
Development and validation of bone-suppressed deep learning classification of COVID-19 presentation in chest radiographs
Introduction
Coronavirus Disease 2019 (COVID-19) is a pandemic disease, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which can result in alveolar damage and respiratory failure (1) and has a fatality rate of 2% (2). New variants of SARS-CoV-2 are associated with increased transmissibility (3), decreased vaccine effectiveness and increased hospital admissions rates (4) than previous variants. In general, the clinical standard for confirmed diagnosis of the disease is reverse-transcriptase polymerase chain reaction (RT-PCR) testing of nasopharyngeal swabs (1,5), with radiology generally being used as a fast auxiliary diagnostic methodology alongside RT-PCR testing (6). Chest X-ray (CXR) imaging is an economical and accessible method for imaging the patient lung (7,8), with mobile systems capable of imaging at patient bedside with less risk of cross-contamination. Rapid and accurate methods to diagnose COVID-19 from CXRs could assist healthcare systems in controlling the spread of the disease.
Deep learning methods—in particular, convolutional neural networks—have shown great success (9) in general image processing applications (10). Such methods have recently been developed as a potential step towards automated diagnosis of COVID-19 from CXRs (8,11-15). Many efforts have been made to improve the diagnostic accuracy of deep learning classification methods, including but not limited to architectural improvements (16) and multi-modality classification (17). Concurrently, many methods for medical image enhancement have been developed, including those for CXR bone suppression (18-25). In this context, bone suppression is the removal of the bone shadows caused by increased attenuation of X-rays in the ribs and clavicles. Bone suppression of CXRs has been demonstrated to improve automated classification and manual diagnosis of lung pathologies (19,26-28). Manji et al. (28) demonstrated that radiologists diagnosed lung nodules in bone-free images acquired from dual energy subtraction radiography faster and more accurately than from standard CXRs. However, dual energy subtraction radiography requires specialized hardware, which may be unavailable for many institutions, and increased patient dose (25). Hence there is interest in the development and use of software-based bone-suppression techniques that could be applied to standard CXRs. Freedman et al. (27) demonstrated that radiologists performed significantly better in detecting lung nodules in CXRs when aided with proprietary visualization software that suppressed the clavicle and ribs, filtered noise and adjusted contrast. Baltruschat et al. (26) found that an ensemble of lung pathology classification models trained on software-based bone-suppressed and lung-cropped images resulted in improved performance when identifying lung pathologies in CXRs compared to an ensemble trained on non-suppressed images. Rajaraman et al. (19) demonstrated that deep learning-based bone suppression of CXRs improved the sensitivity, specificity and accuracy of a tuberculosis classification model significantly by 7%, 4% and 5% respectively for an internal test set.
In this retrospective study, we hypothesize that bone suppression could be used to improve the classification of COVID-19 phenomena in CXRs. The study develops and validates a classification workflow, in which input radiographs are bone-suppressed before classification. The major contributions from this study include: (I) application of bone suppression to improve classification of COVID-19 phenomena; (II) use of an external test set to assess the impact of bone suppression on classification models trained on non-suppressed and bone-suppressed CXRs. This study provides insight into the possibility of further improving deep learning classifier performance via image enhancement. We present the following article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-21-791/rc).
Methods
Overview
The workflow for the study is as follows: first, input images were resized using nearest-neighbor interpolation; second, bone suppression was applied; third, classification was performed. Bone suppression was performed with two methods described in Section Bone suppression of Methods. The methods [‘Gusarev’ (18) and ‘Rajaraman’ (19)] were trained from chest radiographs. These methods are feedforward neural networks, and were chosen for training stability in preliminary work. To examine the effect of bone suppression on image classification performance, two deep learning-based image classification methods [the pre-trained ‘COVID-Net CXR2’ (11) method and the ‘VGG16- Modified’ (19) method] were implemented and used as described in Section Classification of Methods, to classify the images as “COVID” or “Non-COVID”. The performance of the classification models for non-suppressed test CXRs was compared to that for bone-suppressed test CXRs. All bone suppression and classification models were trained and tested on a workstation with a RTX2080Ti GPU (NVIDIA, Santa Clara, CA, USA) using CUDA 11.0. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and with ethics approval from The Hong Kong Polytechnic University Faculty Research Committee (No. HSEARS20201107001). Since only anonymized data was acquired in this study, individual consent for this retrospective study was waived.
Datasets
This retrospective study makes use of anonymized image datasets collected previously for other purposes. Frontal chest radiograph datasets were collected for the purposes of training the bone suppression methods, training the VGG16-Modified classification method, and testing the trained methods. Acquired images included both anteroposterior (AP) and posteroanterior (PA) cases. Imaging methodology included computed radiography and digital radiography from both static and mobile X-ray machines. The Gusarev and Rajaraman bone suppression methods employed in this study were trained using a publicly available dataset of PA chest radiographs (Digital Image Database) from the Japanese Society of Radiological Technology (JSRT) (29). The JSRT dataset was acquired from 13 institutions in Japan and 1 institution in the United States around 1997–1998. Juhász et al. (30) had applied in-house bone suppression algorithms to the JSRT images and publicly released the resulting dataset, which was used in this study as the bone-suppressed “target” dataset to train the bone suppression models. The JSRT and Juhász datasets had been combined and published as the X-ray Bone Shadow Suppression dataset (31) (https://www.kaggle.com/hmchuong/xray-bone-shadow-supression). To the authors’ knowledge, this is the only publicly available set of co-registered non-suppressed and bone-suppressed chest radiographs. A total of 241 non-suppressed and bone-suppressed image pairs were acquired (31), each pair derived from a unique patient, of which 148 images contained at least one lung nodule and 93 images were without nodules. To train the bone suppression models, the dataset was cleaned by removing image pairs where poor suppression (defined as observing at least 1 rib or clavicle with edges visible across the lung tissue) and image artifacts were observed. A total of 217 image pairs (131 with nodule, 86 without nodule) remained after data cleaning. Of those, 20 images (9%) were retained for internal testing; the remaining 197 images were used for 10-fold cross-validation of the bone suppression models.
Second, to train the VGG16-Modified classification model, CXRs from COVID-19 and non-COVID-19 datasets were acquired. A total of 996 CXR images of COVID-19 positive patients were extracted from the publicly available RICORD-1c dataset (32,33), consisting of PA, AP and portable acquisitions from three institutions. Data accrual began in April 2020 and ended before March 5, 2021. A total of 1,125 CXR images of patients with no lung pathology (‘normal’), and 100 CXR images of patients with non-COVID-19 pneumonia, were randomly extracted from the publicly available RSNA Pneumonia Challenge dataset [National Institute of Health Clinical Center, United States (34-36)], which contained patient data that had been acquired between 1992 and 2015. The data was split into an 8-fold cross-validation set (896 COVID, 1,025 normal), and internal testing (100 COVID, 200 non-COVID) sets. Non-COVID pneumonia data was used exclusively in the internal test set to assess the generalizability of the trained models.
Third, an external test dataset was created to test the classification models. An anonymized dataset consisting of 524 COVID-19-positive patients was obtained from Queen Elizabeth Hospital (Hong Kong, China) with ethics approval from the relevant ethics committee (KC/KE-20-0337/ER-4), and combined with a separate anonymized dataset of 564 COVID-19-negative patients that was obtained from Pamela Youde Nethersole Eastern Hospital (Hong Kong, China) with ethics approval from the relevant ethics committee (HKECREC-2020-119). After data cleaning to remove low-resolution, non-AP/PA, and non-chest radiographs, 320 COVID-19 and 518 non-COVID radiographs remained. The earliest CXR from each patient was used for external testing in this study, under the assumption that this was acquired within a day of patient admission. Potentially eligible patients were identified from hospital admissions, forming a consecutive series. Inclusion criteria for this dataset are as follows: patients with age 18 years or older, clinically diagnosed with COVID-19 (using PCR testing), had received chest radiography for COVID-19, and had radiographs acquired between 1st January 2020 and 31st December 2020. Exclusion criteria were patients who had previous pulmonary pneumonia and previous treatment for lung diseases. The proportions of potentially eligible patients who were included or excluded were unavailable due to limitations in the ethical review. Furthermore, due to limitations in the ethical review, this dataset is not available to the public.
The external test dataset for the bone suppression methods and the classification methods was created by combining the data from the two abovementioned Hong Kong hospitals, totaling 518 non-COVID-19 and 320 COVID-19 CXRs from unique patients. From the literature, a test set of this size is suitable (11,19). Known clinical information about the image datasets used in this study is summarized in Table 1.
Table 1
| Dataset | Geographic region | Male/female | Age (years, mean ± SD) | Number of non-pathological/non-COVID pneumonia/COVID-19 patients |
|---|---|---|---|---|
| RSNA Pneumonia Challenge | North America | NA | NA | 1,125/100/0 |
| RICORD-1c | North America | 208/143 | 55±18 | 0/0/996 |
| JSRT (all) | Japan | 116/125 | 58±14 | 93/NA/NA |
| JSRT (for bone suppression) | Japan | 104/113 | NA | 86/NA/NA |
| Hong Kong Hospitals (all) | Hong Kong (China) | 260/264 (QEH); 280/284 (PYNEH) |
46±22 (QEH); 57±25 (PYNEH) |
0/0/524 (QEH); 564/0/0 (PYNEH) |
| Hong Kong Hospitals (for external test set) | Hong Kong (China) | 152/168 (QEH); 265/253 (PYNEH) |
NA | 0/0/320 (QEH); 518/0/0 (PYNEH) |
The authors could not identify the age and sex information of the subjects in the RSNA Pneumonia Challenge dataset. All patients in the JSRT dataset who were not non-pathological had lung nodules. Mean and standard deviation of the patient ages in the PYNEH dataset were derived from 478 out of the 564 unique patients used. SD, standard deviation; JSRT, Japanese Society of Radiological Technology; NA, not applicable/no information found; QEH, Queen Elizabeth Hospital; PYNEH, Pamela Youde Nethersole Eastern Hospital.
Bone suppression
Classification performance on non-suppressed test images was compared to that on bone-suppressed test images. In this study, bone suppression was performed using the following published neural network architectures.
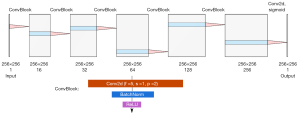
The Gusarev method was modified from that introduced by Gusarev et al. (18), and was instead implemented as shown in Figure 1. Convolutional layer weights were initialized with a normal distribution with a mean of 0 and standard deviation of 0.02, whilst batch normalization weights were initialized with the same standard deviation and a mean of 1. The training batch size was 5, and the network was trained for 400 epochs. An Adam optimizer (38) was used, with an initial learning rate of 0.001 and β1,β2 of 0.9, 0.999 respectively. The learning rate was decreased by 25% every 100 epochs. The loss LGusarev to be minimized was defined in Eq. [1]:
where MSSSIM is the multiscale structural similarity index measure (SSIM) (39) and MSE is the mean-squared error between the predicted bone-suppressed image and the target image.
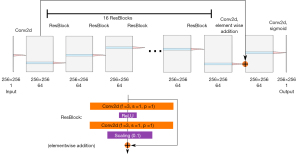
The Rajaraman method was implemented as described by Rajaraman et al. (19) and as shown in Figure 2. Convolutional layer weights were initialized with a normal distribution with a mean of 0 and standard deviation of 0.02. An Adam optimizer (38) was used, with an initial learning rate of 0.001 and β1,β2 of 0.9, 0.999 respectively, to train the model for 200 epochs with a batch size of 8. The learning rate was reduced on plateau by a factor of 0.5 after 10 epochs with no improvement (defined as a change of 0.0001 in the loss). The loss LRajaraman to be minimized was defined in Eq. [2]:
where MAE is the mean absolute error.
The Rajaraman and Gusarev bone suppression networks were trained and evaluated using 10-fold cross validation. The models with the best results were used for further processing. The training images were resized to 256×256 to alleviate the computational cost. Data augmentation operations, performed during training, included: random rotations of up to 10° clockwise and counterclockwise; random translations of up to 10% of image dimensions horizontally and vertically; scaling by a random factor ranging from 0.9 to 1.1; horizontal flipping at a 50% chance; intensity flipping (black to white and white to black) at a 50% chance; contrast stretching (where a random percentage, up to 10%, of the lowest and highest intensity voxels are set to the minimum and maximum intensity allowable for the image, and the remaining pixel intensities in the image are rescaled to fit the range). The bone suppression models were implemented in Python 3.6 and PyTorch 1.10.
The performance of the bone suppression methods was quantified using the peak signal-to-noise ratio (PSNR), SSIM (40) and root-mean-squared error (RMSE) for the internal test set, and was qualitatively assessed using the Hong Kong hospitals external test set. The paired Student t-test was used to assess the statistical significance of the bone suppression methods.
Classification
To identify the effect of bone suppression on classification, two classification models were implemented. The COVID-Net CXR2 model, a publicly available, pre-trained 2-class (‘Not COVID’ and ‘COVID’) classification model (11), was implemented. As shown in the published paper and code (11), all images input into the classifier were processed as follows: (I) the topmost 8% of the image was removed in order to remove metadata in the image; (II) the image was center-cropped by removing both ends of the longer side of the image, such that the remaining image is square; (III) the image was resized using bilinear interpolation to 480×480 pixels; (IV) the image was input into the model’s neural network for classification. This model had been trained on the ImageNet (41) natural image database, then on the COVIDx8B dataset which consisting of approximately 8,700 non-pathological CXRs, 5,900 non-COVID pneumonia CXRs, and 4,600 COVID-19 CXRs obtained from public datasets (32,33,36,42-45). No bone suppression of the dataset had been performed prior to training. The model had then been modified using a proprietary deep learning-based generative synthesis process to create the final model, which was used in this study without modification. COVID-Net had been implemented in TensorFlow 2.6.
COVID-Net had been trained on datasets generated by crowd-sourcing (11). This is noted to be problematic (46), since the submitted images may have been mislabeled, and images may have been submitted to multiple crowd-sourced datasets resulting in duplication within the training data. Therefore, we trained a classification network using datasets sourced from medical archives [RSNA Pneumonia Detection Challenge dataset (36), a subset of the ChestX-ray8 dataset released by the National Institutes of Health Clinical Center, and RICORD-1C (33)]. The network architecture used in this study is inspired by that introduced by Rajaraman et al. (19), and is as follows: a VGG-16 (47) network pretrained on ImageNet (41) was truncated after the last convolutional block to be used as a feature extractor. A global average pooling (48) layer, a dropout layer (49) (50% dropout chance), and then a fully connected output layer with two output nodes was appended in that order. VGG16 had been chosen because it had been used in previous chest disease classification studies (19,50) and had demonstrated improved results over alternatives such as DenseNet-121 (51) and ResNet-50 (52). In this study, our model is denoted “VGG16-Modified”. It was trained as described: (I) all VGG-16 parameters are frozen; (II) the whole network is trained using the Adam optimizer (learning rate = 0.001, beta1 = 0.9, beta2 = 0.999) for 5 epochs; (III) the parameters of the final three convolutional layers of the VGG16 feature extractor are unfrozen; (IV) the network is fine-tuned by training using stochastic gradient descent (learning rate = 5e-6, weight decay = 0.01) for 40 epochs. The training batch size was 8. The network was trained using 8-fold cross-validation.
Three VGG16-Modified models were independently trained using three cross-validation datasets. The first cross-validation dataset consisted of the 896 COVID-19 CXR images and 1,025 non-pathological CXR images introduced in Section Datasets of Methods. The second dataset was generated by Gusarev suppression of the first dataset. The third dataset was generated by Rajaraman suppression of the first dataset. The COVID and non-pathological image data for all three datasets were augmented by four and three times respectively (18), using the same data augmentation operations as those used for bone suppression model training (Section Bone suppression of Methods), resulting in final dataset sizes of 3,584 COVID and 3,075 non-pathological CXR images for each dataset. No augmentation was performed during training. The fold with the best average accuracy was used for further processing. Prior to model input, images were resized and center-cropped to 224×224 pixels (to fit the VGG16 input) using nearest-neighbor interpolation. The VGG16-Modified model was implemented in Python 3.6, PyTorch 1.10 and torchvision 0.11.
After the bone suppression models and VGG16-Modified classifier models had been trained, the performance of the COVID-Net CXR2 and VGG16-Modified classifier models were quantified by sensitivity, specificity, negative predictive value (NPV), accuracy, and area under the receiver operating curve (AUC) on the Hong Kong hospitals external test set. NPV is important because the public health consequences of missing a COVID-19 diagnosis was deemed to be greater than that of mistakenly identifying someone as infected. The performance of COVID-Net CXR2 when classifying a non-suppressed, a Gusarev-suppressed and a Rajaraman-suppressed external test set was compared. The VGG16-Modified model trained on non-suppressed data was assessed using the non-suppressed external test set; the model trained on Gusarev-suppressed data, on the Gusarev-suppressed external test set; and the model trained on Rajaraman-suppressed data, on the Rajaraman-suppressed external test set.
The 95% confidence intervals for AUC were calculated using the DeLong test (53) to assess statistical significance in differences between classifier performance. Due to the nature of this study, the classifiers were not blind-tested. The trained bone suppression models and VGG16-Modified models used in this study are available for use from (https://github.com/danielnflam).
Results
Clinical information
The clinical information of patients whose datasets were used in this study is shown in Table 1. Patients in the JSRT dataset were either non-pathological or had lung nodules.
Bone suppression
Internal test set
Qualitative bone suppression results for a subset of the internal test set of 20 image pairs are shown in Figures 3,4 for Gusarev and Rajaraman suppression, respectively. Quantitative results for all methods are given in Table 2. For both Gusarev and Rajaraman suppression, the suppressed images were significantly (P<0.05) more similar to the target images than the non-suppressed images.
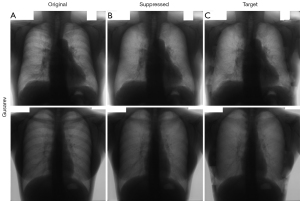
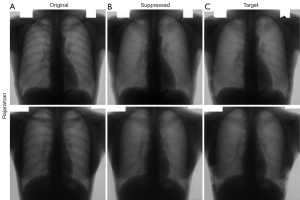
Table 2
| Bone suppression method | PSNR (higher is better, mean ± SD) | SSIM (higher is better, mean ± SD) | RMSE (lower is better, mean ± SD) |
|---|---|---|---|
| None | 32.9±3.0 | 0.982±0.005 | 0.024±0.010 |
| Gusarev | 33.8±2.9 | 0.984±0.004 | 0.022±0.010 |
| Rajaraman | 34.0±3.7 | 0.986±0.004 | 0.021±0.010 |
PSNR, peak signal-to-noise ratio; SD, standard deviation; SSIM, structural similarity index measure; RMSE, root-mean-squared error.
External test set
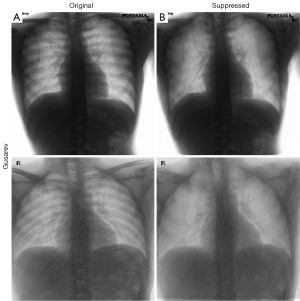
Representative examples of non-suppressed and bone-suppressed CXRs from the Hong Kong hospitals dataset are shown in Figures 5,6 for Gusarev and Rajaraman suppression respectively. As no ground-truth CXR existed for this dataset, no quantitative results could be shown.

Classification
The sensitivity, specificity, NPV, accuracy and AUC results for both classifier architectures processing external test data sets are shown in Table 3.
Table 3
| Bone suppression of external test data | None | Gusarev | Rajaraman |
|---|---|---|---|
| COVID-Net CXR2 | |||
| Sensitivity (%) | 80.93 | 77.81 | 71.25 |
| Specificity (%) | 56.18 | 60.42 | 67.95 |
| NPV (%) | 82.67 | 81.51 | 79.28 |
| Accuracy (%) | 65.63 | 67.06 | 69.21 |
| AUC ± SE95% | 0.686±0.031 | 0.691±0.031 | 0.696±0.032 |
| VGG16-Modified | |||
| Sensitivity (%) | 52.81 | 61.88 | 61.88 |
| Specificity (%) | 86.87 | 78.76 | 84.56 |
| NPV (%) | 74.88 | 76.98 | 78.21 |
| Accuracy (%) | 73.87 | 72.32 | 75.90 |
| AUC ± SE95% | 0.698±0.031 | 0.703±0.032 | 0.732±0.031* |
For the COVID-Net CXR2 architecture, the same model was tested with non-suppressed, Gusarev-suppressed and Rajaraman-suppressed external testing data. For the VGG16-Modified architecture, separate models trained on non-suppressed, Gusarev-suppressed and Rajaraman-suppressed data were each tested with their correspondingly suppressed external test data (e.g., non-suppressed training data model with non-suppressed test data). *, denotes a significant (P<0.05) difference from the non-suppressed external test data. NPV, negative predictive value; AUC, area under the receiver operating curve; SE95%, the error associated with a 95% confidence interval.
For the VGG16-Modified classifier, the AUC (±95% confidence interval) results for the internal test set were 0.887±0.033, 0.828±0.037 and 0.838±0.035 for the non-suppressed, Gusarev-suppressed and Rajaraman-suppressed models respectively. Interestingly, bone suppression resulted in significantly (P<0.05) worse results than no suppression. For the external test set (320 COVID and 518 non-COVID patients), bone suppression improved classifier performance from 0.698±0.031 (no suppression) to 0.703±0.032 (Gusarev suppression) and 0.732±0.031 (Rajaraman suppression). Rajaraman suppression resulted in significantly (P<0.05) improved AUC scores compared to no suppression. NPV increased after bone suppression (from 74.88% to a maximum of 78.21% for Rajaraman suppression).
For COVID-Net CXR2, no significant difference in classifier performance was observed between non-suppressed (0.686±0.031), Gusarev-suppressed (0.691±0.031) and Rajaraman-suppressed (0.696±0.032) external test data sets. NPV decreased after bone suppression (from 82.67% to a minimum of 79.28% for Rajaraman suppression). Interestingly, the AUC for COVID-Net CXR2 did not differ significantly from that for VGG16-Modified, for the non-suppressed external test data set.
Discussion
The Rajaraman-suppressed and Gusarev-suppressed images were significantly (P<0.05) more similar to the target images than the non-suppressed images, suggesting that the bone suppression methods are fit for purpose. Qualitative examination of the Gusarev- and Rajaraman-suppressed radiographs (Figures 5,6, respectively) in this study suggest that rib shadows are reduced post-suppression, albeit imperfectly. A limitation of the bone suppression performed in this study was that the target images were themselves derived from a method by Juhász et al. (30), instead of acquired directly from dual-energy subtraction radiography. Hence, the bone suppression models could have learnt to reproduce the errors produced by Juhász suppression. Published studies have been performed using the JSRT and Juhász et al. datasets to pursue bone suppression of CXRs (19,54), and, to the authors’ knowledge, no other publicly available co-registered non-suppressed and bone-suppressed chest radiograph dataset exists.
The main purpose of the study was to examine whether bone suppression improved the performance of COVID-19 classification models. For the external test set, results demonstrate that Rajaraman suppression significantly (P<0.05) improved AUC for the VGG16-Modified classifier architecture compared to no suppression, whereas no significant improvement was observed for COVID-Net CXR2. This difference may have been because COVID-Net had been trained on a larger and more varied dataset of non-suppressed CXRs (4,600 COVID + 14,600 non-COVID) than VGG16-Modified (896 COVID + 1,025 non-COVID before augmentation). Hence, the COVID-Net CXR2 model could have learned to disregard bone shadow when classifying CXRs. It is interesting to note that, for VGG16-Modified, internal test set results significantly worsened after bone suppression, whilst external test set results demonstrated either no significant change (Gusarev suppression) or significant improvement (Rajaraman suppression). This difference was suspected to arise from minor overfitting in the non-suppressed classifier model. Ultimately, the results suggest that bone suppression could be applied to improve the performance of classifier models, with the worst-case scenario being a statistically insignificant change in performance. Further work could involve acquiring more CXRs to improve classifier performance, and examining the effect of bone suppression on more classifier architectures. Furthermore, it is plausible that, as demonstrated by the research of Rajaraman et al. (19), this technique could be applied to lung pathologies outside of COVID-19 such as lung carcinomas.
Limitations of the study are as follows. Only two classifier architectures were tested in this study, so it is difficult to definitively conclude that bone suppression improves classification performance. In the future, the effect of bone suppression could be tested on a wider variety of classifiers, particularly those based on novel architectures (16) or those using inputs from different imaging modalities (17). Furthermore, bone suppression of the test data has only been tested for two-class classification models. The impact of bone suppression on the performance of three-class models (that classify CXRs into non-pathological, non-COVID-19 pneumonia and COVID-19) (11,12) remains to be seen. The demographics of the training data and external test set are also of interest. The patients of the external test dataset in this study are mostly of East Asian descent, compared to the North American patient data used to train VGG-Rajaraman and the North American and South Asian patient data used to train COVID-Net (11,32-34,36,42,43,45). Possible biological variability in COVID-19 radiological presentation between different demographics may have contributed to poor classifier performance (55,56). Furthermore, standard imaging protocols may differ between nations. Acquisition of patient data from a wider range of institutions could enable further research into whether the results in this study are generalizable.
In general, future work could be pursued in several other ways. Other classification architectures could be tested to see whether bone suppression improved their performance. More image data could be acquired to improve both the classifier performance and the generality of the external test set. Other image enhancement techniques beyond bone suppression could be pursued to improve classification performance for other clinical conditions imaged using other modalities. Radiomics analysis of the enhanced images, and analysis of other non-deep learning features (57), could be combined with deep learning features to further improve classification performance.
Conclusions
Rajaraman bone suppression of external test data was found to significantly (P<0.05) improve the AUC of the VGG16-Modified classifier. In general, bone suppression significantly improved, or did not worsen, classifier performance for an external test data set. Further research into bone suppression and other image enhancement techniques could significantly improve deep learning image classification performance.
Acknowledgments
We would like to acknowledge the patients and staff of Pamela Youde Nethersole Eastern Hospital and Queen Elizabeth Hospital for providing the data with which this study was performed.
Funding: This work was supported by Health and Medical Research Fund (No. HMRF COVID190211), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region.
Footnote
Reporting Checklist: The article is presented in accordance with the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-21-791/rc
Conflicts of Interest: All authors have completed the ICMJE Uniform Disclosure Form (Available at https://qims.amegroups.com/article/view/10.21037/qims-21-791/coif). FMK has research grants from Varian Medical Systems, Merck Pharmaceutical (through Chinese Society of Clinical Oncology), and speaker’s honorarium from AstraZeneca. NFDL was employed by an institution and worked under a supervisor that received money from the Health and Medical Research Fund (No. HMRF COVID190211), the Food and Health Bureau, The Government of the Hong Kong Special Administrative Region. JC has received research grant (No. HMRF COVID190211) for this study. YXJW serves as the Editor-In-Chief of Quantitative Imaging in Medicine and Surgery. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and with ethics approval from The Hong Kong Polytechnic University Faculty Research Committee (No. HSEARS20201107001). Since only anonymized data was acquired and used in this study, individual consent for this retrospective study was waived. Data from Queen Elizabeth Hospital was acquired with ethics approval from the Kowloon Central/Kowloon East Research Ethics Committee (No. KC/KE-20-0337/ER-4). Data from Pamela Youde Nethersole Eastern Hospital was acquired with ethics approval from the Hong Kong East Cluster Research Ethics Committee (No. HKECREC-2020-119).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420-2. [Crossref] [PubMed]
- Campbell F, Archer B, Laurenson-Schafer H, Jinnai Y, Konings F, Batra N, Pavlin B, Vandemaele K, Van Kerkhove MD, Jombart T, Morgan O, le Polain de Waroux O. Increased transmissibility and global spread of SARS-CoV-2 variants of concern as at June 2021. Euro Surveill 2021; [Crossref] [PubMed]
- Sheikh A, McMenamin J, Taylor B, Robertson C. Public Health Scotland and the EAVE II Collaborators. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet 2021;397:2461-2. [Crossref] [PubMed]
- Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med 2020;382:1708-20. [Crossref] [PubMed]
- Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of Chest CT and RT-PCR Testing for Coronavirus Disease 2019 (COVID-19) in China: A Report of 1014 Cases. Radiology 2020;296:E32-40. [Crossref] [PubMed]
- Islam N, Ebrahimzadeh S, Salameh JP, Kazi S, Fabiano N, Treanor L, et al. Thoracic imaging tests for the diagnosis of COVID-19. Cochrane Database Syst Rev 2021;3:CD013639. [PubMed]
- Pandit MK, Banday SA, Naaz R, Chishti MA. Automatic detection of COVID-19 from chest radiographs using deep learning. Radiography (Lond) 2021;27:483-9. [Crossref] [PubMed]
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Li FF. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vis 2015;115:211-52. [Crossref]
- Voulodimos A, Doulamis N, Doulamis A, Protopapadakis E. Deep Learning for Computer Vision: A Brief Review. Comput Intell Neurosci 2018;2018:7068349. [Crossref] [PubMed]
- Wang L, Lin ZQ, Wong A. COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep 2020;10:19549. [Crossref] [PubMed]
- Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. Appl Intell (Dordr) 2020. [Epub ahead of print]. doi:
10.1007/s10489-020-01829-7 .10.1007/s10489-020-01829-7 - Padma T, Kumari CU. Deep Learning Based Chest X-Ray Image as a Diagnostic Tool for COVID-19. In: 2020 International Conference on Smart Electronics and Communication (ICOSEC). Trichy: IEEE, 2020:589-92.
- Fontanellaz M, Ebner L, Huber A, Peters A, Löbelenz L, Hourscht C, Klaus J, Munz J, Ruder T, Drakopoulos D, Sieron D, Primetis E, Heverhagen JT, Mougiakakou S, Christe A. A Deep-Learning Diagnostic Support System for the Detection of COVID-19 Using Chest Radiographs: A Multireader Validation Study. Invest Radiol 2021;56:348-56. [Crossref] [PubMed]
- Hwang EJ, Kim H, Yoon SH, Goo JM, Park CM. Implementation of a Deep Learning-Based Computer-Aided Detection System for the Interpretation of Chest Radiographs in Patients Suspected for COVID-19. Korean J Radiol 2020;21:1150-60. [Crossref] [PubMed]
- Wang SH, Khan MA, Govindaraj V, Fernandes SL, Zhu Z, Zhang YD. Deep Rank-Based Average Pooling Network for Covid-19 Recognition. Computers, Materials, & Continua 2022;70:2797-813.
- Zhang YD, Zhang Z, Zhang X, Wang SH. MIDCAN: A multiple input deep convolutional attention network for Covid-19 diagnosis based on chest CT and chest X-ray. Pattern Recognit Lett 2021;150:8-16. [Crossref] [PubMed]
- Gusarev M, Kuleev R, Khan A, Ramirez Rivera A, Khattak AM. Deep learning models for bone suppression in chest radiographs. In: 2017 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB). Manchester: IEEE, 2017:1-7.
- Rajaraman S, Zamzmi G, Folio L, Alderson P, Antani S. Chest X-ray Bone Suppression for Improving Classification of Tuberculosis-Consistent Findings. Diagnostics (Basel) 2021;11:840. [Crossref] [PubMed]
- Ren G, Xiao H, Lam SK, Yang D, Li T, Teng X, Qin J, Cai J. Deep learning-based bone suppression in chest radiographs using CT-derived features: a feasibility study. Quant Imaging Med Surg 2021;11:4807-19. [Crossref] [PubMed]
- Oh DY, Yun ID. Learning Bone Suppression from Dual Energy Chest X-rays using Adversarial Networks. arXiv 2018. arXiv:1811.02628.
- Zhou Z, Zhou L, Shen K. Dilated conditional GAN for bone suppression in chest radiographs with enforced semantic features. Med Phys 2020;47:6207-15. [Crossref] [PubMed]
- von Berg J, Young S, Carolus H, Wolz R, Saalbach A, Hidalgo A, Giménez A, Franquet T. A novel bone suppression method that improves lung nodule detection: Suppressing dedicated bone shadows in radiographs while preserving the remaining signal. Int J Comput Assist Radiol Surg 2016;11:641-55. [Crossref] [PubMed]
- Suzuki K, Abe H, MacMahon H, Doi K. Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN). IEEE Trans Med Imaging 2006;25:406-16. [Crossref] [PubMed]
- Vock P, Szucs-Farkas Z. Dual energy subtraction: principles and clinical applications. Eur J Radiol 2009;72:231-7. [Crossref] [PubMed]
- Baltruschat IM, Steinmeister LA, Ittrich H, Adam G, Nickisch H, Saalbach A, von Berg J, Grass M, Knopp T. When Does Bone Suppression And Lung Field Segmentation Improve Chest X-Ray Disease Classification? In: 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019). Venice: IEEE, 2019:1362-6.
- Freedman MT, Lo SC, Seibel JC, Bromley CM. Lung nodules: improved detection with software that suppresses the rib and clavicle on chest radiographs. Radiology 2011;260:265-73. [Crossref] [PubMed]
- Manji F, Wang J, Norman G, Wang Z, Koff D. Comparison of dual energy subtraction chest radiography and traditional chest X-rays in the detection of pulmonary nodules. Quant Imaging Med Surg 2016;6:1-5. [PubMed]
- Shiraishi J, Katsuragawa S, Ikezoe J, Matsumoto T, Kobayashi T, Komatsu K, Matsui M, Fujita H, Kodera Y, Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologist’' detection of pulmonary nodules. AJR Am J Roentgenol 2000;174:71-4. [Crossref] [PubMed]
- Juhász S, Horváth Á, Nikházy L, Horváth G, Horváth Á. Segmentation of Anatomical Structures on Chest Radiographs. In: Bamidis PD, Pallikarakis N. editors. XII Mediterranean Conference on Medical and Biological Engineering and Computing 2010. Berlin, Heidelberg: Springer Berlin Heidelberg, 2010:359-62.
- Huynh MC. Dataset: X-ray Bone Shadow Supression, 2018. Available online: https://www.kaggle.com/hmchuong/xray-bone-shadow-supression
- Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, et al. Data from Medical Imaging Data Resource Center (MIDRC)–- RSNA International COVID Radiology Database (RICORD) Release 1c–- Chest x-ray, Covid+ (MIDRC-RICORD-1c). The Cancer Imaging Archive 2021; [Crossref]
- Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, et al. The RSNA International COVID-19 Open Radiology Database (RICORD). Radiology 2021;299:E204-13. [Crossref] [PubMed]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. Chestxray8: hospital-scale chest X-ray database and benchmarks on weakly supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition. Hawaii, USA: IEEE, 2017:2097-106.
- Shih G, Wu CC, Halabi SS, Kohli MD, Prevedello LM, Cook TS, Sharma A, Amorosa JK, Arteaga V, Galperin-Aizenberg M, Gill RR, Godoy MCB, Hobbs S, Jeudy J, Laroia A, Shah PN, Vummidi D, Yaddanapudi K, Stein A. Augmenting the National Institutes of Health Chest Radiograph Dataset with Expert Annotations of Possible Pneumonia. Radiol Artif Intell 2019;1:e180041. [Crossref] [PubMed]
- Radiological Society of North America. RSNA Pneumonia Detection Challenge, 2019. Available online: https://www.kaggle.com/c/rsna-pneumonia-detection-challenge
- Ioffe S, Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on International Conference on Machine Learning, 2015 (ICML'15). PMLR 2015;37:448-56.
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. In: Bengio Y, LeCun Y. editors. 3rd International Conference for Learning Representations. San Diego, CA, USA: ICLR, 2015.
- Wang Z, Simoncelli EP, Bovik AC. Multiscale structural similarity for image quality assessment. In: The Thrity-Seventh Asilomar Conference on Signals, Systems & Computers, 2003. Pacific Grove, CA, USA: IEEE, 2003:1398-402.
- Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004;13:600-12. [Crossref] [PubMed]
- Deng J, Dong W, Socher R, Li LJ, Li K, Li FF. ImageNet: A large-scale hierarchical image database. In: 2009 IEEE Conference on Computer Vision and Pattern Recognition. Miami, FL, USA: IEEE, 2009:248-55.
- Chung AG. Figure 1 COVID-19 Chest X-ray Dataset Initiative, 2020. Available online: https://github.com/agchung/Figure1-COVID-chestxray-dataset
- Chung AG. Actualmed COVID-19 Chest X-ray Dataset Initiative, 2020. Available online: https://github.com/agchung/Actualmed-COVID-chestxray-dataset
- Chowdhury MEH, Rahman T, Khandakar A, Mazhar R, Kadir MA, Mahbub ZB, Islam KR, Khan MS, Iqbal A, Emadi NA, Reaz MBI, Islam MT. Can AI Help in Screening Viral and COVID-19 Pneumonia? IEEE Access 2020;8:132665-76.
- Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Abul Kashem SB, Islam MT, Al Maadeed S, Zughaier SM, Khan MS, Chowdhury MEH. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput Biol Med 2021;132:104319. [Crossref] [PubMed]
- Roberts M, Driggs D, Thorpe M, Gilbey J, Yeung M, Ursprung S, Aviles-Rivero AI, Etmann C, McCague C, Beer L, Weir-McCall JR, Teng Z, Gkrania-Klotsas E. AIX-COVNET, Rudd JHF, Sala E, Schönlieb CB. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nature Machine Intelligence 2021;3:199-217. [Crossref]
- Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In: International Conference on Learning Representations (ICLR) 2015. ICLR 2015.
- Lin M, Chen Q, Yan S. Network In Network. In: International Conference on Learning Representations (ICLR) 2014. ICLR 2014.
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: A simple way to prevent neural networks from overfitting. J Mach Learn Res 2014;15:1929-58.
- Nishio M, Noguchi S, Matsuo H, Murakami T. Automatic classification between COVID-19 pneumonia, non-COVID-19 pneumonia, and the healthy on chest X-ray image: combination of data augmentation methods. Sci Rep 2020;10:17532. [Crossref] [PubMed]
- Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Honolulu, HI, USA: IEEE, 2017:4700-8.
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2016:770-8.
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45. [Crossref] [PubMed]
- Huynh MC, Nguyen TH, Tran MT. Context Learning for Bone Shadow Exclusion in CheXNet Accuracy Improvement. In: 2018 10th International Conference on Knowledge and Systems Engineering (KSE). Ho Chi Minh City: IEEE, 2018:135-40.
- Navar AM, Purinton SN, Hou Q, Taylor RJ, Peterson ED. The impact of race and ethnicity on outcomes in 19,584 adults hospitalized with COVID-19. pLoS One 2021;16:e0254809. [Crossref] [PubMed]
- Romano SD, Blackstock AJ, Taylor EV, El Burai Felix S, Adjei S, Singleton CM, Fuld J, Bruce BB, Boehmer TK. Trends in Racial and Ethnic Disparities in COVID-19 Hospitalizations, by Region–- United States, March-December 2020. MMWR Morb Mortal Wkly Rep 2021;70:560-5. [Crossref] [PubMed]
- Zhang X, Wang D, Shao J, Tian S, Tan W, Ma Y, Xu Q, Ma X, Li D, Chai J, Wang D, Liu W, Lin L, Wu J, Xia C, Zhang Z. A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography. Sci Rep 2021;11:3938. [Crossref] [PubMed]


