
Repeatability of MR fingerprinting in normal cervix and utility in cervical carcinoma
Introduction
Conventional quantitative magnetic resonance imaging (MRI) techniques that aim to estimate MR parameters—such as T1, T2, and apparent diffusion coefficient—are sensitive to variations in the content of metabolites and tissue components. Diffusion-weighted imaging is routinely used in the diagnosis and assessment of pelvic diseases, but quantitative T1 and T2 mappings have not been adopted in routine clinical practice. These are challenging to image in moving regions, including the abdomen and pelvis, due to long acquisition times (1,2).
Magnetic resonance fingerprinting (MRF) is a fast imaging acquisition technique that can overcome the limitations of conventional T1 and T2 mapping (3). It is based on the premise that MR parameters can be reliably estimated from the evolution of MR signals, known as MR fingerprints, that are acquired with pseudorandomised acquisition parameters, such as repetition times and flip angles (3). To estimate MR parameters, MRF is matched against a precomputed dictionary, which contains a library of MRI signal evolution of different biological tissues, on a voxel-by-voxel basis. MRF thus permits the estimation of quantitative and co-registered parametric maps from a single sequence (1).
Conventional T1- and T2-weighted imaging are part of the routine set of sequences for diagnosis and assessment of various cancers. However, these sequences do not directly provide quantitative assessment of pathophysiological changes in tissues. Additionally, early subtle changes within the tumours could not be readily appreciated even by experienced radiologists unless the tumours change sufficiently in size, morphology or intensity (4). Thus, these conventional sequences without quantitative measurements have a limited role in the detection of early changes in the tumour microenvironment. On the other hand, MRF may be a promising alternative by providing quantitative metrics that could evaluate the biological characteristics of tumours based on the various contrast mappings which can be acquired in a single sequence. Given these advantages, MRF has the potential to estimate multiple and useful imaging parameters, which can be implemented for cancer characterisation and treatment response assessment and potentially incorporated in radiomics and machine-learning (4,5).
In order to be clinically useful, quantitative parameters, such as T1 and T2 values derived from MRF, need to have high repeatability and reproducibility (6). Initiatives like the Quantitative Imaging Biomarkers Alliance (QIBA) investigate the standardisation and reliability testing of such quantitative techniques in order to translate these biomarkers into clinical practice (7). Numerous studies have reported good to excellent repeatability and reproducibility of MRF in vivo even across MRI platforms of different field strengths (8-13). Recent studies have demonstrated the application of MRF in the brain (4,8,9,13-16), and preliminary investigations in other body parts, including breast, liver, kidney, prostate and ovaries (2,5,10,17-23). Furthermore, MRF has shown the potential ability in differentiating cancers from normal tissues and distinguishing primary tumours from metastases (15,20,21). Nevertheless, MRF in cervical carcinoma (CC) has hitherto not been investigated. The aims of this feasibility study were to evaluate the agreement between MRF and phantom reference values, scan-rescan repeatability of MRF in normal cervix and to explore the ability of MRF in discriminating CC from normal cervical tissues.
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This prospective study was approved by the local Institutional Review Board, and informed consent were obtained from all the study subjects.
Study subjects recruitment
Twelve healthy volunteers were recruited for pelvic MRI and MRF between December 2019 and April 2020. The inclusion criteria for healthy volunteers were those (I) without clinical symptoms and (II) no prior history of hormone therapy, gynaecological conditions or pelvic surgery.
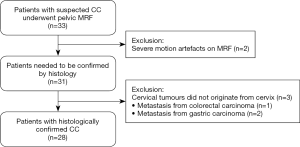
Initially, 33 patients with clinically visible cervical masses were prospectively recruited and underwent pelvic MRI and MRF between June 2019 and March 2020. The inclusion criterion for patients was histologically confirmed CC. Exclusion criteria included (I) prior history of pelvic surgery, chemoradiation or other malignancy, (II) cervical tumour that did not originate from the cervix, and (III) severe motion artefacts observed on MRI or MRF maps.
Clinicopathological assessment
Cervical biopsies from clinically suspected CC were assessed by a pathologist with more than 10 years’ experience in gynaecological malignancy; and reviewed at multi-disciplinary team meetings. The assessed histopathological markers included histological subtypes and tumour grades, according to the WHO Classification of Tumours of Female Reproductive Organs (24). All cases were staged using the revised 2018/2019 International Federation of Gynecology and Obstetrics (FIGO) staging for CC based on MRI by a board-certified radiologist (R1: >10 years’ experience in pelvic MRI) (25).
MRF acquisition and processing
We have implemented an in-house single-slice inversion-recovery fast imaging protocol with steady-state free precession for MRF acquisition and an in-house MRF reconstruction pipeline, which consisted of spiral reconstruction using non-uniform Fourier Transform (26), dictionary generation using the extended phase graph algorithm (27), and in-house dictionary matching written in MATLAB (R2017b, The MathWorks, Natick, MA, USA), similar to references (3,16). All MRI examinations were performed using a 3.0 T MRI scanner (Achieva 3T TX, Philips Healthcare Best, the Netherlands). Dictionary matching was done on a machine equipped with an Intel Xeon Gold 6428 (20 cores @ 2.50 GHz) and 500 GB of random access memory (RAM), and matching took approximately 6 minutes per slice. The generated T1 and T2 maps were co-registered, as the maps were reconstructed from the same base MRF signal in postprocessing.
MRF in phantom
MRF was performed on the standard International Society of Magnetic Resonance in Medicine/National Institute of Standards and Technology (ISMRM/NIST) MRI system using an 8-channel phase array brain and dorsal coils for signal reception as previously described in reference (28). MRF protocol and dictionary details may be found in Table 1. An additional 5 s delay was needed before each acquisition to ensure the net magnetization to be in its thermal equilibrium state, the resulting scan time per slice was 19 s. The phantom was scanned 5 times per session over 15 sessions throughout 65 days.
Full table
Circular regions of interest (ROIs) of 6 T1 and 11 T2 fiducial spheres from all repetitions in each session were contoured. The mean and standard deviation of T1and T2 values over all voxels in the ROI were measured for all 15 sessions. Measured T1 and T2 values from MRF were averaged across the 15 acquisitions. T1 spheres had reference values ranging from 367 to 1,838 ms, while T2 spheres had reference values ranging from 15 to 646 ms. As we aimed to evaluate the T1 and T2 values with the ranges observed in biological tissues, T1 spheres 7 to 14 and T2 spheres 12 to 14 which contained lower T1 and T2 values were excluded from our measurements.
MRF in-vivo
MRI was performed with a 16-channel phased-array torso coil for signal reception. All the subjects fasted for 6 hours and received 20 mg intravenous hyoscine butylbromide (Buscopan, Boehringer Ingelheim, Germany) before MRI examinations to reduce peristaltic artefacts. Conventional MRI and MRF protocols were standardised for all subjects and the MRF protocol was identical to that used in the phantom experiment (Tables 1,2). The dictionary was computed with range and increments tabulated in Table 1 (5). For healthy volunteers, the same MRF protocol was performed twice (MRF1 and MRF2) with a 10-minute break interval (4,10). For patients with CC, MRF was only performed once after conventional sequences. Eight to fourteen slices of MRF sequence were performed for each subject, the scanning range covered the whole normal cervix or the entire tumour of all patients.
Full table
Normal cervix and CC delineation
First, a second radiologist (R2; board-certified with 3 years’ experience in pelvic MRI) delineated the volumes of interest (VOIs) of normal cervical tissues and CC using open-source ImageJ software (1.53d, National Institutes of Health, USA) and the VOIs were subsequently verified by R1. In cases of disagreement, VOIs were revised in consensus. As axial T2-weighted images had corresponding imaging slices to MRF maps, axial and sagittal T2-weighted images were taken as references when drawing VOIs on the T1 and T2 maps obtained from MRF. VOIs were individually delineated on T1 maps first and then copied to T2 maps as these maps were co-registered. Mean T1 and T2 values of healthy volunteers and patients with CC were computed over all voxels in the VOIs using an in-house MATLAB script.
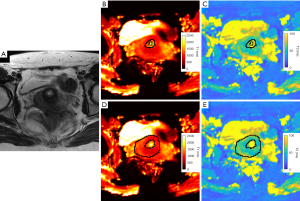
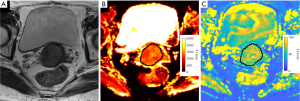
For healthy volunteers, the VOIs of cervical mucosa and stroma were individually delineated on the T1 and T2 maps of MRF1 (Figure 1). The central canal was excluded from the delineation of cervical mucosa. The same procedure was repeated on the T1 and T2 maps obtained from MRF2 (Figure 2). For patients with CC, the VOIs of CC were drawn by strictly delineating the border of CC from adjacent normal tissue on each slice to cover the full extent of the tumours on the T1 and T2 maps (Figure 3).
Statistical analysis
Linear regression was performed, and the coefficient of determination (R2) was calculated to assess the agreement of T1 and T2 values between MRF and phantom reference values. Bland-Altman plots were used to evaluate the repeatability of MRF scan-rescan on healthy volunteers. The bias and limits of agreement of Bland-Altman plots represent the mean difference and the range of variation between the two measurements, respectively. Within-subject coefficients of variation (wCV), two-way mixed, single score, agreement intraclass correlation coefficients {ICC [2, 1]} were respectively defined in Eq. [1] and Eq. [2] (29,30):
where σ represents measurement standard deviation, µ measurement mean, and τ measurement variance. The repeatability of MRF with ICC of 0.50 was regarded as poor, 0.50–0.75 moderate, 0.75–0.90 good or >0.90 excellent (30). Continuous variables were tested for normality using Shapiro-Wilk test. Mann-Whitney U tests were used for comparing the difference in T1 and T2 values between CC and normal cervical tissues, and the Benjamini-Hochberg procedure was used for multiple comparisons correction. A P value <0.05 was considered as statistical significance. Receiver operating characteristic (ROC) analysis was performed, and the area under the curve (AUC), sensitivity and specificity were used for the evaluation of the diagnostic efficiency and cut-off values. AUC of 0.7–0.8 was considered acceptable, 0.8–0.9 was considered excellent and >0.9 was considered as an outstanding accuracy (31). All the statistical analyses were performed using in-house R scripts (3.6.2, R Core Team, Vienna, Austria).
Results
Phantom experiment
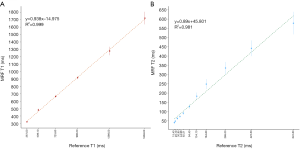
Coefficient of determination between MRF-derived T1 and T2 values with reference values was R2=0.999 for T1 and 0.981 for T2 (28). The correlation plots were shown in Figure 4.
Demographics and clinical characteristics
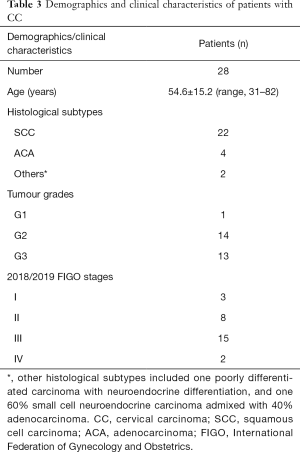
In total, 12 healthy volunteers (28.7±5.1 years; range, 24–43 years) were enrolled. Of the 33 patients, 5 were excluded, thus 28 patients with histologically confirmed CC (54.6±15.2 years; range, 31–82 years) were analysed. A diagram of patient selection was shown in Figure 5. The demographics and clinical characteristics of patients with CC were shown in Table 3.
Full table
MRF scan-rescan repeatability of normal cervix
The wCV of MRF-derived T1 and T2 values in the mucosa were 1.90% and 4.25%, respectively, and those in the stroma were 2.44% and 3.47%, respectively. The ICC for T1 and T2 in the mucosa were 0.96 and 0.94, respectively, and in the stroma, they were 0.92 and 0.95, respectively. Bland-Altman analysis showed that T1 values of mucosa and stroma had similar biases and limits of agreement. Furthermore, T2 values of mucosa and stroma had similar biases, but T2 stroma had tighter limits of agreement. Bland-Altman plots were shown in Figure 6.
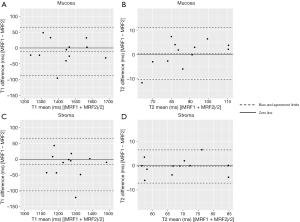
Comparison between CC and normal cervical tissues
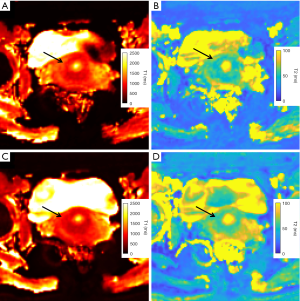
CC had an average T1 value of 1,529±112 ms and was significantly higher than normal mucosa (P=0.031, compared with MRF1 and MRF2) and stroma (P<0.001, compared with MRF1 and MRF2). CC had an average T2 value of 69±9 ms and was significantly lower than normal mucosa (P<0.001, compared with MRF1 and MRF2). A full tabulation of the T1 and T2 values of CC and normal cervical tissues, as well as the cut-offs, AUC, sensitivity, and specificity, can be found in Table 4 with corresponding box and whisker plots depicted in Figure 7.
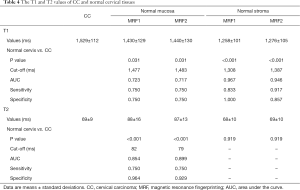
Full table
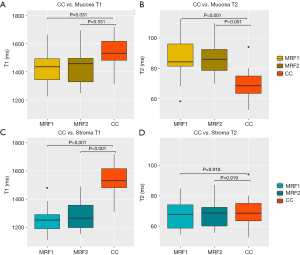
Discussion
Preliminary studies of MRF have been applied in abdomen and pelvis, including liver, kidney, prostate and ovaries (2,5,10,17-23). However, the repeatability of MRF in normal cervix and the utility of MRF in CC have never been evaluated. In this feasibility study, strong correlations between MRF and phantom reference values and excellent scan-rescan repeatability of MRF in normal cervical tissues were demonstrated. Furthermore, the quantitative T1 and T2 parameters obtained from MRF were able to differentiate between CC and normal cervical tissues.
Our study evaluated the agreement between MRF and reference values provided by the ISMRM/NIST phantom. Strong agreements were observed with R2=0.999 for T1, 0.981 for T2. A recent MRF study reported similar results of R2=0.996 for T1, 0.997 for T2, which used the same ISMRM/NIST MRI system phantom (12). R2 of T2 values was slightly lower in our measurements, which may be explained by the fact that B1 varies from day to day, thereby inducing greater variation in T2 values compared to T1 values (32). Moreover, it is expected that B1 inhomogeneity on 3.0 T would affect the accuracy of relaxation property mapping with more bias on estimated T2 than T1 (2,33).
High repeatability is paramount for any imaging techniques, such as quantitative MRI, that is intended for routine clinical use (6). It is, therefore, imperative that the repeatability of MRF for cervical imaging be evaluated. Our results showed that the wCV and ICC of MRF scan-rescan were all excellent for both T1 and T2 values, concordant with recent MRF studies in brain, breast and knee (8-10,13,34).
Considering that CC could arise from the endocervical canal, commonly found in the elderly population as the squamocolumnar junction recedes within the cervical canal, and could present as endophytic growth (35), we have performed separate analyses on mucosa and stroma to better understand the underlying changes in cervical tissues when CC develops. Our results showed that the T1 value of CC was significantly higher than that of normal mucosa and stroma, and the T2 value of CC was lower than that of normal mucosa. Amongst the factors that could affect T1, variations in the concentration of mucin, a T1-shortening macromolecule, may contribute to a lower T1 value in normal cervical tissues than CC (36). The level of mucin production is variable in both normal cervix and CC. In normal cervical tissues, the mucin level of endocervical secretion changes within menstrual cycle (37). However, we did not investigate the effect of menstrual cycle and menopausal status on T1 and T2 values due to the limited sample size. In CC, mucin production is decreased in squamous cell carcinomas and increased in adenocarcinoma and adenosquamous carcinoma (38). As the majority of our cohort were squamous cell carcinomas, T1 values of CC would be expected to be higher than normal cervical tissues. These findings suggested that differences in mucin content may explain the variation in T1 values in cancerous and normal cervical tissues, though further investigations are required to elucidate the cause of T1 changes.
For T2 values, that of CC was lower than normal mucosa, and not different from that of normal stroma. The normal stroma consists of fibromuscular tissues; the inner stroma contains a large amount of fibroblasts and smooth muscle cells, whereas the outer stroma has lower concentration of smooth muscle cells. The signal difference between inner stroma and outer stroma is conspicuous in young women and may be absent in the elderly (39). In our study, the majority of the healthy volunteers were young women, potentially affecting the T2 values of normal stroma. Furthermore, high vascularisation in the outer stroma and inflow phenomena may attribute to an increased T2 value (40). Both factors could be accounting for the similar T2 values between normal stroma and CC. Nevertheless, the lack of difference in T2 values of CC and normal stroma may suggest MRF-derived T2 value may have limited value in differentiating the normal stroma that surrounds the CC and could impact on longitudinal comparison during and after treatment.
To determine whether CC can be distinguished from normal cervical tissues based on T1 and T2 values, we have performed ROC analysis. The AUCs for T1 and T2 were acceptable to outstanding, demonstrating their potential in differentiating CC from normal cervical tissues. These results concurred with findings in previous MRF studies in other tumour types which demonstrated that MRF could distinguish cancerous and normal tissues in the prostate and breast (20-22). It is, however, worth noting that the variations in T1 and T2 values in different tissue types were different compared to other studies, indicating that these variations may likely be organ-specific and disease-dependent.
Our study has several limitations. First, we did not perform a comparison in the T1 and T2 values between conventional quantitative MRI versus MRF. However, we have demonstrated good agreement between MRF and phantom reference values. A number of studies have reported high agreements in T1 and T2 values between MRF and conventional quantitative MRI (2,17,18). Second, our current MRF model assumed that all tissues are static and does not account for fluid flow (3). The present MRF sequence uses 2D slice selective excitation and the presence of fluid flow across the bladder renders the estimation of relaxation times in this region unreliable. Third, scan-rescan repeatability was only conducted on healthy volunteers; further investigation of scan-rescan for patients will be needed. Fourth, the interval between scan-rescan may not be sufficient in a clinical scenario. However, there have been previous MRF studies that used the same length interval (4,10); more clinically appropriate time intervals will be explored in follow-up studies. Fifth, all the MRF sequences were conducted on a single 3T MRI scanner, reproducibility of MRF for cervical imaging on multiple scanners would be necessary to evaluate the transferability to other MRI units and vendors in the future. Sixth, the sample sizes of both patients and healthy volunteers were limited, larger cohorts would be needed to validate our results. Lastly, patients with both SCC and ACA were recruited and the healthy volunteers were not age-matched to the patients with CC, both these factors and variabilities may further confound our results due to the inhomogeneity of the data.
Conclusions
Our study demonstrated that the agreement of MRF with reference in phantom, scan-rescan repeatability of MRF in normal cervix were excellent, and T1 and T2 values from MRF could differentiate CC from normal cervical tissues.
Acknowledgments
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1382). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (HKU/HA HKWC IRB), IRB Reference No. UW 19-683. Informed consent was taken from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Poorman ME, Martin MN, Ma D, McGivney DF, Gulani V, Griswold MA, Keenan KE. Magnetic resonance fingerprinting Part 1: Potential uses, current challenges, and recommendations. J Magn Reson Imaging 2020;51:675-92. [Crossref] [PubMed]
- Chen Y, Jiang Y, Pahwa S, Ma D, Lu L, Twieg MD, Wright KL, Seiberlich N, Griswold MA, Gulani V. MR fingerprinting for rapid quantitative abdominal imaging. Radiology 2016;279:278-86. [Crossref] [PubMed]
- Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495:187-92. [Crossref] [PubMed]
- Lu L, Chen Y, Shen C, Lian J, Das S, Marks L, Lin W, Zhu T. Initial assessment of 3D magnetic resonance fingerprinting (MRF) towards quantitative brain imaging for radiation therapy. Med Phys 2020;47:1199-214. [Crossref] [PubMed]
- Kaggie JD, Deen S, Kessler DA, McLean MA, Buonincontri G, Schulte RF, Addley H, Sala E, Brenton J, Graves MJ, Gallagher FA. Feasibility of quantitative magnetic resonance fingerprinting in ovarian tumors for T1 and T2 mapping in a PET/MR setting. IEEE Trans Radiat Plasma Med Sci 2019;3:509-15. [Crossref] [PubMed]
- O'Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14:169-86. [Crossref] [PubMed]
- Shukla-Dave A, Obuchowski NA, Chenevert TL, Jambawalikar S, Schwartz LH, Malyarenko D, Huang W, Noworolski SM, Young RJ, Shiroishi MS, Kim H, Coolens C, Laue H, Chung C, Rosen M, Boss M, Jackson EF. Quantitative imaging biomarkers alliance (QIBA) recommendations for improved precision of DWI and DCE-MRI derived biomarkers in multicenter oncology trials. J Magn Reson Imaging 2019;49:e101-21. [Crossref] [PubMed]
- Buonincontri G, Biagi L, Retico A, Cecchi P, Cosottini M, Gallagher FA, Gómez PA, Graves MJ, McLean MA, Riemer F, Schulte RF, Tosetti M, Zaccagna F, Kaggie JD. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0 T. Neuroimage 2019;195:362-72. [Crossref] [PubMed]
- Körzdörfer G, Kirsch R, Liu K, Pfeuffer J, Hensel B, Jiang Y, Ma D, Gratz M, Bär P, Bogner W, Springer E, Lima Cardoso P, Umutlu L, Trattnig S, Griswold M, Gulani V, Nittka M. Reproducibility and repeatability of MR fingerprinting relaxometry in the human brain. Radiology 2019;292:429-37. [Crossref] [PubMed]
- Panda A, Chen Y, Ropella-Panagis K, Ghodasara S, Stopchinski M, Seyfried N, Wright K, Seiberlich N, Griswold M, Gulani V. Repeatability and reproducibility of 3D MR fingerprinting relaxometry measurements in normal breast tissue. J Magn Reson Imaging 2019;50:1133-43. [Crossref] [PubMed]
- Hamilton JI, Pahwa S, Adedigba J, Frankel S, O'Connor G, Thomas R, Walker JR, Killinc O, Lo WC, Batesole J, Margevicius S, Griswold M, Rajagopalan S, Gulani V, Seiberlich N. Simultaneous mapping of T1 and T2 using cardiac magnetic resonance fingerprinting in a cohort of healthy subjects at 1.5T. J Magn Reson Imaging 2020;52:1044-52. [Crossref] [PubMed]
- Kato Y, Ichikawa K, Okudaira K, Taoka T, Kawaguchi H, Murata K, Maruyama K, Koerzdoerfer G, Pfeuffer J, Nittka M, Naganawa S. Comprehensive evaluation of B1+-corrected FISP-based magnetic resonance fingerprinting: accuracy, repeatability and reproducibility of T1 and T2 relaxation times for ISMRM/NIST system phantom and volunteers. Magn Reson Med Sci 2020;19:168-75. [Crossref] [PubMed]
- Ma D, Jones SE, Deshmane A, Sakaie K, Pierre EY, Larvie M, McGivney D, Blümcke I, Krishnan B, Lowe M, Gulani V, Najm I, Griswold MA, Wang ZI. Development of high-resolution 3D MR fingerprinting for detection and characterization of epileptic lesions. J Magn Reson Imaging 2019;49:1333-46. [Crossref] [PubMed]
- Keil VC, Bakoeva SP, Jurcoane A, Doneva M, Amthor T, Koken P, Mädler B, Block W, Fimmers R, Fliessbach K, Hattingen E. MR fingerprinting as a diagnostic tool in patients with frontotemporal lobe degeneration: a pilot study. NMR Biomed 2019;32:e4157 [Crossref] [PubMed]
- Badve C, Yu A, Dastmalchian S, Rogers M, Ma D, Jiang Y, Margevicius S, Pahwa S, Lu Z, Schluchter M, Sunshine J, Griswold M, Sloan A, Gulani V. MR fingerprinting of adult brain tumors: initial experience. AJNR Am J Neuroradiol 2017;38:492-9. [Crossref] [PubMed]
- Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med 2015;74:1621-31. [Crossref] [PubMed]
- Hermann I, Chacon-Caldera J, Brumer I, Rieger B, Weingärtner S, Schad LR, Zöllner FG. Magnetic resonance fingerprinting for simultaneous renal T1 and T2* mapping in a single breath-hold. Magn Reson Med 2020;83:1940-8. [Crossref] [PubMed]
- Jaubert O, Arrieta C, Cruz G, Bustin A, Schneider T, Georgiopoulos G, Masci PG, Sing-Long C, Botnar RM, Prieto C. Multi-parametric liver tissue characterization using MR fingerprinting: simultaneous T1, T2, T2*, and fat fraction mapping. Magn Reson Med 2020;84:2625-35. [Crossref] [PubMed]
- Cloos MA, Knoll F, Zhao T, Block KT, Bruno M, Wiggins GC, Sodickson DK. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun 2016;7:12445. [Crossref] [PubMed]
- Chen Y, Panda A, Pahwa S, Hamilton JI, Dastmalchian S, McGivney DF, Ma D, Batesole J, Seiberlich N, Griswold MA, Plecha D, Gulani V. Three-dimensional MR fingerprinting for quantitative breast imaging. Radiology 2019;290:33-40. [Crossref] [PubMed]
- Panda A, Obmann VC, Lo WC, Margevicius S, Jiang Y, Schluchter M, Patel IJ, Nakamoto D, Badve C, Griswold MA, Jaeger I, Ponsky LE, Gulani V. MR fingerprinting and ADC mapping for characterization of lesions in the transition zone of the prostate gland. Radiology 2019;292:685-94. [Crossref] [PubMed]
- Yu AC, Badve C, Ponsky LE, Pahwa S, Dastmalchian S, Rogers M, Jiang Y, Margevicius S, Schluchter M, Tabayoyong W, Abouassaly R, McGivney D, Griswold MA, Gulani V. Development of a combined MR fingerprinting and diffusion examination for prostate cancer. Radiology 2017;283:729-38. [Crossref] [PubMed]
- Shiradkar R, Panda A, Leo P, Janowczyk A, Farre X, Janaki N, Li L, Pahwa S, Mahran A, Buzzy C, Fu P, Elliott R, MacLennan G, Ponsky L, Gulani V, Madabhushi A. T1 and T2 MR fingerprinting measurements of prostate cancer and prostatitis correlate with deep learning-derived estimates of epithelium, lumen, and stromal composition on corresponding whole mount histopathology. Eur Radiol 2021;31:1336-46. [Crossref] [PubMed]
- Kurman RJ, Carcangiu ML, Herrington CS, Young RH. editors. WHO classification of tumours of female reproductive organs. 4th ed. Lyon: International Agency for Research on Cancer, 2014.
- Bhatla N, Berek JS, Cuello Fredes M, Denny LA, Grenman S, Karunaratne K, et al. Revised FIGO staging for carcinoma of the cervix uteri. Int J Gynaecol Obstet 2019;145:129-35. [Crossref] [PubMed]
Michigan Image Reconstruction Toolbox (MATLAB) 2020 . Available online: https://web.eecs.umich.edu/~fessler/code/- Weigel M. Extended phase graphs: dephasing, RF pulses, and echoes - pure and simple. J Magn Reson Imaging 2015;41:266-95. [Crossref] [PubMed]
- Vardhanabhuti V, Au HT, Ding J, Lee EY, Cao P, Hui ES. Repeatability of magnetic resonance fingerprinting using ISMRM/NIST MRI phantom in Philips 3T MRI Scanner. 2020. Available online: https://archive.ismrm.org/2020/3760.html
- Jones R, Brian P. Clinical investigation and statistics in laboratory medicine. London: ACB Venture Publications, 1997.
- Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 2016;15:155-63. [Crossref] [PubMed]
- Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol 2010;5:1315-6. [Crossref] [PubMed]
- Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med 2017;78:1452-7. [Crossref] [PubMed]
- Brink WM, Gulani V, Webb AG. Clinical applications of dual-channel transmit MRI: a review. J Magn Reson Imaging 2015;42:855-69. [Crossref] [PubMed]
- Sharafi A, Zibetti MVW, Chang G, Cloos M, Regatte RR. MR fingerprinting for rapid simultaneous T1, T2, and T1ρ relaxation mapping of the human articular cartilage at 3T. Magn Reson Med 2020;84:2636-44. [Crossref] [PubMed]
- Okamoto Y, Tanaka YO, Nishida M, Tsunoda H, Yoshikawa H, Itai Y. MR imaging of the uterine cervix: imaging-pathologic correlation. Radiographics 2003;23:425-45; quiz 534-5. [Crossref] [PubMed]
- Lee NK, Kim S, Kim HS, Jeon TY, Kim GH, Kim DU, Park DY, Kim TU, Kang DH. Spectrum of mucin-producing neoplastic conditions of the abdomen and pelvis: cross-sectional imaging evaluation. World J Gastroenterol 2011;17:4757-71. [Crossref] [PubMed]
- Lapertosa G, Baracchini P, Fulcheri E, Tanzi R. Patterns of mucous secretion in normal and pathological conditions of the endocervix. Eur J Gynaecol Oncol 1986;7:113-9. [PubMed]
- Hüsniye Dilek F, Küçükali T. Mucin production in carcinomas of the uterine cervix. Eur J Obstet Gynecol Reprod Biol 1998;79:149-51. [Crossref] [PubMed]
- Lien HH. MR imaging of invasive carcinoma of the uterine cervix. Acta Radiol 1999;40:236-45. [Crossref] [PubMed]
- deSouza NM, Hawley IC, Schwieso JE, Gilderdale DJ, Soutter WP. The uterine cervix on in vitro and in vivo MR images: a study of zonal anatomy and vascularity using an enveloping cervical coil. AJR Am J Roentgenol 1994;163:607-12. [Crossref] [PubMed]


