
Research on obtaining pseudo CT images based on stacked generative adversarial network
Introduction
As one of the most common gynecological tumors, cervical cancer seriously endangers women’s health and lives (1,2). The current treatment methods include surgery or a combination of radiotherapy and chemotherapy, of which radiotherapy is suitable for the treatment of various cancer stages, and its therapeutic effect has been confirmed. Radiotherapy is becoming increasingly accurate due to the gradual advancement of precision techniques from two-dimensional (2D) traditional to image-guided radiotherapy.
Image guided devices used in the medical linear accelerator for the radiotherapy of cervical cancer include an electronic portal imaging device (EPID), cone beam CT (CBCT), and ultrasound (US). EPID is a two-dimensional image guidance device with the advantages of fast scanning speed and convenient operation, but the resolution of soft tissue is low, and its MV level energy increases radiation exposure (3,4). CBCT image guidance mainly uses rigid or affine registration between the CT image obtained in the simulation stage and the CBCT image obtained in the treatment stage based on bony landmarks or gray image information to verify positioning between the fractional treatment. However, it too has the disadvantage of the low resolution of soft tissue and radiation, and the artifacts caused by the radiation hardening effect and electron scattering reduce the image quality after image reconstruction (5-7). Compared with EPID- and CBCT-based image-guided devices, US-based image-guided devices offer the advantages of portability, non-radiation, and real-time operation, while effectively locating the position of the applicator (8) and have a good effect in the application of cervical cancer radiotherapy (9). Presently, the US can only assist CBCT images or independently carry out positioning verification before radiotherapy and cannot formulate and modify radiotherapy plans directly on US images. Therefore, in this study, we introduced a pseudo CT image synthesis technology based on US images to solve the problems mentioned above.
The conventional methods for obtaining pseudo CT images based on US images are mainly divided into two categories, the deformation-field registration method, and the image-synthesis method. The deformation field is obtained via registration between the US images scanned in the simulation stage, and those scanned in the treatment stage, and it is applied to the CT images scanned in the simulation stage to obtain the virtual CT image (10-12). However, the deformation field obtained using this method is discontinuous, resulting in the inaccurate connection of tissues and organs at the critical points of different regions of interest (ROIs). Although this problem can be solved through smooth filter processing, it reduces the image definition, yet the deformation-field method’s limitation can be overcome via image synthesis. The image-synthesis method involves the use of machine-learning or deep-learning algorithms to extract the image features of CT and other modal images, pair the features of the same region, consider the matching results as the training set, add the images of other modes in the test phase, and predict the corresponding pseudo CT images. Presently, scholars mostly use magnetic resonance imaging (MRI) and positron emission tomography (PET) images to synthesize pseudo CT images, and existing research on the synthesis of pseudo CT images based on US images is not profound (13-15).
Owing to the increased prevalence of deep learning, Gatys et al. proposed the neural style transfer (NST) method based on the convolutional neural network (CNN) and has been gradually used in image synthesis. The working principle of the method is to define the image-content-loss function and style-loss function. Simultaneously, both the content abstract-feature and style abstract-feature of two kinds of modal images are extracted using the deep-learning network model. Subsequently, one of the modal images is used as the reference image, and the other is reconstructed via iterative optimization (16). This method utilizes the advantage of CNNs to extract high-level features of images, artificially separates image style and content, and performs iterative optimization. Following Johnson et al. proposed the image method named fast NSF, wherein the forward convolution neural network replaced back propagation to expedite the optimization of the model to achieve real-time image-style transfer (17). Although the fast NSF method can stylize images in real time using GPU, owing to obvious differences among the styles of different modal images, a separate model must be trained for each style in advance, causing many manual operations to suffer from significant limitations. However, because the NSF method takes the pixel of the image as input, it judges the loss of each pixel one by one in the pixel space then averages the total loss of all the pixels to obtain blurred synthetic images. A method based on the generative adversarial network (GAN) model initially proposed by Goodfellow et al. has been developed for medical-image synthesis to achieve satisfactory results (18-21). This method utilizes the idea of confrontation to learn the generative model, automatically defines the potential loss function, and learns the original real sample set’s data distribution so that the sample distribution generated using the generator can approach the real-number sample distribution and possesses satisfactory generalization ability. As the training mode is antagonistic, the demand for training samples for the model is considerably reduced (22). The methods of synthesizing medical images based on GAN are mainly divided into supervised and unsupervised methods. The former method is based on the GAN model that contains conditional information. By learning the correspondence between the source images and target images, the mapping method of GAN through random vector is changed (23). Ben-cohen et al. established a model to synthesize PET images obtained by abdominal CT using the conditional GAN (cGAN) that contained the full convolutional network (FCN). The experimental results showed that the model could detect malignant tumors in the liver region and that it could replace the conventional PET–CT scan as the evaluation image for estimating the efficacy of drug treatment (24).
The latter is based on a coupled GAN model, which searches for the nonlinear mapping relationship between the two image domains or the nonlinear mapping relationship from different image domains to the same shared potential intermediate domain. The weight sharing, image reconstruction, and cyclic consistency constraints are used to replace the constraints on the synthesized images’ contents, thereby limiting the learning range of the generator. Owing to the lack of explicit supervision information in the training process, this method can be used to synthesize unpaired source images and target images. Usually, the DualGAN or CycleGAN models are used in this process (25-28). Wolterink et al. used the CycleGAN model to train unpaired brain CT and MRI data by using a pair of mirror-symmetric GANs to establish a mapping relationship between the two modal images’ data. The results showed that the pseudo CT images synthesized using the model were similar to real brain CT images in terms of anatomical structure (29). Also, Wang et al. added a deform convolutional network layer to the classical CycleGAN generator. They normalized the mutual information loss to the entire circulation network’s loss function, thereby proving that the improved CycleGAN could be used for the mutual synthesis of the brain or abdominal MRI images and CT images with inconsistent imaging range (30). However, in CycleGAN-based image synthesis, the mapping difficulty between the two modal images is not consistent; therefore, it is difficult to completely reverse the entire cyclic network, which complicates the total objective function’s optimization. On this basis, the synthesis model between the two modal images also requires more time for training.
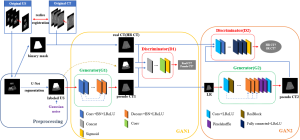
A single GAN cannot effectively process tasks, so we used Zhang et al.’s idea to synthesize high-resolution pseudo images based on stacked generative adversarial network (sGAN), and used multiple GANs to independently solve different tasks in the image-synthesis process (31). In this study, the synthetic process of pseudo CT was divided into two stages. In the first stage, labeled US images were input to the improved cGAN, and the corresponding mapping relationship with the CT image was established. This meant that both the main texture information and grayscale information of the target images were obtained, and low-resolution (LR) blurred pseudo CT images were synthesized. In the second stage, the LR blurred pseudo CT images obtained in the first stage were taken as the input of the super-resolution GAN (SRGAN). Also, ResNet and FCN were used to correct the results of the first stage obtaining pseudo CT images with clear texture and accurate grayscale information.
Methods
To the best of our knowledge, this is the first application of GANs to synthesize US and CT images. We adopted cGAN in the supervised mode for image synthesis. A considerable advantage to this method is that by simultaneously conducting both procedures, the patient does not have to be moved. This means the relative position of the abdominal organs, including organs at risk (OARs), remains the same, reducing image guidance error and satisfying conditions for pairing training sets in image synthesis. In this study, the labeled US images obtained using the U-Net segmentation method were used as the input of cGAN. Also, pseudo CT images were synthesized using the generator of the encoder–decoder model that was based on both residual networks (ResNet) and the discriminator of FCN. Subsequently, the improved SRGAN was used further to enhance the imaging quality of the pseudo CT images, and the 3D pseudo CT images were synthesized using a reconstruction algorithm. The flow chart of the synthesis of pseudo CT images is depicted in Figure 1.
Data acquisition and image preprocessing
The image data selected in the experiment were all 3D volume data of preoperative cervical cancer patients under Volumetric Modulated Arc Therapy (VMAT), of which 75 cases were used for model training and 10 used for model testing. The US and CT images obtained were collected during the patients’ normal treatment, and informed consent was obtained from all individual participants. The ethics association approved the study of the Second People’s Hospital of Changzhou, Nanjing Medical University (2017-002-01). The CT images were obtained by Optima CT520 produced by the GE Corporation (United States). The scanning conditions were as follows: tube voltage 120 kv, tube current 220 mA, image size 512×512×(196–239), voxel spacing 0.97653×0.97653×1 mm3. The CT images of 85 patients were scanned with the Clarity Ultrasound device produced by the Elekta Corporation (Sweden). The scanning conditions were as follows: C5-2/60 US probe, center frequency 3.5 MHz, image size 400×400×(220–295), and voxel spacing 1×1×1 mm3. Each patient was required to empty their bladder then 300 mL of water was delivered through a catheter to ensure the bladder was full before scanning. Before collecting US data, technicians calibrated the indoor laser lamp coordinate system and US probe with a calibration phantom. The 3D US probe has an infrared light refraction device similar to the shape of Octopus which reflects infrared light through eight different points and is received by the infrared device above the scanning bed. The position of the probe, which represents the position of the scanned single frame image, is calculated according to its infrared receiving frequency. The 3D US is then reconstructed by interpolation based on each frame image’s position by clarity system image. These calibrations ensure that the coordinate position of each 3D US voxel collected by the system is known relative to the indoor laser light coordinate system. When collecting patients’ US data, the US probe is installed on the self-developed ultrasonic robotic arm, which can avoid the pressing error caused by manual data acquisition.
A US device is different from a CT device in terms of its scanning conditions and imaging principles. The original US and CT images required preprocessing before they could be used for pseudo image synthesis. While the advantage of US imaging is that it can satisfactorily distinguish the contour and relative position of OARs, the image noise is high, and it does not form a linear correction with the grayscale information of the CT image. This means it cannot be directly used in the synthesis of pseudo CT images and the region of the US image must be divided on the basis of semantic information. In addition, we performed affine registration between the original 3D US images and CT images to ensure their sizes and resolutions were consistent. The size of the processed US and CT image data sets was 256×256×100, and the voxel resolution was 1×1×1 mm3. Subsequently, we created labeled US images using the semantic information as the input of cGAN. The labeled US images refer to the results obtained by labeling different OARs in the original US images based on the image segmentation method. Each labeled region’s grayscale value is a certain number, but the values are different among different labeled regions. The image quality of the original US images was low, and the grayscale information in the images could not form a linear correlation with tissue density. Therefore, we referred to the method of Isola et al. (32), which sees the original US images replaced by the labeled US images with Gaussian noise as the input of the sGAN model. The labeled US images retain the edge information of tissues and organs and have relative position information between different OARs. There were four acquisition steps in the labeling of US images. In the first step, the ImageJ software was used to reslice the 2D slices of the 3D US and CT images after registration to ensure that the 2D slices of the two modal images within the registration range were aligned one by one. The 3D volume data was then divided into 100 2D layers. In the second step, binary masks were made, and the 2D US and CT images of the same slice were multiplied by pixels to obtain the overlapping region. The pixel value of the overlapping region was set to 1, while the remaining region was the background, whose pixel value was set to 0. The third step saw the binary masks obtained in the second step, respectively processed with the original US and CT images for pixel calculation. The redundant pixels in the original images were then removed to ensure that the two modalities’ imaging regions were the same. Using the U-Net method, the US images were segmented into multi-target regions to obtain labeled US images in the fourth step. Also, to reduce the bladder region’s deformation due to US probe compression, we connected the upper two vertices of the bladder region in the US images to compensate for the partial absence of the region from the images. The flow chart of the acquisition of the labeled US images is depicted in Figure 2.
Supervised deep conditional convolutional adversarial network
CNN with ResNet has been proven to perform satisfactorily in a variety of image-processing tasks (33-35). Through identity shortcut connection, the network’s output is expressed as a linear superposition between the input and a nonlinear transformation thereof. Notably, the most significant difference between ResNet and direct-connected CNN is that the former provides some protection to information integrity because it can directly transmit it from input to output along with the shortcut. This makes the target of model learning more concise and solves the problem of gradient disappearance in the training process (36).
The generator is an improved U-Net to which a residual connection has been added. This means each deconvolution layer’s input is the linear superposition between the previous layer’s output and the output of the symmetric convolutional layer, effectively doubling the feature maps (37). This method retains the image feature information at different resolutions, ensuring the encoder’s feature information can be continuously re-remembered during the decoder operation and that the synthesized pseudo images can retain the feature information of the original input images to the greatest extent. Also, z is the input of random Gaussian noise distribution. The parameters of the network model after training have been determined. If the inputs are the labeled US images without adding noise, the output results of pseudo CT images will be unchanged, making them different from the real CT images. The structure of the generator with residual connections is depicted in Figure 3.
The input of the network is the labeled US images. The nodes from each network layer in each encoder part to the next include the convolution layer, batch-normalization (BN) layer, and LReLU activation layer. Also, each node to the next in the decoder part includes the deconvolution layer, BN layer, and ReLU activation layer. The network layers of both the encoder and decoder are symmetrically distributed, and the loss function of the generator includes content loss and counter loss. The mean square error (MSE) loss function can be used to train the network to obtain a high peak signal-to-noise ratio (PSNR), so the calculation of the MSE is determined by the MSE (LMSE) between the pseudo CT images [G(x, z)] synthesized using the US images (x) and ground truth CT (CTgt) images (y), where z is the noise. Calculation of the PSNR uses the binary cross entropy (LBCE) as the loss where α is the LBCE loss term’s weight coefficient in the loss function. In following the experimental results of Isola et al., we set this to 0.5 (32). The loss function of the generator is given as follows:
[1]
This study also used a Markov discriminator, an FCN that comprises five convolutional layers and one Sigmoid activation layer. The input passes through one node, including the convolution and LReLU activation layers, then through three nodes, including the convolution, BN, and ReLU activation layers, and the output is finally obtained through the convolution layer and fully connected layer. The discriminator’s output size is a probability matrix with the dimensions of 30×30×1 mm3. The result of averaging the matrix is the final output of the discriminator. This architecture can make the model retain more image details during training. The network structure of the discriminator is depicted in Figure 4.
To calculate the loss function, the discriminator includes two inputs and two corresponding outputs. It takes as one input the labeled US image (x) and pseudo CT image [G(x, z)] generated using the generator to obtain the output {D[x, G(x,z)]} and takes as another output the labeled US image (x) and CTgt images (y) to obtain the output [D(x,y)]. The loss function of the discriminator is given as follows:
[2]
Super-resolution reconstruction of pseudo CT images
LR CT images are converted into high-resolution ones via certain algorithms, and the resulting images are super-resolution CT images. Super-resolution CT images have high pixel density and detailed information. As the resolution of the pseudo CT images obtained using cGAN was not high, their texture information was blurred, so we used the SRGAN to obtain more abundant texture information of the pseudo CT images.
The generated network contained five residual blocks, and each contained two convolution layers with a kernel size of 3×3. Each convolution layer was connected to a BN layer and PReLU activation layer. Subsequently, four deconvolution layers were connected, and each contained deconvolution, BN, and ReLU activation operations. The second to fourth deconvolution layers’ inputs were the linear superposition of the output of the previous layer and output of the first deconvolution layer. Subsequently, two Pixelshuffle layers and one deconvolution layer were connected, and the Pixelshuffle layer improved the image resolution via sub-pixel operation. The feature maps of r2 channels were obtained via convolution, then high-resolution pseudo CT images were obtained via periodic screening, wherein r was the upscaling factor, i.e., the expansion rate of the images (38). The network structure of the generator is depicted in Figure 5.
The loss function of the SRGAN generator also includes content loss and adversarial loss. The content loss is the MSE (LMSE) between the super-resolution pseudo CT image [G(a)] synthesized using the LR pseudo CT image and CTgt image (b). The adversarial loss is the binary cross entropy (LBCE) between probability 1, and probability D[G(a)] that G(a) is identified as a real image (39) and β is the weight coefficient of LBCE loss term in the loss function. This is an empirical value, which we also set to 0.5. The loss function of the generator is given as follows:
[3]
The discriminator is a CNN with 10 convolution layers, three fully connected layers, and one sigmoid activation layer. The convolution layer includes convolution, BN, and LReLU activation operations. The first two fully connected layers use LReLU as the activation function, and the third fully connected layer uses the Sigmoid activation function to obtain the judgment result of the discriminator. For a convolutional layer, the size of the convolutional kernel is 3×3; the number of convolutional kernels is 32, 32, 64, 128, 128, 256, 512, and 512; the step size is 1, 2, 1, 2, 1, 2, 1, 2, and 2, and the Padding is SAME. The output obtained after convolution is reconstructed into a 1D tensor and taken as the fully connected layer’s input. Also, the number of nodes in all three fully connected layers is 1024, 512, and 1, respectively. The structure of the discriminator is depicted in Figure 6. This discriminator also includes two inputs and two outputs. It takes as one input the G(a) of the super-resolution pseudo CT image generated using the generator to obtain the output D[G(a)], and takes b (CTgt image) as another input to obtain the output D(b). The loss function of the discriminator is given as follows:
[4]
Technical details
All networks are optimized with the Adam optimizer, where cGAN is trained from scratch with a learning rate of 10−6 with a batch size of 1. SRGAN is trained from scratch with a learning rate of 10−4, and the batch size is also 1. The two GAN networks’ epochs are both 100, and the code is implemented using the TensorFlow library. We performed experiments on a personal computer equipped with a GPU (Nvidia GeForce RTX 2080Ti) and a CPU (Intel Core i9-9900K).
Evaluation
To verify the model’s performance, a five-fold cross-validation technique was used in the training and testing steps. The 75 cases were randomly divided into five groups, and four groups (including 60 cases) were selected for each experiment to test the trained model. After training, the model was applied to the CBCT images of the remaining 15 subjects to generate pseudo CT. To verify the accuracy of the pseudo CT images obtained based on the improved sGAN, pseudo CT synthesized based on the NSF and CycleGAN methods were selected as control experiments, and the CTgt was the actual CT image.
The accuracy of the HU value of pseudo CT and real CT of each subject is evaluated by calculating the mean absolute error (MAE) of voxels in the pelvic region:
[5]
where N is the total number of voxels in the CT pelvic region. CTre is a real image scanned by a CT machine, and CTps is the pseudo CT obtained by different methods. The smaller the MAE value, the closer the pseudo CT images’ HU value to the real CT images.
To test the results of 10 cervical cancer patients in the testing phase, the accuracy of the image-synthesis algorithm was evaluated using the following three quantitative metrics: NMI (40), SSIM (41), and PSNR (42).
The first metric is NMI, which is used to evaluate the similarity between CTgt images and pseudo CT images, and was obtained using different methods. The function expression is as follows:
[6]
[7]
where I(CTgt,CTps) denotes the mutual information value between the pseudo CT and CTgt images, and H(CTgt) and H(CTps) denote the information entropy. The closer the NMI metric value is to 1, the better the registration result.
The second metric is SSIM, and its mathematical definition is as follows:
[8]
where
The mathematical definition of the PSNR measurement method is as follows:
[9]
where Igt and Ips denote the CTgt and pseudo CT images, respectively. Also, X, Y, and Z denote the size of the image, and MAXI denotes the maximum grayscale value of the CT images. The lower the MAE value and higher the PSNR value, the more similar the synthetic pseudo CT image is to the CTgt image. The calculation of the three measurements mentioned above methods is related to the grayscale of CT images, and the grayscale range of the CT images is unified into the range of [0, 4,000] (43).
The DSC was used to evaluate bladder and uterine regions’ segmentation accuracy in the pseudo CT images (44). In this study, the distinct curve guided FCN proposed was used to segment the OARs in the pelvic region of the pseudo and real CT images (45). Calculating the volume overlap of bladder and uterus between CT images, the accurate segmentation results should have a high organ volume overlap rate. Notably, DSC is given as follows:
[10]
where
To verify the accuracy of the pseudo-CT image based on US image synthesis in dose calculation, this study conducted a phantom experiment and selected real scanned CT images as the ground truth images for dosimetry verification. Medical physicists define and outline the planning target volume (PTV) and organs at risk (OARs) on CTgt images and copy them to the pseudo CT images synthesized based on different deep learning methods. The VMAT radiotherapy plan was made on the CTgt images, and the Monte Carlo algorithm calculated a dose of 4,500 cGy/25 F. After the optimized plan met the clinical requirements, the plan was copied to different pseudo CT images. To compare the difference between pseudo CT and CTgt images in the radiotherapy plan, the 4,500 cGy prescription dose with 95% PTV was used as the plan’s passing criterion. The doses in PTV and OARs of cervical cancer patients, which were obtained based on pseudo CT and CTgt images under the same optimization conditions of VMAT treatment in the Monacao planning system, were compared. Dosimetry evaluation indexes mainly include dose volume histogram (DVH), maximum dose (Dmax), average dose (Dmean), and minimum dose (Dmin).
Results
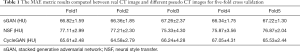
Table 1 shows the MAE measurement results between the real CT images and the pseudo CT images synthesized by different methods. Compared with NSF’s method, sGAN has a lower MAE value, which indicates that the pseudo CT image synthesized by SGAN is closer to the real CT image. Compared with CycleGAN, SGAN does not have obvious advantages, although on the premise of ensuring image quality, the SGAN method has more stable training results than the CycleGAN method and is also universal for training volume data.
Full table
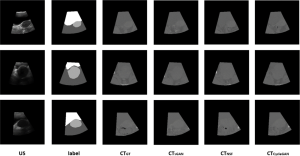
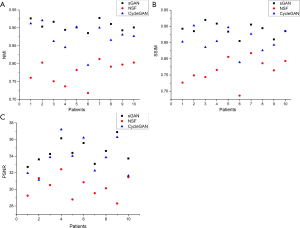
Figure 7 depicts the pseudo CT image obtained using three different image-synthesis methods. Although the NSF-based method can roughly obtain pseudo CT images, it blurs the edge information between different tissues and organs, resulting in the mismatching of CT values in different imaging regions. Compared with the NST method, the pseudo CT images obtained using CycleGAN are more accurate; however, the resulting images are not stable because of the difference between the two modal images to be synthesized. If the difference is considerable, the synthesized image data is poor; otherwise, the result is satisfactory. The sGAN method proposed in this study ensures that each organ’s relative position in the image remains unchanged and provides accurate organ contour and grayscale information. Also, the result of image synthesis is relatively stable, indicating a more favorable outcome compared with the other methods. The NMI, SSIM, and PSNR measurement results for the pseudo CT images synthesized using three different image-synthesis methods, and CTgt images are depicted in Figure 8. According to the comparison results, the CTgt and pseudo CT images obtained using the method proposed in this study are significantly different from those obtained using the other two methods (tNMI(sGAN-NSF) =13.594, tNMI(sGAN-CycelGAN) =3.053, tSSIM(sGAN-NSF) =10.621, tSSIM(sGAN-CycelGAN) =2.914, tPSNR(sGAN-NSF) =8.999, tPSNR(sGAN-CycleGAN) =2.530, and P<0.05).
The difference between the contour of the OARs in the CTgt image and pseudo CT image is depicted in Figure 9. The contour is obtained via the automatic segmentation method of region growth. The yellow region in Figure 9A represents the contour of the OARs in the pseudo CT image obtained using SGAN; the blue region in Figure 9B represents the contour of the OARs in the pseudo CT image obtained using NSF, and the white region in Figure 9C represents the contour of the OARs in the pseudo CT image obtained using CycleGAN. The red region in each of these figures represents the contour of the OARs in the CTgt image, and the closer the red and other-color contour regions are, the more similar the regions are to one another. Table 2 lists the registration accuracy of the OARs evaluated via DSC values between CTgt and pseudo CT images synthesized using different methods. Compared with the other two methods, the registration results of the OARs between these images obtained using the sGAN method have a higher DSC value.

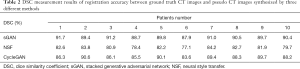
Full table
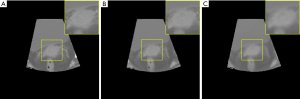
Also, to prove the effectiveness of SGAN in synthesizing pseudo CT images compared with cGAN alone, we compared the pseudo CT image results obtained by sGAN and cGAN separately, and the comparison results are shown in Figure 10. Figure 10A shows the real CT images; Figure 10B shows the pseudo CT images obtained based on sGAN, and Figure 10C shows the pseudo CT images obtained based on cGAN. The yellow square is a local enlarged image. This demonstrates that cGAN alone cannot obtain high-resolution pseudo CT images, and the results need to be input into SRGAN for resolution reconstruction, to obtain a more clear texture and better visual effects.
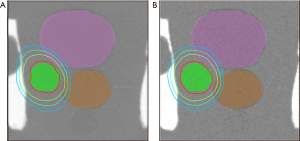
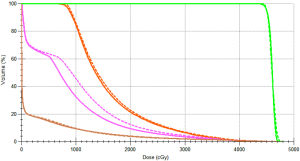
We used an ultrasound phantom for dosimetry verification. The dose distribution results of real CT and pseudo CT images obtained based on the SGAN method are shown in Figure 11, respectively. The green region is PTV, the pink and orange regions are OAR1 and OAR2, respectively, and the brown line is the outer contour. In Figure 11, the dose lines around the PTV region are 4,500 cGy, 4,000 cGy, 3,500 cGy, and 3,000 cGy from inside to outside. This shows that the dose distribution of CTgt and pseudo CT obtained based on sGAN is different, and the pseudo CT has a better conformal degree. The difference is that the relative position between OARs and PTV in the synthesized pseudo CT image is different from that of CTgt. Figure 12 shows the comparison results of the DVH dose difference of the same OARs and PTV in different CT images. The solid line is CTgt, and the dashed line is the pseudo CT obtained based on sGAN. This shows that with the exception of OAR1, the remaining dose lines in the DVH chart have relatively small differences, and their specific dose differences are shown in Table 3. Table 3 also lists the Dmax, Dmean, and Dmin values of PTV and OARs under pseudo-CT and CTgt radiotherapy plans. The difference in most index values between the two CT images is within 50 cGy.
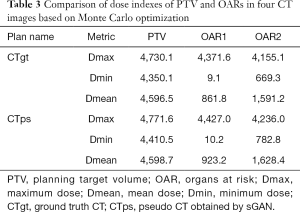
Full table
Discussion
In this study, 3D US and CT images were registered in the image pre-processing stage, and the 2D images were rearranged according to the patient’s axial direction. Therefore, in training data, the target was to search for the mapping relationship between continuous 2D images in ultrasound and CT image domains, reducing the problem of mismatch during training. Also, following the pseudo CT image synthesis task based on sGAN, we use 3D image reconstruction technology to reconstruct a 2D pseudo CT image sequence. The reconstructed 3D volume data were compared with ground truth CT images (real scanning CT), which showed good similarity in anatomy and dosimetry. From the experimental results, it is evident that compared with other methods, sGAN can more effectively synthesize pseudo CT images. Within the same imaging range, the organs’ anatomical structure in the pseudo CT image obtained using the sGAN method was more similar to that in the CTgt image. The sGAN method used the image-synthesis network and the image super-resolution reconstruction network and trained these different GAN networks separately, thereby reducing the complexity of the total loss function and obtaining satisfactory training results in the local network. In the cGAN-based image synthesis, labeled images based on the US images were added to constrain the synthetic region of different organs in the pseudo CT images. Simultaneously, the content of different labels was locally matched with the corresponding region in the target CT image to avoid semantic mismatch. In the SRGAN-based image synthesis, a series of residual blocks and skip connection were added to the generator to reconstruct the LR pseudo CT image and further improve its texture clarity. The pseudo CT images obtained using the sGAN method based on the two networks were similar to the real CT images. Also, the NSF method suffers from semantic-mismatching problems, such as background-texture migration of the style image to the foreground of the target image without maintaining semantic consistency during the transfer process, and no global linear matching relationship exists between US image and CT image (46). Therefore, the pseudo CT images obtained using this method differed from the CTgt images in terms of grayscale information and anatomical structure. Also, CycleGAN achieved satisfactory results in unsupervised image-synthesis tasks. However, for the image synthesis based on the complexity of different textures and anatomical structures, the consistent loss coefficient of the cycle loss used by CycleGAN was invariant. Therefore, although the grayscale information of the pseudo CT synthesized using this method was similar to that of the CTgt, it could not effectively and stably synthesize the pseudo CT image. When OARs are irregular and texture information complex, the pseudo CT image synthesized using this method would be distorted, and the contour of the organs within the imaging range show greater deviation from the real image.
In the cGAN- based image synthesis, the generator’s structure was designed as the encoder–decoder mode with residual concatenation. First, the input labeled US image was down-sampled to reduce the image’s feature resolution, then the LR image features were learned via feature transformation. Finally, the LR image features were amplified in the spatial dimension via up-sampling, and the image was restored to the input dimension. The mode can compress redundant feature information and effectively extract it. In the decoder part of the generator, the previous layer’s output features and those of the image layer of the decoder were spliced as the input of the deconvolution layer, thereby ensuring that the finally generated pseudo CT image integrated more shallow information and provided accurate gradient information. The generated CT image had a high coincidence degree with the corresponding organ contour in the US image. In terms of discriminators, we used a six-layer full CNN. The labeled US image was connected to the pseudo CT image synthesized using the generator and real CT image, respectively, as two discriminator inputs. The discriminator’s output was a matrix of dimension 30×30 pixels, and each element of the matrix corresponding to a receptive field of 70×70 pixels of the original input image. This means the input image was divided into 900 image blocks, each of 70×70 pixels, to identify true and false. The discriminator’s output in the traditional GAN comprised the single probability of judging whether the input image was true or false. This method takes the mean value of each element in the matrix as the output of true/false and judges the local region of the input image block-by-block so that the discriminator can accurately judge the input image’s details. This improves the final generation quality of the pseudo CT image. Also, in the generator and discriminator, the LReLU activation function was used to avoid highly sparse gradients, and BN was used to normalize the output of the intermediate hidden layer, thereby stabilizing the distribution of input data in each layer of the network and expediting the learning speed of the model (47).
In SRGAN-based image super-resolution reconstruction, the second to sixth layers of the generator used residual blocks. Except for the first ResNet layer, each ResNet layer’s input was the sum of the outputs of the first two layers. We also used skip connection to transfer the input layer’s shallow information to deeper deconvolution layers in the network to ensure that the gradient information of CT images could be effectively transmitted with the deepening of the number of network layers. This enhanced the robustness of SRGAN and avoided the gradient-diffusion phenomenon in the back-propagation process. The VGG19 network structure was adopted in the discrimination network, and a BN layer was added to expedite the training. Also, the max-pooling layer in the network was removed to avoid the feature-information loss. In taking the pseudo CT images obtained using cGAN as the input of SRGAN, their resolution can be improved, and after the SRGAN-based reconstruction, the texture information of the pseudo CT image became more significant and is more similar to the CTgt image.
The supervised-mode-based sGAN also has limitations. Because this method requires paired data sets in pseudo CT image synthesis, mismatched CT–US image pairs may fail synthetic images. Also, inaccurate label production in the preprocessing would affect the subsequent image-synthesis results. In subsequent experiments, we will use the improved CycleGAN network to establish the nonlinear mapping relationship between US and CT images and carry out the pseudo CT image synthesis task. By adjusting the network parameters and loss function, we want to make the CycleGAN network training stable. Also, the image-synthesis model used in this study is based on 2D resliced image data. Compared with 3D image synthesis, the training time is longer and spatial information is ignored. Since this study is a preliminary one on applying the deep-learning method in the US- and CT-image synthesis, in the following experiments, we will use more patient data and introduce the GAN based on 3D convolution kernel further to improve the accuracy of pseudo CT image synthesis. We will also evaluate the application of pseudo CT images of real patients in radiotherapy from the perspective of dosimetry.
Conclusions
We proposed a novel method to perform the step-by-step synthesis of pseudo CT images based on sGAN. In this method, LR pseudo CT images were synthesized using labeled US images as the input to cGAN. Subsequently, the pseudo CT images’ resolution was further improved via SRGAN, and images with more significant texture information were obtained. The experimental results showed that the pseudo CT images obtained using the proposed method had high similarity with CTgt images and had a satisfactory radiotherapy application potential.
Acknowledgments
We would like to thank Dr. Li Xiaoqin from the ultrasound department of Changzhou Second People’s Hospital for her guidance on ultrasound imaging in the paper, and also thank Prof. Dai Jianrong from the Institute of Cancer Research, Chinese Academy of Medical Sciences for his guidance in this work.
Funding: This work was supported in part by the Changzhou Key Laboratory of medical physics, Jiangsu Province, China (Grant no.CM20193005) and in part by the Innovation Foundation for Doctor Dissertation of Northwestern Polytechnical University (Grant no.CX202039).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-1019). The authors have no conflicts of interest to declare.
Ethical Statement: The Medical Ethics Committee of the Second People’s Hospital of Changzhou, Nanjing Medical University approved the study (2017-002-01), and informed consent was obtained from all individual patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Eichenauer DA, Aleman BMP, André M, Federico M, Hutchings M, Illidge T. Hodgkin lymphoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2018;29:iv19-29. [Crossref]
- Zhang XQ, Zhao SG. Cervical image classification based on image segmentation preprocessing and a CapsNet network model. Int J Imaging Syst Technol 2019;29:19-28. [Crossref]
- Delaby N, Bouvier J, Sorel S, Jouyaux F, Chajon E, Castelli J. PO-0889: Validation of transit EPID and application for Head & Neck adaptive radiotherapy. Radiother Oncol 2018;127:S471-2. [Crossref]
- Anvari A, Poirier Y, Sawant A. Kilovoltage transit and exit dosimetry for a small animal image-guided radiotherapy system using built-in EPID. Med Phys 2018;45:4642-51. [Crossref] [PubMed]
- Yan H, Zhen X, Cervino L, Jiang SB, Jia X. Progressive cone beam CT dose control in image-guided radiation therapy. Med Phys 2013;40:060701. [Crossref] [PubMed]
- Meroni S, Mongioj V, Giandini T, Bonfantini F, Cavallo A, Carrara M. EP-1822: limits and potentialities of the use of CBCT for dose calculation in adaptive radiotherapy. Radiother Oncol 2016;119:S854-5. [Crossref]
- Zhang J, Zhan GW, Lu J. SU-E-I-05: A Correction Algorithm for Kilovoltage Cone-Beam Computed Tomography Dose Calculations in Cervical Cancer Patients. Med Phys 2015;42:3242. [Crossref]
- Su L, Ng SK, Zhang Y, Ji T, Ding K. Feasibility Study of Real‐Time Ultrasound Monitoring for Abdominal Stereotactic Body Radiation Therapy. Med Phys 2016;43:3727. [Crossref]
- Li M, Ballhausen H, Hegemann N, Nina S. Hegemann. A comparative assessment of prostate positioning guided by three-dimensional ultrasound and cone beam CT. Radiat Oncol 2015;10:82-3. [Crossref] [PubMed]
- Camps S, Meer SVD, Verhaegen F, Fontanarosa D. Various approaches for pseudo-CT scan creation based on ultrasound to ultrasound deformable image registration between different treatment time points for radiotherapy treatment plan adaptation in prostate cancer patients. Biomed Phys Eng Express 2016;2:035018. [Crossref]
- van der Meer S, Camps SM, van Elmpt WJ, Podesta M, Sanches PG, Vanneste BG, Fontanarosa D, Verhaegen F. Simulation of pseudo CT images based on deformable image registration of ultrasound images: A proof of concept for transabdominal ultrasound imaging of the prostate during radiotherapy. Med Phys 2016;43:1913-20. [Crossref] [PubMed]
- Sun H, Lin T, Xie K, Sui J, Ni X. Imaging study of pseudo-CT images of superposed ultrasound deformation fields acquired in radiotherapy based on step-by-step local registration. Med Biol Eng Comput 2019;57:643-51. [Crossref] [PubMed]
- Andreasen D, Van LK, Edmund JM. A patch-based pseudo-CT approach for MRI-only radiotherapy in the pelvis. Med Phys 2016;43:4742-50. [Crossref] [PubMed]
- Cao X, Yang J, Gao Y, Shen D. Region-adaptive deformable registration of CT/MRI pelvic images via learning-based image synthesis. IEEE Trans Image Process 2018;27:3500-12. [Crossref] [PubMed]
- Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q, Shen D. Medical image synthesis with deep convolutional adversarial networks. IEEE Trans Biomed Eng 2018;65:2720-30. [Crossref] [PubMed]
- Gatys LA, Ecker AS, Bethge M. Image style transfer using convolutional neural networks. Computer Vision Pattern Recognition, 2016;2414-23.
- Johnson J, Alahi A, Li FF. Perceptual losses for real-time style transfer and super-resolution. Proc European Conference Computer Vision. Springer Press, 2016;694-711.
- Goodfellow IJ, Pouget-Abadie J, Mirza M, et al. Generative adversarial nets. In: Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2 (NIPS' 14). Cambridge, MA, USA: MIT Press, 2014:2672-80.
- Jin CB, Kim H, Liu M, Jung W, Joo S, Park E, Ahn YS, Han IH, Lee JI, Cui X. Deep CT to MR Synthesis Using Paired and Unpaired Data. Sensors (Basel) 2019;19:2361. [Crossref] [PubMed]
- Bi L, Kim J, Kumar A, Feng D, Fulham M. Synthesis of positron emission tomography (PET) images via multi-channel generative adversarial networks (GANs). Springer, Cham, 2017:43-51.
- Dar SU, Yurt M, Karacan L, Erdem A, Erdem E, Cukur T. Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Trans Med Imaging 2019;38:2375-88. [Crossref] [PubMed]
- Costa P, Galdran A, Meyer MI, Niemeijer M, Abramoff M, Mendonca AM, Campilho A. End-to-End Adversarial Retinal Image Synthesis. IEEE Trans Med Imaging. 2018;37:781-91. [Crossref] [PubMed]
- Lin J, Xia Y, Qin T, Chen Z, Liu TY. Conditional Image-to-Image Translation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2018:5524-32.
- Ben-Cohen A, Klang E, Raskin S, Amitai MM, Greenspan H. Virtual PET images from CT data using deep convolutional networks: Initial results. International workshop on simulation and synthesis in medical imaging. Springer, Cham, 2017;49-57.
- Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. Proceedings of the IEEE international conference on computer vision 2017:2223-32.
- Liu MY, Breuel T, Kautz J. Unsupervised image-to-image translation networks. Adv Neural Inf Process Syst 2017.700-8.
- Yi Z, Zhang H, Tan P, Gong M. Dual GAN: Unsupervised dual learning for image-to-image translation. Proceedings of the IEEE international conference on computer vision 2017;2849-57.
- Liu MY, Huang X, Mallya A, Karras T, Aila T, Lehtinen J. Few-shot unsupervised image-to-image translation. Proceedings of the IEEE International Conference on Computer Vision 2019;10551-60.
- Wolterink JM, Dinkla AM, Savenije MHF, Seevinck PR, Berg C, Isgum I. Deep MR to CT synthesis using unpaired data. International workshop on simulation and synthesis in medical imaging. Springer, Cham, 2017;14-23.
- Wang C, Macnaught G, Papanastasiou G, Macgillivray T, Newby D. Unsupervised learning for cross-domain medical image synthesis using deformation invariant cycle consistency networks. International Workshop on Simulation and Synthesis in Medical Imaging. Springer, Cham, 2018;52-60.
- Zhang H, Xu T, Li H, Zhang S, Wang X, Huang X. StackGAN: Text to photo-realistic image synthesis with stacked generative adversarial networks. Proceedings of the IEEE international conference on computer vision 2017;5907-15.
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. Proceedings of the IEEE conference on computer vision and pattern recognition 2017;1125-34.
- Kudo Y, Aoki Y. Dilated convolutions for image classification and object localization. Fifteenth IAPR International Conference Machine Vision Applications, 2017.
- Qiao Z, Cui Z, Niu X, Geng S, Qiao Y. Image segmentation with pyramid dilated convolution based on ResNet and U-Net. International Conference Neural Information Processing 2017.
- Chen H, Dou Q, Yu L, Qin J, Heng PA. VoxResNet: Deep voxelwise residual networks for brain segmentation from 3D MR images. Neuroimage 2018;170:446-55. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J, Deep residual learning for image recognition. Proceedings of the IEEE conference on computer vision and pattern recognition 2016;770-8.
- Zeiler MD, Taylor GW, Fergus R. Adaptive deconvolutional networks for mid and high level feature learning. 2011 International Conference on Computer Vision. IEEE, 2011:2018-25.
- Shi W, Caballero J, Huszár F, Totz J, Aitken AP, Bishop R, Rueckert D, Wang Z. Real-time single image and video super-resolution using an efficient sub-pixel convolutional neural network. Proceedings of the IEEE conference on computer vision and pattern recognition 2016;1874-83.
- Dong C, Loy CC, He K, Tang X. Image Super-Resolution Using Deep Convolutional Networks. IEEE Trans Pattern Anal Mach Intell 2016;38:295-307. [Crossref] [PubMed]
- Nithiananthan S, Schafer S, Mirota DJ, Stayman JW, Zbijewski W, Reh DD, Gallia GL, Siewerdsen JH. Extra-dimensional demons: A method for incorporating missing tissue in deformable image registration. Med Phys 2012;39:5718-31. [Crossref] [PubMed]
- Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Process 2004;13:600-12. [Crossref] [PubMed]
- Hore A, Ziou D. Image quality metrics: PSNR vs. SSIM. 2010 20th international conference on pattern recognition. IEEE, 2010;2366-9.
- Cao X, Yang J, Gao Y, Guo Y, Shen D. Dual-core steered non-rigid registration for multi-modal images via bi-directional image synthesis. Med Image Anal 2017;41:18-31. [Crossref] [PubMed]
- Lee J, Nishikawa R, Reiser I, Boone J. WE‐G‐207‐05: Relationship between CT image quality, segmentation performance, and quantitative image feature analysis. Med Phys 2015;42:3697. [Crossref]
- He K, Cao X, Shi Y, Nie D, Gao Y, Shen D. Pelvic Organ Segmentation Using Distinctive Curve Guided Fully Convolutional Networks. IEEE Trans Med Imaging 2019;38:585-95. [Crossref] [PubMed]
- Zhao H, Rosin PL, Lai YK, Wang YN. Automatic semantic style transfer using deep convolutional neural networks and soft masks. Vis Comput 2020;36:1307-24. [Crossref]
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. arXiv preprint arXiv:1502.03167, 2015.







