24.80) | 85 (39.91) | 0.003 |
| Positive | 131 (51.57) | 88 (41.31) | ||
| Null | 60 (23.63) | 40 (18.78) | ||
| PR | Negative | 70 (27.56) | 99 (46.48) | 0.001 |
| Positive | 123 (48.43) | 74 (34.74) | ||
| Null | 61 (24.01) | 40 (18.78) | ||
| HER2 | Negative | 143 (56.30) | 77 (36.15) | 0.034 |
| Positive | 51 (20.08) | 96 (45.07) | ||
| Null | 60 (23.62) | 40 (18.78) | ||
| Ki-67 level | Low | 150 (59.06) | 89 (41.78) | 0.091 |
| High | 44 (17.32) | 84 (39.44) | ||
| Null | 60 (23.62) | 40 (18.78) | ||
| Lymphatic metastasis | No | 254 (100) | 206 (96.71) | 0.008 |
| Yes | 0 (0.00) | 7 (3.29) | ||
| Gland background | Homogeneous | 146 (57.48) | 106 (49.77) | 0.096 |
| Heterogeneous | 108 (42.52) | 107 (50.23) | ||
| Phenotype | Mass | 112 (44.09) | 75 (35.21) | 0.050 |
| Other types | 142 (55.91) | 138 (64.79) | ||
| Calcification | Absent | 132 (51.97) | 49 (23.00) | <0.001 |
| Present | 122 (48.03) | 164 (77.00) |
DCIS, ductal carcinoma in situ; MIC, microinvasion; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2.
Deep learning models
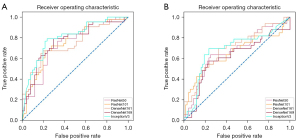
The network architecture of the Inception-v3 model is shown in Table S1. The AUCs, classification accuracy, sensitivity, and specificity of the DNN model performance are shown in Table 3. The ROC curves of the internal and external datasets using the subsets of the DNN models are shown in Figure 3. In the internal evaluation, the 5 DNN models achieved classification results with AUCs ranging from 0.740 to 0.803. The Inception-v3 model showed the best classification performance with an AUC of 0.803 [95% confidence interval (CI): 0.709 to 0.878] and a higher classification accuracy than that of the other four deep learning models. The DenseNet-169 model had a higher sensitivity of 79.1%. The specificities of the ResNet-101, DenseNet-161, and Inception-v3 models reached 76.5%. The AUC of the logistic regression model in the internal test set was 0.740 (95% CI: 0.685 to 0.757). The classification ability of the logistic regression model was consistent with that of ResNet-50. The accuracy and specificity of the logistic regression model were lower than the other five deep learning models; however, the sensitivity was higher than that of ResNet-101 and DenseNet-161 (72.5% vs. 67.4% vs. 67.4%). In the external test, the AUCs of the 5 DNN models ranged from 0.614 to 0.696. The DenseNet-161 model showed a better classification performance AUC of 0.696 (95% CI: 0.597–0.784). Inception-v3 had a classification accuracy of 74.3% and a sensitivity of 69.7%. Both ResNet-50 and DenseNet-161 had a specificity of 79.4%.
Table 3
| Models | Accuracy | Sensitivity | Specificity | F1 | AUC |
|---|---|---|---|---|---|
| Internal validation | |||||
| ResNet-50 | 0.713 | 0.767 | 0.667 | 0.769 | 0.740 (0.639–0.825) |
| ResNet-101 | 0.723 | 0.674 | 0.765 | 0.704 | 0.757 (0.657–0.839) |
| DenseNet-161 | 0.723 | 0.674 | 0.765 | 0.704 | 0.746 (0.646–0.830) |
| DenseNet-169 | 0.745 | 0.791 | 0.706 | 0.795 | 0.768 (0.669–0.849) |
| Inception-v3 | 0.766 | 0.767 | 0.765 | 0.781 | 0.803 (0.709–0.878) |
| Logistic regression | 0.691 | 0.725 | 0.651 | 0.712 | 0.740 (0.685–0.757) |
| External validation | |||||
| ResNet-50 | 0.713 | 0.545 | 0.794 | 0.642 | 0.670 (0.570–0.761) |
| ResNet-101 | 0.693 | 0.545 | 0.765 | 0.640 | 0.653 (0.551–0.745) |
| DenseNet-161 | 0.703 | 0.515 | 0.794 | 0.618 | 0.696 (0.597–0.784) |
| DenseNet-169 | 0.703 | 0.576 | 0.765 | 0.665 | 0.614 (0.512–0.709) |
| Inception-v3 | 0.743 | 0.697 | 0.765 | 0.761 | 0.685 (0.585–0.774) |
AUC, the area under the curve.
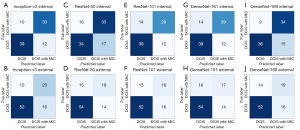
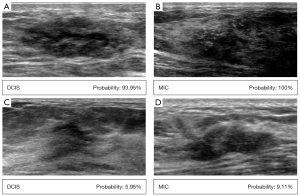
The confusion matrices of the internal validation of 94 patients from West China Hospital of Sichuan University and the external validation of 101 patients from Sichuan Provincial People’s Hospital are shown in Figure 4, including the correct classification and error classification numbers of each DNN model for DCIS with and without MIC. Inception-v3 showed the best performance in the internal test set. Of the 22 cases that were incorrectly classified by Inception-v3, 12 manifested hypoechoic areas, 10 manifested mass structures on the ultrasound images (8 solid; 2 cystic solid), and 3 MIC of those 10 had calcification. Figure 5 shows some examples of Inception-v3 identifying the probability of MIC.
Discussion
This study established that a DNN model can be used to identify MIC in DCIS based on ultrasound images. The most accurate DNN model was Inception-v3, which showed 76.6% accuracy and an AUC of 0.803. In the internal test, the AUC of Inception-v3 was higher than that of the logistic model (0.803 vs. 0.740). Internal validation showed the AUCs of the 5 DNN models to range from 0.740 to 0.803, while the AUCs of the external validation ranged from 0.614 to 0.696. The best AUC in the external validation (0.696; DenseNet-161) was surpassed by that of Inception-v3 in the internal validation (0.803). The better performance of Inception-v3 was mainly because the multiple operations in the Inception block (e.g., convolution kernel sizes of 1×1, 3×3, and 5×5) facilitated the extraction of variable features on multiple scales.
Deep learning has been used to detect orthotopic cancer lesions, predict the risk of invasive cancer, and identify high-risk populations based on imaging features. Recently, deep learning has been used to differentiate atypical ductal hyperplasia from DCIS (36,37) with AUC values of 0.86 (using a total of 298 images from 149 patients) and 0.90 (a total of 280 images from 140 patients). Mutasa et al. (38) predicted DCIS and invasive cancer with an AUC of 0.71 (246 images from 123 patients), and Shi et al. (39) predicted occult invasion from pure DCIS with an AUC of 0.70 (99 patients). These findings were based on mammography alone and have not been externally verified. The present study is the first to report the use of artificial intelligence, specifically deep learning, to identify MIC on ultrasound images. DNN models were found to have great potential for diagnostic utility.
The MIC type of cancer is a minimally invasive breast cancer representing approximately 0.9% of breast cancer diagnoses (40). Previous investigations identifying MIC have been based on clinicopathological factors or imaging characteristics. Overall, the histological grade of DCIS with MIC was higher than that of DCIS without MIC (13,22,41,42). Calcification on ultrasound images is more often seen in cases of MIC than in DCIS without MIC (22,41). The expression of molecular markers varies across different studies. Treatment modalities and prognoses of MIC patients were found to be similar to those in invasive breast cancer (42). In the present study, high nuclear grade, necrosis, ER negativity, PR negativity, HER2 positivity, lymph node metastasis, and calcification were all found to be more common in MIC patients (all P<0.05). We used a logistic model combined with clinicopathological factors and ultrasound features to identify MIC with an accuracy comparable to that of DNN and an AUC of 0.740 (95% CI: 0.685 to 0.757).
For invasive carcinoma or extensive DCIS patients, SLNB has been a standard surgical technique (43). Axillary lymph node positivity was found almost exclusively in patients with MIC (13,41,44). Based on a large dataset, 7.6% of MIC had lymph node metastases (45). In the present study, 7 of 213 MIC patients had lymph node metastasis, representing a rate of 3.3%. Future studies of surgical planning may assess the use of SLNB in MIC patients identified by DNN models.
As reported in this study, many DCIS patients exhibit a heterogeneous glandular background and diverse imaging manifestations, most of which are non-mass structures. If these features are extracted manually using conventional methods, it may be difficult to determine the tumor boundary and range; this decreases the accuracy of the extracted contour and the tumor size estimation. The DNN models perform reliably as they do not require artificial computing features. They can be used to objectively identify important tumor features while eliminating misinterpretation and human error, thereby reducing the clinical workload (46,47). The DNN algorithms are more robust and direct due to the omission of unnecessary steps during the learning period. Subsequent studies may use new algorithms and larger sample sizes to renew interest in machine learning (48). Further, an ultrasound is easy to operate, and images are easily acquired; therefore, it is vital that research can support the suitability of ultrasonography for applying a DNN system.
There are several limitations to this study. First, only ultrasound data were used, while mammography and MRI data were excluded. Second, the dataset was relatively small, and the classification performance of the experimental model did not achieve the expected effect (the classification accuracy was less than 80%). However, the DNN models benefit greatly from a large validation data set; more data from multiple hospitals may improve the performance of the deep learning model. Third, as this was a retrospective study, the number of ultrasound images was uneven. This study only selected four images of each patient as input, which cannot fully represent all the features of the tumor. Future studies can use full-automatic volume imaging and three-dimensional ultrasound to obtain more comprehensive characteristics. Finally, the external validation did not use a logistic regression model due to the unavailability of clinicopathological data.
Conclusions
We implemented deep learning to detect MIC on ultrasound images. The DNN models provided accurate and robust performance. These findings suggest that DNN models may accurately identify MIC in DCIS using ultrasound images and have the potential to provide an objective auxiliary diagnostic method for clinicians. In particular, networks based on the Inception-v3 model may perform the best for these and similar applications.
Acknowledgments
Funding: This work was supported by a grant from the 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (No. ZYGD18012) and the Chengdu Science and Technology Plan (No. 2017-CY02-00027-GX-PDF).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-46/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-46/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee on Biomedical Research of West China Hospital, Sichuan University (No. 2020-1219). The committee waived individual consent for this retrospective analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Narod SA, Iqbal J, Giannakeas V, Sopik V, Sun P. Breast Cancer Mortality After a Diagnosis of Ductal Carcinoma In Situ. JAMA Oncol 2015;1:888-96. [Crossref] [PubMed]
- Namm JP, Mueller J, Kocherginsky M, Kulkarni S. The utility of sentinel lymph node biopsy in patients with ductal carcinoma in situ suspicious for microinvasion on core biopsy. Ann Surg Oncol 2015;22:59-65. [Crossref] [PubMed]
- Lillemoe TJ, Tsai ML, Swenson KK, Susnik B, Krueger J, Harris K, Rueth N, Grimm E, Leach JW. Clinicopathologic analysis of a large series of microinvasive breast cancers. Breast J 2018;24:574-9. [Crossref] [PubMed]
- Phantana-Angkool A, Voci AE, Warren YE, Livasy CA, Beasley LM, Robinson MM, Hadzikadic-Gusic L, Sarantou T, Forster MR, Sarma D, White RL Jr. Ductal Carcinoma In Situ with Microinvasion on Core Biopsy: Evaluating Tumor Upstaging Rate, Lymph Node Metastasis Rate, and Associated Predictive Variables. Ann Surg Oncol 2019;26:3874-82. [Crossref] [PubMed]
- Rakovitch E, Sutradhar R, Lalani N, Nofech-Mozes S, Gu S, Goldberg M, Hanna W, Fong C, Paszat L. Multiple foci of microinvasion is associated with an increased risk of invasive local recurrence in women with ductal carcinoma in situ treated with breast-conserving surgery. Breast Cancer Res Treat 2019;178:169-76. [Crossref] [PubMed]
- Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC cancer staging manual. 7th ed. New York, NY: Springer; 2010.
- Amin MB, Edge SB, Greene FL, et al, eds. AJCC Cancer Staging Manual. 8th ed. New York, NY: Springer; 2017.
- Kalli S, Semine A, Cohen S, Naber SP, Makim SS, Bahl M. American Joint Committee on Cancer's Staging System for Breast Cancer, Eighth Edition: What the Radiologist Needs to Know. Radiographics 2018;38:1921-33.
- Virnig BA, Tuttle TM, Shamliyan T, Kane RL. Ductal carcinoma in situ of the breast: a systematic review of incidence, treatment, and outcomes. J Natl Cancer Inst 2010;102:170-8. [Crossref] [PubMed]
- Rakovitch E, Nofech-Mozes S, Narod SA, Hanna W, Thiruchelvam D, Saskin R, Taylor C, Tuck A, Sengupta S, Elavathil L, Jani PA, Done SJ, Miller N, Youngson B, Kong I, Paszat L. Can we select individuals with low risk ductal carcinoma in situ (DCIS)? A population-based outcomes analysis. Breast Cancer Res Treat 2013;138:581-90. [Crossref] [PubMed]
- Prasad ML, Osborne MP, Giri DD, Hoda SA. Microinvasive carcinoma (T1mic) of the breast: clinicopathologic profile of 21 cases. Am J Surg Pathol 2000;24:422-8. [Crossref] [PubMed]
- Shatat L, Gloyeske N, Madan R, O'Neil M, Tawfik O, Fan F. Microinvasive breast carcinoma carries an excellent prognosis regardless of the tumor characteristics. Hum Pathol 2013;44:2684-9. [Crossref] [PubMed]
- Kim M, Kim HJ, Chung YR, Kang E, Kim EK, Kim SH, Kim YJ, Kim JH, Kim IA, Park SY. Microinvasive Carcinoma versus Ductal Carcinoma In Situ: A Comparison of Clinicopathological Features and Clinical Outcomes. J Breast Cancer 2018;21:197-205. [Crossref] [PubMed]
- Sopik V, Sun P, Narod SA. Impact of microinvasion on breast cancer mortality in women with ductal carcinoma in situ. Breast Cancer Res Treat 2018;167:787-95. [Crossref] [PubMed]
- National Comprehensive Cancer Network. Clinical Practice Guidelines in Oncology: breast. Available online: https://www.nccn.org/professionals/physician_gls/pdf/breast.pdf. Published 2017. Accessed October 20, 2017.
- Mandelson MT, Oestreicher N, Porter PL, White D, Finder CA, Taplin SH, White E. Breast density as a predictor of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst 2000;92:1081-7. [Crossref] [PubMed]
- Melnikow J, Fenton JJ, Whitlock EP, Miglioretti DL, Weyrich MS, Thompson JH, Shah K. Supplemental Screening for Breast Cancer in Women With Dense Breasts: A Systematic Review for the U.S. Preventive Services Task Force. Ann Intern Med 2016;164:268-78. [Crossref] [PubMed]
- Tagliafico AS, Calabrese M, Mariscotti G, Durando M, Tosto S, Monetti F, Airaldi S, Bignotti B, Nori J, Bagni A, Signori A, Sormani MP, Houssami N. Adjunct Screening With Tomosynthesis or Ultrasound in Women With Mammography-Negative Dense Breasts: Interim Report of a Prospective Comparative Trial. J Clin Oncol 2016;34:1882-8. [Crossref] [PubMed]
- Su X, Lin Q, Cui C, Xu W, Wei Z, Fei J, Li L. Non-calcified ductal carcinoma in situ of the breast: comparison of diagnostic accuracy of digital breast tomosynthesis, digital mammography, and ultrasonography. Breast Cancer 2017;24:562-70. [Crossref] [PubMed]
- Hooley RJ, Greenberg KL, Stackhouse RM, Geisel JL, Butler RS, Philpotts LE. Screening US in patients with mammographically dense breasts: initial experience with Connecticut Public Act 09-41. Radiology 2012;265:59-69. [Crossref] [PubMed]
- Shen S, Zhou Y, Xu Y, Zhang B, Duan X, Huang R, Li B, Shi Y, Shao Z, Liao H, Jiang J, Shen N, Zhang J, Yu C, Jiang H, Li S, Han S, Ma J, Sun Q. A multi-centre randomised trial comparing ultrasound vs mammography for screening breast cancer in high-risk Chinese women. Br J Cancer 2015;112:998-1004. [Crossref] [PubMed]
- Jin ZQ, Lin MY, Hao WQ, Jiang HT, Zhang L, Hu WH, Zhang M. Diagnostic evaluation of ductal carcinoma in situ of the breast: ultrasonographic, mammographic and histopathologic correlations. Ultrasound Med Biol 2015;41:47-55. [Crossref] [PubMed]
- Vieira CC, Mercado CL, Cangiarella JF, Moy L, Toth HK, Guth AA. Microinvasive ductal carcinoma in situ: clinical presentation, imaging features, pathologic findings, and outcome. Eur J Radiol 2010;73:102-7. [Crossref] [PubMed]
- Davis KL, Barth RJ Jr, Gui J, Dann E, Eisenberg B, Rosenkranz K. Use of MRI in preoperative planning for women with newly diagnosed DCIS: risk or benefit? Ann Surg Oncol 2012;19:3270-4. [Crossref] [PubMed]
- Fancellu A, Turner RM, Dixon JM, Pinna A, Cottu P, Houssami N. Meta-analysis of the effect of preoperative breast MRI on the surgical management of ductal carcinoma in situ. Br J Surg 2015;102:883-93. [Crossref] [PubMed]
- Yoon GY, Choi WJ, Kim HH, Cha JH, Shin HJ, Chae EY. Surgical Outcomes for Ductal Carcinoma in Situ: Impact of Preoperative MRI. Radiology 2020;295:296-303. [Crossref] [PubMed]
- Park AY, Gweon HM, Son EJ, Yoo M, Kim JA, Youk JH. Ductal carcinoma in situ diagnosed at US-guided 14-gauge core-needle biopsy for breast mass: preoperative predictors of invasive breast cancer. Eur J Radiol 2014;83:654-9. [Crossref] [PubMed]
- Chou SS, Romanoff J, Lehman CD, Khan SA, Carlos R, Badve SS, et al. Preoperative Breast MRI for Newly Diagnosed Ductal Carcinoma in Situ: Imaging Features and Performance in a Multicenter Setting (ECOG-ACRIN E4112 Trial). Radiology 2021;301:66-77. [Crossref] [PubMed]
- Canelo-Aybar C, Taype-Rondan A, Zafra-Tanaka JH, Rigau D, Graewingholt A, Lebeau A, Pérez Gómez E, Rossi PG, Langendam M, Posso M, Parmelli E, Saz-Parkinson Z, Alonso-Coello P. Preoperative breast magnetic resonance imaging in patients with ductal carcinoma in situ: a systematic review for the European Commission Initiative on Breast Cancer (ECIBC). Eur Radiol 2021;31:5880-93. [Crossref] [PubMed]
- Ciritsis A, Rossi C, Eberhard M, Marcon M, Becker AS, Boss A. Automatic classification of ultrasound breast lesions using a deep convolutional neural network mimicking human decision-making. Eur Radiol 2019;29:5458-68. [Crossref] [PubMed]
- Zhou LQ, Wu XL, Huang SY, Wu GG, Ye HR, Wei Q, Bao LY, Deng YB, Li XR, Cui XW, Dietrich CF. Lymph Node Metastasis Prediction from Primary Breast Cancer US Images Using Deep Learning. Radiology 2020;294:19-28. [Crossref] [PubMed]
- Wan KW, Wong CH, Ip HF, Fan D, Yuen PL, Fong HY, Ying M. Evaluation of the performance of traditional machine learning algorithms, convolutional neural network and AutoML Vision in ultrasound breast lesions classification: a comparative study. Quant Imaging Med Surg 2021;11:1381-93. [Crossref] [PubMed]
- Qi X, Zhang L, Chen Y, Pi Y, Chen Y, Lv Q, Yi Z. Automated diagnosis of breast ultrasonography images using deep neural networks. Med Image Anal 2019;52:185-98. [Crossref] [PubMed]
- Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2016. pp. 2818–26.
- Zeiler MD. Adadelta: an adaptive learning rate method. arXiv preprint arXiv:1212. 5701, 2012.
- Ha R, Mutasa S, Sant EPV, Karcich J, Chin C, Liu MZ, Jambawalikar S. Accuracy of Distinguishing Atypical Ductal Hyperplasia From Ductal Carcinoma In Situ With Convolutional Neural Network-Based Machine Learning Approach Using Mammographic Image Data. AJR Am J Roentgenol 2019; [Epub ahead of print]. [Crossref] [PubMed]
- Mutasa S, Chang P, Nemer J, Van Sant EP, Sun M, McIlvride A, Siddique M, Ha R. Prospective Analysis Using a Novel CNN Algorithm to Distinguish Atypical Ductal Hyperplasia From Ductal Carcinoma in Situ in Breast. Clin Breast Cancer 2020;20:e757-60. [Crossref] [PubMed]
- Mutasa S, Chang P, Van Sant EP, Nemer J, Liu M, Karcich J, Patel G, Jambawalikar S, Ha R. Potential Role of Convolutional Neural Network Based Algorithm in Patient Selection for DCIS Observation Trials Using a Mammogram Dataset. Acad Radiol 2020;27:774-9. [Crossref] [PubMed]
- Shi B, Grimm LJ, Mazurowski MA, Baker JA, Marks JR, King LM, Maley CC, Hwang ES, Lo JY. Prediction of Occult Invasive Disease in Ductal Carcinoma in Situ Using Deep Learning Features. J Am Coll Radiol 2018;15:527-34. [Crossref] [PubMed]
- Hoda SA, Chiu A, Prasad ML, Giri D, Hoda RS. Are microinvasion and micrometastasis in breast cancer mountains or molehills? Am J Surg 2000;180:305-8. [Crossref] [PubMed]
- Yao JJ, Zhan WW, Chen M, Zhang XX, Zhu Y, Fei XC, Chen XS. Sonographic Features of Ductal Carcinoma In Situ of the Breast With Microinvasion: Correlation With Clinicopathologic Findings and Biomarkers. J Ultrasound Med 2015;34:1761-8. [Crossref] [PubMed]
- Champion CD, Ren Y, Thomas SM, Fayanju OM, Rosenberger LH, Greenup RA, Menendez CS, Hwang ES, Plichta JK. DCIS with Microinvasion: Is It In Situ or Invasive Disease? Ann Surg Oncol 2019;26:3124-32. [Crossref] [PubMed]
- Gooch JC, Schnabel F, Chun J, Pirraglia E, Troxel AB, Guth A, Shapiro R, Axelrod D, Roses D. A Nomogram to Predict Factors Associated with Lymph Node Metastasis in Ductal Carcinoma In Situ with Microinvasion. Ann Surg Oncol 2019;26:4302-9. [Crossref] [PubMed]
- Costarelli L, Cianchetti E, Corsi F, Friedman D, Ghilli M, Lacaria M, Menghini L, Murgo R, Ponti A, Rinaldi S, Del Turco MR, Taffurelli M, Tinterri C, Tomatis M, Fortunato L. Microinvasive breast carcinoma: An analysis from ten Senonetwork Italia breast centres. Eur J Surg Oncol 2019;45:147-52. [Crossref] [PubMed]
- Wang W, Zhu W, Du F, Luo Y, Xu B. The Demographic Features, Clinicopathological Characteristics and Cancer-specific Outcomes for Patients with Microinvasive Breast Cancer: A SEER Database Analysis. Sci Rep 2017;7:42045. [Crossref] [PubMed]
- Gao Y, Liu B, Zhu Y, Chen L, Tan M, Xiao X, Yu G, Guo Y. Detection and recognition of ultrasound breast nodules based on semi-supervised deep learning: a powerful alternative strategy. Quant Imaging Med Surg 2021;11:2265-78. [Crossref] [PubMed]
- Rodríguez-Ruiz A, Krupinski E, Mordang JJ, Schilling K, Heywang-Köbrunner SH, Sechopoulos I, Mann RM. Detection of Breast Cancer with Mammography: Effect of an Artificial Intelligence Support System. Radiology 2019;290:305-14. [Crossref] [PubMed]
- Erickson BJ, Korfiatis P, Akkus Z, Kline TL. Machine Learning for Medical Imaging. Radiographics 2017;37:505-15. [Crossref] [PubMed]


