
Practical methods for segmentation and calculation of brain volume and intracranial volume: a guide and comparison
Introduction
Accurate segmentation and calculation of total brain volume (BV) and intracranial volume (ICV) (further—volumetry) may serve research studies in neuroscience. Since these volumetric measurements can be used to predict clinical outcomes in space-occupying pathology such as stroke or tumor, such data can be invaluable to clinicians as well. For example, in patients with malignant middle cerebral artery infarcts (a space-occupying pathology), the ability to accommodate swelling in the cranium has prognostic implications. Over time, growing cerebral edema leads to transtentorial herniation and brainstem compression with a high mortality rate (1). Often the survivors are left severely disabled (2). In theory, a patient with a smaller brain will have more room to accommodate the mass effect from the edema than a patient with the same size infarct and intracranial space, but larger brain (3). Therefore, knowing BV and ICV is important in research studies that model spatial compensation reserve as it relates to pathological volumes.
Manual segmentation of BV and ICV is extremely time consuming. There is a relative lack of published broad recommendations for automated volumetry, especially for users without expertise in computer science and for settings with limited resources or when neuroimaging quality is suboptimal due to clinical circumstances. In addition to a relative lack of instructions in medical literature for how to collect these data, there also exists a knowledge gap regarding the accuracy and validation of existing volumetry methods (4). Therefore, we recognize the need to decrease the barrier to entry for research and clinical groups to perform volumetric cranial imaging analysis using free and reliable software tools while providing the comparative guidance and instructions that can be used efficiently without special programming knowledge. Though we also recognize that many automated and manual software exist for volumetric imaging analysis of the brain and skull, we have narrowed our focus to include free non-commercial platforms which are cited in neuroscience literature and are easy to use—FreeSurfer, 3D Slicer, and volBrain.
FreeSurfer
FreeSurfer is a free, open-source software developed in 2012 by scientists at the Laboratory for Computational Engineering at the Athinoula A. Martinos Center for Biomedical Engineering (Charlestown, MA, USA) and is available at https://surfer.nmr.mgh.harvard.edu for Linux and Mac OS platforms (5). FreeSurfer automatically performs all the steps of brain segmentation along with labeling of anatomical brain structures, and statistical volumetry analysis using T1 magnetic resonance imaging (MRI) scans (4-6). The FreeSurfer pipeline performs segmentation automatically from a command line (6). Of existing free tools, we chose FreeSurfer because it is a widely used research tool for volumetric analysis in neurodegenerative diseases (7) and is a common standard to which other clinical software are compared (4,8-10). Though FreeSurfer is easy to use, it requires downloading and installation which can be a complicated step in implementing the software package. It also requires high computational power and is not certified for clinical use (4). It can only utilize T1-MRI scans of the brain.
3D Slicer
3D Slicer (Slicer) is another free, open-source software originally developed between the Massachusetts Institute of Technology Artificial Intelligence Laboratory, the Surgical Navigation and Robotics Laboratory, and Surgical Planning Laboratory at the Brigham and Women’s Hospital (Boston, MA, USA) in 1998–1999 (11-18). Slicer is available at https://www.slicer.org for Linux, Mac, and Windows. Slicer is used for biomedical image informatics, processing, and visualization and has been under continuous development by The Slicer Community since conception. We chose to include Slicer in this study because it is a relatively versatile, user friendly software with a robust online discussion forum for user help. In contrast to FreeSurfer, Slicer can use a variety of imaging modalities, including MRI and computed tomography (CT) scans of the brain and other organs. It provides skull stripping, thresholding, and manual editing tools through a simple graphical user interface (GUI). These tools often require additional modules or “extensions”, many of which are made by third parties. Slicer does not provide the degree of granular segmentation and structure labeling that FreeSurfer and volBrain do. However, Slicer gives easier control to the user for fine-tuning the output of the software.
volBrain
volBrain is a free, online platform developed by José V. Manjón (Valencia Polytechnic University, Valencia, Spain) and Pierrick Coupé (University of Bordeaux, Bordeaux, France) and is available at (https://volbrain.upv.es). The volBrain pipeline provides a fully automated method of obtaining BV and ICV from T1 MRI data and does not require any installation, configuration, or training (19). volBrain also conveniently provides easy instructions on its website interface for using the pipeline. To use volBrain, the user must register for access to the system and upload a single anonymized T1 MRI study in neuroimaging informatics technology initiative (NIFTI) format (other compression formats also accepted). The jobs uploaded to the volBrain web server are then distributed across multiple available machines, reducing the computational load. volBrain then generates CSV and PDF reports containing results, which are then emailed to the user. In some brain regions and study populations, volBrain results are comparable to, if not better than, state of the art methods such as FreeSurfer (19).
This study was designed with the intention to investigate the inter-method reliability, accuracy, and practicality of using these volumetry tools. We present the following article in accordance with the MDAR checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-958/rc).
Methods
Subjects
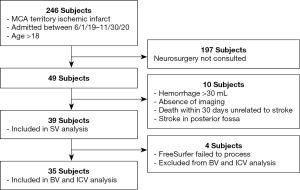
A cohort of patients with space-occupying pathology was chosen to test the performance of volumetry tools in a clinical setting. Our cohort (determined retrospectively) included 39 patients (subjects) treated at a university hospital between June 1st, 2019 and November 30th, 2020 with ischemic middle cerebral artery territory brain infarcts in the acute stage. MRI and CT scans were obtained within 72 h of the last known well time as part of a routine ischemic stroke workup. Subject characteristics can be found in Table 1 and the inclusion/exclusion criteria can be found in Figure 1. All scans were acquired in digital imaging and communications in medicine (DICOM) format. Radiographic scans are completely anonymized and there are no patient identifiers mentioned within the text. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board at the University of Arkansas for Medical Sciences and individual consent for this retrospective analysis was waived.
Table 1
| Gender | Number of subjects (percentage of total) | Average age (years) ± standard deviation |
|---|---|---|
| Male | 16 (45.7%) | 60.06±15.60 |
| Female | 19 (54.3%) | 58.84±17.50 |
BV, brain volume; ICV, intracranial volume.
Volumetry
Volumetry from CT/MRI scans was accomplished using 3D Slicer (v. 4.11.0), FreeSurfer (v. 7.1.1), and volBrain (v. 1.0).
We employed FreeSurfer for T1 MRI analysis using the recon-all function. The graphic representations of segmented volumes were imported into the FreeView visualization module for visual inspection. See Figure 2A,2B for examples of FreeSurfer brain segmentations.
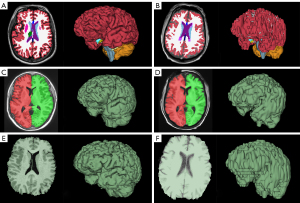
We used the volBrain pipeline for T1 MRI analysis. We extracted the Brain (BV) and Intracranial Cavity (ICV) from the PDF data report and visually checked the Intracranial cavity extraction and Macrostructures result images (see Figure 2C,2D for examples).
We used 3D Slicer for T1 MRI, non-contrast head CT, and head CT angiogram (CTA) analysis. Slicer segmentations of BV and ICV were created using a thresholding tool and specialized segmentation extensions such as SwissSkullStripper (20) and Wrap Solidify (21) with minimal use of manual tools such as painting and erasing. In our study, Slicer segmentations of BV and ICV were performed by an investigator (TH) who was blinded to the results from FreeSurfer (performed by different investigator; JH) and volBrain (performed later). Visual inspection for accuracy was performed in the Slicer viewer (see Figure 2E,2F for examples).
Each subject in our cohort had specific pathology (ischemic stroke). In each case, we measured the size of the stroke (stroke volumes, SV) on B1000 diffusion-weighted MRI scans (see Appendix 1). Our data processing resulted in segmentation and calculations of BV, ICV, SV for each subject.
For every volumetry tool used in this study, we exported a sample segmentation into Slicer for 3D visualization and to show large scale differences in accuracy between methods and scan quality (Figures 2,3). The exception was ICV segmentation using FreeSurfer. At the time of this writing, we are aware of no graphic export tools in the current FreeSurfer version for visual verification of intracranial segmentations.
Data analysis
Results were analyzed in StatPlus in Microsoft Excel. Data analysis included coefficient of determination (R2) with 95% confidence interval (CI) and P value between segmentation methods for BV and ICV. Table 2 provides R2 values that can be detected with the given sample sizes at 80% and 90% power. Bland-Altman (B-A) plots were generated to evaluate agreement between methods. In the B-A plots (Figures 4,5), the solid horizontal line represents the mean difference between the two methods. The middle-dashed lines represent the 95% CI of ±1.96 standard errors of the mean difference between methods. The outside solid lines represent the limits of normal, which are ±1.96 standard deviations from the mean.
Table 2
| Power | n=39 | n=35 | n=22 | n=13 |
|---|---|---|---|---|
| 80% | 0.1862 | 0.2058 | 0.3120 | 0.4846 |
| 90% | 0.2389 | 0.2627 | 0.3883 | 0.5776 |
Practical user experience
A multifaceted comparison of volumetry tools from a practical user perspective was performed to compile a list of distinguishing features which has been summarized in Table 3.
Table 3
| Software | FreeSurfer | 3D Slicer | volBrain |
|---|---|---|---|
| Authors and developers | Laboratory for Computational Engineering at the Athinoula A. Martinos Center for Biomedical Engineering (Charlestown, MA, USA) | Massachusetts Institute of Technology Artificial Intelligence Laboratory, the Surgical Navigation and Robotics Laboratory, and Surgical Planning Laboratory at the Brigham and Women’s Hospital (Boston, MA, USA) | José V. Manjón (Valencia Polytechnic University, Valencia, Spain) and Pierrick Coupé (University of Bordeaux, Bordeaux, France) |
| Internet address | https://surfer.nmr.mgh.harvard.edu | https://www.slicer.org | https://volbrain.upv.es |
| Requires download and installation | Yes | Yes | No (i.e., the tool is online) |
| OS platform | Linux, Mac OS | Linux, Mac OS, Microsoft Windows | Not applicable |
| Input image modalities | T1-MRI | T1-MRI, DWI-MRI, CT, CTA | T1-MRI |
| Input file type | DICOM | DICOM | NIFTI |
| Need for interaction with human user | Minimal | Significant | Minimal |
| Need for software add-ons | No | Yes | No |
| Processing speed | Slower | Fast | Fast |
| Brain anatomical structure labeling | Automatic | Not available | Automatic |
| Volumetry of distinct pathological imaging findings | Not available | Semi-automated | Not available |
MRI, magnetic resonance imaging; DWI, diffusion weighted imaging; CT, computed tomography; CTA, computed tomography angiography; DICOM, digital imaging and communications in medicine; NIFTI, neuroimaging informatics technology initiative.
Instructions
Based on our explorations we were able to consolidate the workflows into detailed instructions for FreeSurfer and Slicer in the Appendix 1. Our aim in providing these instructions are to increase the accessibility for clinical groups regarding volumetric analysis. Highly helpful instructions for using volBrain are included on the volBrain website with video tutorials (https://volbrain.upv.es/instructions.php).
Results
Processed scans
In this study, we analyze the inter-method reliability of FreeSurfer, Slicer, and volBrain using real-world scans obtained in a clinical setting where subjects could have been critically ill and/or uncooperative, requiring faster diagnostic scans with MRI slice intervals sometimes greater than 1.1 mm. Therefore, there were two major categories of MRI in this study: 14 subjects had high quality sagittal MRIs with 1.1 mm slice intervals (3D scans), 25 subjects had lower quality axial MRIs with 3–6 mm slice intervals (2D scans); 1 subject (7.1%) in the 3D group and 3 subjects (12.0%) in 2D group failed to complete the recon-all function on FreeSurfer for unknown reasons and were excluded from analysis. Three of these 4 subjects also failed to complete the volBrain pipeline. To keep the sample size consistent, the remaining subject was also removed from all other analysis. This left 13 3D subjects and 22 2D subjects for analysis.
For every subject, segmentation of the intracranial space on Slicer was performed using high resolution CT scans (both non-contrast and CTAs, with slice thickness 1 mm or less). This method was 100% successful and resulted in a visually accurate segmentation of ICV for every subject.
Volumetry
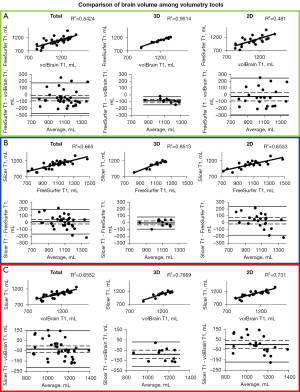
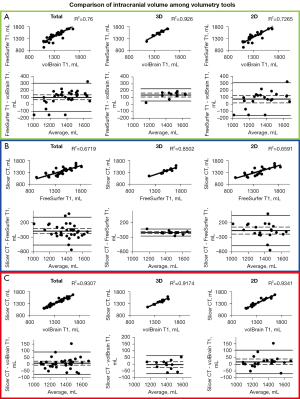
Results of the BV correlations between each method are displayed in Figure 4. ICV correlation results are displayed in Figure 5. Coefficient of determination, 95% CI, and P value for each correlation are shown in Table 4. Table 2 shows R2 values that can be detected with the given sample sizes at 80% and 90% power. For almost every comparison between methods, our group of subjects with 3D scans showed higher BV and ICV correlations compared to our group of subjects with 2D scans. All correlations were statistically significant (P≤0.05).
Table 4
| Quality | Comparison | R2 | 95% CI | P value |
|---|---|---|---|---|
| ICV | ||||
| Total | Slicer_CT vs. FreeSurfer | 0.6719 | 0.4543 to 0.7743 | <0.0001 |
| FreeSurfer vs. volBrain | 0.7600 | 0.5867 to 0.8355 | <0.0001 | |
| Slicer_CT vs. volBrain | 0.9307 | 0.8739 to 0.9522 | <0.0001 | |
| 3D | Slicer_CT vs. FreeSurfer | 0.8502 | 0.5224 to 0.9041 | <0.0001 |
| FreeSurfer vs. volBrain | 0.926 | 0.7231 to 0.9484 | <0.0001 | |
| Slicer_CT vs. volBrain | 0.9174 | 0.7980 to 0.9632 | <0.0001 | |
| 2D | Slicer_CT vs. FreeSurfer | 0.6591 | 0.3462 to 0.7818 | <0.0001 |
| FreeSurfer vs. volBrain | 0.7265 | 0.4532 to 0.8262 | <0.0001 | |
| Slicer_CT vs. volBrain | 0.9341 | 0.8525 to 0.9578 | <0.0001 | |
| BV | ||||
| Total | Slicer_T1 vs. FreeSurfer | 0.6650 | 0.4351 to 0.7648 | <0.0001 |
| FreeSurfer vs. volBrain | 0.5424 | 0.2766 to 0.6770 | <0.0001 | |
| Slicer_T1 vs. volBrain | 0.6552 | 0.4316 to 0.7631 | <0.0001 | |
| 3D | Slicer_T1 vs. FreeSurfer | 0.8513 | 0.5640 to 0.9140 | <0.0001 |
| FreeSurfer vs. volBrain | 0.9514 | 0.8410 to 0.9713 | <0.0001 | |
| Slicer_T1 vs. volBrain | 0.7669 | 0.3980 to 0.8712 | <0.0001 | |
| 2D | Slicer_T1 vs. FreeSurfer | 0.6553 | 0.3325 to 0.7756 | <0.0001 |
| FreeSurfer vs. volBrain | 0.481 | 0.1337 to 0.6610 | <0.0004 | |
| Slicer_T1 vs. volBrain | 0.7310 | 0.4605 to 0.8291 | <0.0001 | |
The methods being compared are described in the Comparison column of the table. P values are from tests comparing the observed R2 value to 0 (zero; no association). CI, confidence intervals; ICV, intracranial volume; Slicer, 3D Slicer; CT, computed tomography; BV, brain volume; T1, T1-weighted magnetic resonance imaging.
BV analysis
FreeSurfer and volBrain with 3D scans show the best correlation (R2=0.9514) and agreement for all BV methods we compared (Figure 4A). In Figure 2 we display the visual difference in BV segmentation between two subjects (left-3D MRI; right-2D MRI). It is clear that using 3D MRIs on all three of our reported platforms produces more accurate BV segmentations. This is most evident in the differences in quality of the three-dimensional segmentations between Figure 2A,2C,2E (3D scans) and Figure 2B,2D,2F (2D scans).
Interestingly, there is a relatively wide discrepancy in correlation between 3D and 2D scans in our FreeSurfer-volBrain comparison (Figure 4A). FreeSurfer-Slicer and volBrain-Slicer comparisons showed a smaller difference between 3D and 2D scans in both cases (Figure 4B,4C respectively). We suggest that good inter-method reliability for BV can only be shown between FreeSurfer and volBrain using 3D T1 MRIs. However, compared to FreeSurfer and Slicer, volBrain might provide a better segmentation when 2D scans are used. By comparing Figure 2D (volBrain) with Figure 2B (FreeSurfer) and Figure 2F (Slicer), volBrain provides better large-scale accuracy when using 2D scans.
ICV analysis
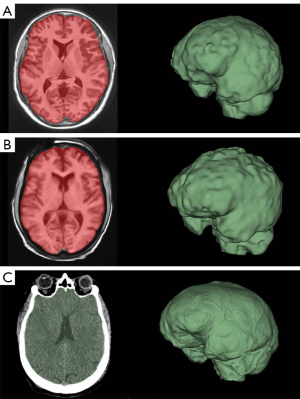
FreeSurfer ICV segmentations correlate well with Slicer and volBrain when using 3D scans (R2>0.850 in both cases) (Figure 5A,5B). However, though these are strong correlations, our B-A plots indicate that FreeSurfer consistently overestimated ICV compared to Slicer and volBrain (Figure 5A,5B). Using 3D scans, FreeSurfer overestimates ICV by an average of 143.43 mL (10.40%) compared to volBrain (T1) (P<0.0001) (95% CI: 115.72–171.13) and 148.09 mL (10.78%) compared to Slicer (CT) (P<0.0001) (95% CI: 111.14–185.03). Figure 3 does not include a three-dimensional segmentation for ICV using FreeSurfer, because FreeSurfer does not provide an ICV segmentation.
The Slicer method of segmenting the intracranial space is not split into 3D and 2D scans because CT scans were used; however, we compared this method with FreeSurfer and volBrain for each respective subject (Figure 5B,5C). ICV segmentations from CT scans using Slicer were visually accurate (Figure 3C). We also show that volBrain creates convincing segmentations of ICV (Figure 3A,3B), which correlate and agree very well with this Slicer method (R2=0.9307) regardless of scan quality (Figure 5C). Due to high correlation, agreement, and visual accuracy, we report excellent inter-method reliability between volBrain (T1) and Slicer (CT) segmentations of ICV.
Discussion
Though there is no study that confirms the accuracy of these methods compared to “ground truth”, there are some important considerations in acquiring an accurate segmentation. A method’s accuracy can be visually assessed by its ability to differentiate a region of interest from other parts of the scan (small-scale) and its ability to create an acceptable segmentation (large-scale), which is inherently limited by scan quality and slice interval (for examples, see the segmentations overlaid on T1 axial MRI sections and the differences in segmentation quality between 2D and 3D scans in Figures 2,3).
Visualization of segmentations
Though FreeSurfer provides a visualization GUI, Freeview, which allows for checking of segmentations in orthogonal planes, it does not provide a user-friendly method of assessing the large-scale accuracy of the total segmentation in three-dimensional view.
An advantage of using Slicer in this study is its ability for quality checking of segmentations in three dimensions. Not only were we able to check our Slicer-generated segmentations using the Slicer viewer, but we also report methods of importing FreeSurfer and volBrain results into this viewer. In this study, these methods were employed to generate Figures 2,3. Such simple methods of visualization can be invaluable to a research group with limited expertise in computer science (see Appendix 1 for more information).
FreeSurfer
Though some studies have raised questions about the accuracy of FreeSurfer volume calculations (22,23), FreeSurfer is often used as a standard against which other automated volumetry software are often compared (4,8-10). Using the described methods, the results of BV calculations in our study are similar between FreeSurfer, Slicer, and volBrain when good quality 3D T1-MRI scans are used. However, the inter-method reliability between these methods is lesser when using 2D T1-MRIs with higher slice thickness. It is important to note, that the FreeSurfer website does recommend using MRI data with thickness no larger than 1.5 mm (24). This means that any research group interested in reliable volumetric analysis of the brain must ensure that high-quality 3D T1 MRIs are acquired. This may be a limiting factor for volumetric imaging analysis in certain clinical settings.
It has been suggested by the developers of FreeSurfer that it would be best to calculate ICV by directly counting voxels; however, the difficulty of distinguishing between cortical bone and CSF on T1 images due to similar MR intensity prevents them from doing so (25). Instead, FreeSurfer utilizes a relationship between the ICV and the BV following the approach of Buckner et al. (26) and scales the BV to correspond to that of a known atlas image, and then uses the corresponding scaling factor to estimate ICV (22,23). According to a study by Klasson et al. (22), this FreeSurfer method of estimating ICV is biased by total BV. Another study published in 2015 showed that FreeSurfer calculates an ICV which correlates to manual segmentation with an R2=0.801 (23). In this study, though FreeSurfer ICV showed high correlation with other methods, it appears to overestimate the ICV by a significant margin compared to other methods which show strong agreement. These overestimations and the inability to check the FreeSurfer ICV segmentation cast significant doubt upon the accuracy of FreeSurfer ICV calculations.
FreeSurfer offers an advantage by providing a segmentation and volume for every anatomical brain structure (Figure 2A,2B), which can be useful for further analysis if needed.
3D Slicer
Slicer consistently provides a visually convincing segmentation of the total intracranial space when high resolution CT scans are used—in contrast to FreeSurfer which only provides an estimated ICV without a visual segmentation, thus precluding visual inspection for accuracy. If high resolution non-contrast CT is not available, a modern CTA can be used for the above-described method of ICV segmentation but may require more manual editing (see instructions in Appendix 1 for further information). Notably, we identified a commonly encountered ICV segmentation error using CT scans (Figure 6A), but this was overcome using the procedure we provide in the Appendix 1.
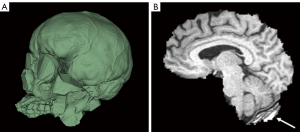
ICV from CT is decidedly accurate compared to MRI because of the contrast between tissue and bone and the minimal geometric distortions inherent to CT technology as opposed to MRI (27). We suggest comparing this method to other commercial volumetry software in future studies and potentially using Slicer and modern volumetric CT scans as a method of choice for accurate ICV calculations.
BV segmentation using Slicer with T1 MRIs requires skull stripping and thresholding of the brain. In our experience, this is a less accurate and more tedious segmentation method compared to FreeSurfer and volBrain which are fully automated. At times, Slicer removes part of the brain when stripping the skull. Other times, Slicer leaves part of the skull “un-stripped” (Figure 6B). These errors can be fixed using manual tools, but the process can be tedious in many cases (see Appendix 1).
volBrain
In this study, we found volBrain to be the most efficient and user-friendly method we explored. The instructions found online are easy and self-explanatory, which overcomes some of the most difficult aspects of implementing volumetric imaging analysis. To convert the subject DICOM to NIFTI, the user must simply install dcm2nii and “click and drag” the DICOM file into the GUI. volBrain results include screenshots in axial, coronal, and sagittal planes (examples of axial screenshots are included in Figure 2C,2D). Additionally, volBrain provides processed NIFTI files on its website which can be loaded as data into Slicer (see Appendix 1) or MRIcron (volBrain recommended) for 3D viewing. volBrain results also include an analysis of the asymmetry of the brain, the percentage of the intracranial cavity occupied by brain tissue (see Intracranial reserve analysis in the Appendix 1), and subcortical structure labeling.
In Figure 2C,2D, we show that volBrain provides sufficiently accurate visual segmentations of the brain even when 2D scans are used. Similarly, Figure 3A,3B show that volBrain produces good intracranial cavity segmentations regardless of scan quality. However, these segmentations are not as accurate as those derived from Slicer using CT scans (Figure 3C). Nevertheless, of the methods we tested with T1 MRI data, volBrain provides the most reliable segmentations of BV and ICV if T1 MRI quality is limited. In contrast to FreeSurfer, there is no explicit recommendation regarding T1 MRI slice thickness on the volBrain website.
Supra- and infratentorial volume
Another advantage to FreeSurfer and volBrain are their ability to automatedly separate the supratentorial and infratentorial brain compartments. There are clinical situations where interplay between supratentorial and infratentorial volumes becomes a critical factor in determining spatial and temporal dynamics of brain shift (which are upward and downward transtentorial brain herniations). A few common clinical examples are posterior fossa tumors, malignant middle cerebral artery strokes, supratentorial intra- and extra-axial hematomas, and obstructive hydrocephalus. Therefore, it may be important for investigators studying intracranial space-occupying lesions to know accurate supra- and infratentorial compartment volumes.
Since the tentorium cerebelli is dome-shaped, segmenting these volumes using Slicer would require tedious manual correction in three dimensions. If investigators require an estimation of BV above or below the tentorium cerebelli, FreeSurfer consistently provides visually accurate segmentations using high quality scans. volBrain can also provide an estimation of the supratentorial volume if the user subtracts the cerebellum and brainstem (both provided in result file) from the total BV. Therefore, it may be practical to use FreeSurfer or volBrain to calculate these volumes in a study involving space-occupying pathology.
Workflow efficiency
It is important to consider the efficiency of a software in clinical practice or research setting. FreeSurfer data processing is time consuming and requires substantial computing power (FreeSurfer processing took roughly 6 h on our PC workstation running Ubuntu 20.04.1 with an AMD Ryzen 5 1600 processor and 32 GB of DDR3 memory), which may not suit well to time-sensitive clinical practice today. Its practicality will likely improve following Moore’s law, which observes that transistor density on computer chips roughly doubles each year.
Slicer was significantly faster than FreeSurfer in our experience and could provide ICV and BV segmentations of one subject in ≈30 minutes on our workstation. Slicer, however, requires more user interaction through a GUI—in contrast to FreeSurfer which performs roughly 6 h of unsupervised calculations after typing a few simple commands.
volBrain was the most efficient and easy to use. Due to online nature of volBrain, it avoids the need for a dedicated workstation but, at the same time, requires imaging study anonymization in order to comply with patient confidentiality. In addition, volBrain cannot directly process DICOM format images and requires a simple file conversion using a free program (dcm2nii) followed by an upload to the volBrain website. Unlike FreeSurfer, volBrain provides quicker processing time and allows for multiple subject processing at once (up to 10 scans daily with the current version) (19).
Conclusions
This study provides results of volumetric imaging analysis of the brain and intracranial space in a clinically representative cohort of subjects. All three tools—3D Slicer, FreeSurfer, and volBrain—are free, reliable, require no complex programming, but still have certain limitations and significant differences.
Using high-resolution non-contrast CT scans and CTAs, Slicer provides a simple and visually accurate method of segmenting the intracranial cavity. volBrain also provides a fully automated, visually accurate, and easy method, which agrees with Slicer. Both of these methods may be more trustworthy than T1 MRI-derived FreeSurfer ICV calculations, which may overestimate the true value even when using high-quality 3D T1 scans. For BV, all three of our methods showed good correlation and agreement, with visually accurate segmentations when using 3D scans. volBrain may provide a better segmentation of the brain when T1 MRI quality is limited.
Slicer allows for processing of multiple types of scans, while FreeSurfer and volBrain only use T1 MRI scans. Slicer also allows for easy manual editing of the output and 3D visualization of the results, while FreeSurfer and volBrain require complex procedures to edit and visualize their segmentations in three dimensions. However, unlike Slicer, both FreeSurfer and volBrain allow for fully automated BV and ICV calculations.
Our study highlights the importance of acquiring high quality volumetric (3D) T1 MRI scans if these methods of segmentation are to be implemented for research or integrated into clinical workflow. volBrain provides the easiest method for segmentation, whereas Slicer provides the most user involved segmentation experience. We provide a practical guide to decrease the barrier to entry for research and clinical groups to perform volumetric imaging analysis with these methods. The readers should be able to select the right volumetry tool based on our study findings and then follow our step-by-step instructions to accomplish specific volumetry tasks.
Acknowledgments
We would like to thank Kim Gates for providing our subject list with the required radiographic scans for our analysis. We also thank Andriy Fedorov and Andras Lasso for their helpful answers to our questions about FreeSurfer and 3D Slicer, respectively. Also, thank you to Horace J. Spencer III from the University of Arkansas for Medical Sciences, Department of Biostatistics in the College of Public Health for his valuable advice on statistical analysis.
Funding: This work was supported by the Fund to Cure Stroke Grant from the University of Arkansas for Medical Sciences Department of Neurology in the College of Medicine, which provided student stipends to TH, DB, JH, and HS ($3,000 total). Therefore, the authors would like to thank Dr. William Culp and Dr. Robert Skinner, for the review and acceptance of our grant proposal.
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-958/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-21-958/coif). VP reports that he is a consultant for Medtronic. Consulting fees are paid directly to University of Arkansas for Medical Sciences. VP did not receive any funds. Medtronic Navigation provided equipment (laptop computer) and software (StealthViz) for the study. VP owns Medtronic Plc (MDT) stocks. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Radiographic scans are completely anonymized and there are no patient identifiers mentioned within the text. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional board of the University of Arkansas for Medical Sciences and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hacke W, Schwab S, Horn M, Spranger M, De Georgia M, von Kummer R. 'Malignant' middle cerebral artery territory infarction: clinical course and prognostic signs. Arch Neurol 1996;53:309-15. [Crossref] [PubMed]
- Hofmeijer J, Kappelle LJ, Algra A, Amelink GJ, van Gijn J, van der Worp HBHAMLET investigators. Surgical decompression for space-occupying cerebral infarction (the Hemicraniectomy After Middle Cerebral Artery infarction with Life-threatening Edema Trial [HAMLET]): a multicentre, open, randomised trial. Lancet Neurol 2009;8:326-33. [Crossref] [PubMed]
- Johnson RD, Maartens NF, Teddy PJ. Decompressive craniectomy for malignant middle cerebral artery infarction: evidence and controversies. J Clin Neurosci 2011;18:1018-22. [Crossref] [PubMed]
- Pemberton HG, Zaki LAM, Goodkin O, Das RK, Steketee RME, Barkhof F, Vernooij MW. Technical and clinical validation of commercial automated volumetric MRI tools for dementia diagnosis-a systematic review. Neuroradiology 2021;63:1773-89. [Crossref] [PubMed]
- Fischl B. FreeSurfer. Neuroimage 2012;62:774-81. [Crossref] [PubMed]
- Reuter M, Schmansky NJ, Rosas HD, Fischl B. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage 2012;61:1402-18. [Crossref] [PubMed]
- Vernooij MW, Pizzini FB, Schmidt R, Smits M, Yousry TA, Bargallo N, Frisoni GB, Haller S, Barkhof F. Dementia imaging in clinical practice: a European-wide survey of 193 centres and conclusions by the ESNR working group. Neuroradiology 2019;61:633-42. [Crossref] [PubMed]
- Ochs AL, Ross DE, Zannoni MD, Abildskov TJ, Bigler EDAlzheimer's Disease Neuroimaging Initiative. Comparison of Automated Brain Volume Measures obtained with NeuroQuant and FreeSurfer. J Neuroimaging 2015;25:721-7. [Crossref] [PubMed]
- Reid MW, Hannemann NP, York GE, Ritter JL, Kini JA, Lewis JD, Sherman PM, Velez CS, Drennon AM, Bolzenius JD, Tate DF. Comparing Two Processing Pipelines to Measure Subcortical and Cortical Volumes in Patients with and without Mild Traumatic Brain Injury. J Neuroimaging 2017;27:365-71. [Crossref] [PubMed]
- Ross DE, Ochs AL, Tate DF, Tokac U, Seabaugh J, Abildskov TJ, Bigler ED. High correlations between MRI brain volume measurements based on NeuroQuant® and FreeSurfer. Psychiatry Res Neuroimaging 2018;278:69-76. [Crossref] [PubMed]
- Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In: Jolesz FA. editor. Intraoperative Imaging and Image-Guided Therapy. New York, NY, USA: Springer New York, 2014:277-89.
- Kikinis R, Pieper S, Fillion-Robin JC. 3D Slicer. 4.10.2. 2019.
- Kapur T, Pieper S, Fedorov A, Fillion-Robin JC, Halle M, O'Donnell L, et al. Increasing the impact of medical image computing using community-based open-access hackathons: The NA-MIC and 3D Slicer experience. Med Image Anal 2016;33:176-80. [Crossref] [PubMed]
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the medical image computing community. 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro, 2006. Arlington, VA, USA: IEEE, 2006.
- Pieper S, Halle M, Kikinis R. 3D Slicer. 2004 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821). Arlington, VA, USA: IEEE, 2004.
- Gering DT, Nabavi A, Kikinis R, Hata N, O'Donnell LJ, Grimson WE, Jolesz FA, Black PM, Wells WM 3rd. An integrated visualization system for surgical planning and guidance using image fusion and an open MR. J Magn Reson Imaging 2001;13:967-75. [Crossref] [PubMed]
- Gering DT, Nabavi A, Kikinis R, Grimson WEL, Hata N, Everett P, Jolesz F, Wells WM. editors. An Integrated Visualization System for Surgical Planning and Guidance Using Image Fusion and Interventional Imaging. Berlin, Heidelberg: Springer Berlin Heidelberg, 1999.
- Manjón JV, Coupé P. volBrain: An Online MRI Brain Volumetry System. Front Neuroinform 2016;10:30. [Crossref] [PubMed]
- Lorensen B, Bauer S, Fejes T, Reyes M, Gelas A. Documentation/Nightly/Modules/SwissSkullStripper. Slicer Wiki. Available online: https://www.slicer.org/wiki/Documentation/Nightly/Modules/SwissSkullStripper
- Weidert S, Andress S, Linhart C, Suero EM, Greiner A, Böcker W, Kammerlander C, Becker CA. 3D printing method for next-day acetabular fracture surgery using a surface filtering pipeline: feasibility and 1-year clinical results. Int J Comput Assist Radiol Surg 2020;15:565-75. [Crossref] [PubMed]
- Klasson N, Olsson E, Eckerström C, Malmgren H, Wallin A. Estimated intracranial volume from FreeSurfer is biased by total brain volume. Eur Radiol Exp 2018;2:24. [Crossref]
- Malone IB, Leung KK, Clegg S, Barnes J, Whitwell JL, Ashburner J, Fox NC, Ridgway GR. Accurate automatic estimation of total intracranial volume: a nuisance variable with less nuisance. Neuroimage 2015;104:366-72. [Crossref] [PubMed]
- Busa E, Schmansky N. FreeSurfer Beginners Guide. FreeSurferWiki. Available online: https://surfer.nmr.mgh.harvard.edu/fswiki/FreeSurferBeginnersGuide
- Schmansky N, Reuter M. eTIV - estimated Total Intracranial Volume, aka ICV. FreeSurferWiki. Available online: https://surfer.nmr.mgh.harvard.edu/fswiki/eTIV
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage 2004;23:724-38. [Crossref] [PubMed]
- Sumanaweera T, Glover G, Song S, Adler J, Napel S. Quantifying MRI geometric distortion in tissue. Magn Reson Med 1994;31:40-7. [Crossref] [PubMed]


