
Prediction of neoadjuvant chemotherapy response in high-grade osteosarcoma: added value of non-tumorous bone radiomics using CT images
Introduction
Osteosarcoma is the most common primary malignant bone tumor originated from the primitive bone-forming mesenchymal cells (1). When diagnosed, around 90% of osteosarcoma patients are classified as high-grade osteosarcoma (HOS) (2). Preoperative neoadjuvant chemotherapy combined with definitive surgical resection, as the standard treatment option, has significantly improved the five-year survival rate from less than 20% to more than 60% (3). Unfortunately, there still remains a considerable amount of osteosarcoma patients that might not benefit from chemotherapy, especially those with poor histologic responses from chemotherapy (4,5). Therefore, accurate pre-treatment pathological response prediction to chemotherapy in patients with HOS is essential in determining the best treatment option. If poor response could be predicted, it might be helpful for modifying treatment regimen for chemotherapy and/or surgery (6,7).
Radiomics analysis based on medical images has been used to predict outcomes of osteosarcoma treated with chemotherapy in previous studies (8-10). Song et al. found that tumor texture features of baseline 18F-fluorodeoxyglucose uptake measured by positron emission tomography (PET) images could predict pathological response to chemotherapy in patients with localized osteosarcoma of the extremities (10). Davis et al. reported that the percentage change of maximum standardized uptake during treatment could be used as metabolic predictors of pathological response in osteosarcoma (9). Wu et al. illustrated that the percentage change of maximum standardized uptake during treatment could be used as metabolic predictors of pathological response in osteosarcoma (11). Our previous study demonstrated that delta CT imaging features could be used for individualized pathologic response evaluation after chemotherapy in osteosarcoma (12). These findings showed that imaging features extracted from tumors have the potential to predict chemotherapy outcomes.
Several previous studies suggested that texture features from normal parenchymal structures of solid tumors could reflect the biologic factors associated with tumor development, which could be interpreted by the important role of normal parenchymal stromal cells in tumor formation and development (13-15). Sala et al. demonstrated that parenchymal texture features could be used to evaluate or predict the stage and subtype in breast cancer (16). Li et al. found that combining quantitative radiomics features from tumors with contralateral parenchyma characterizations can improve the diagnostic accuracy for breast cancer (17). The idea of combined features for breast cancer intrigued our present study of looking into the correlation between non-tumorous bone characteristics and tumor development in osteosarcoma.
This study was developed with a hypothesis that the combination of imaging features extracted from non-tumorous bone region and tumor region on diagnostic CT images could improve the accuracy of pathological response prediction to chemotherapy. Quantitative CT imaging features from both tumor and non-tumorous bone regions were extracted and analyzed. The prediction performance of pathological response classifiers developed based on the combination features was assessed and compared with the classifiers developed based on the tumor features alone.
Methods
Patient data set
This retrospective study was approved by the Institutional Research Ethics Board. Patient informed consent was waived for the study. All medical images and baseline characteristics were obtained from the Second Affiliated Hospital, Zhejiang University School of Medicine (Zhejiang, China) between November 2013 and November 2017.
The focus of the study was on HOS in extremities, considering that large tumor size can impact the accuracy of non-tumorous bone region delineation. Patients’ inclusion criteria are: (I) patients were diagnosed as HOS in extremities; (II) patients underwent diagnostic CT scans before chemotherapy; (III) patients underwent surgical resections after chemotherapy; (IV) patients had confirmed pathological response to chemotherapy by pathological examination; (V) patients had complete baseline characteristics and histologic information. The baseline characteristics were obtained from the Electronic Medical Record System (EMRS), including age, gender, and location of primary tumor, tumor stage, pathologic subtype, any pulmonary metastasis, and type of chemotherapy regimens. The flowchart of patient exclusion criteria was presented in Figure 1.
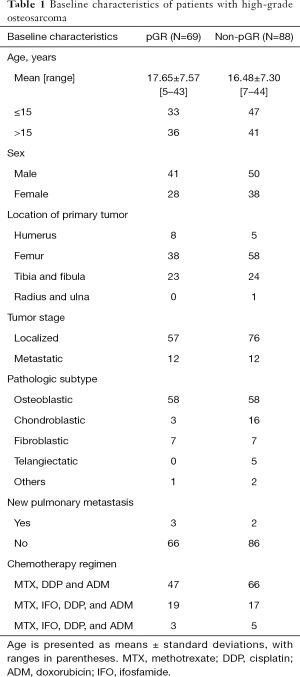
The baseline characteristics of patients with pathological good response (pGR) and non-pGR were described in Table 1. A total of 157 patients with HOS were involved in this study, 69 of whom showed pGR to chemotherapy and 88 showed non-pGR. Specifically, tumor volumes for patients with pGR ranged from 15.53 to 372.70 cm3 (mean ± standard deviation, 134.82±84.45 cm3), and 7.02 to 1,142.96 cm3 (173.57±160.13 cm3) for patients with non-pGR. Patient dataset was stratified to the training dataset and independent validation dataset randomly at a ratio of 3:1.
Full table
CT image acquisition
All images were acquired on five CT scanners, including a 16 multi-slice CT (MSCT) (Siemens Sensation 16, Siemens Medical Systems, Erlangen, Germany), a 64-MSCT (Philips Brilliance 64, Philips Medical Systems, Cleveland, USA), a 128-MSCT (Definition Flash, Siemens Medical Systems), a 160-MSCT (Volume Zoom, Siemens Medical Systems) and a 320-MSCT (Aquilion ONE, Toshiba Medical Systems, Otawara, Japan). All CT scan parameters were as follows: tube voltage of 100–140 kVp, tube current of 80–480 mAs, slice thickness of 3–6 mm, and pixel size of 0.3×0.3–1.0×1.0 mm2. Standard DICOM format was used for all medical images in this study.
Neoadjuvant chemotherapy, pathological response assessment and surgery
All patients received preoperative chemotherapy followed by surgical resection. The neoadjuvant chemotherapy schema was designed according to the National Comprehensive Cancer Network guidelines. The total duration of neoadjuvant chemotherapy ranged from 8 to 10 weeks.
The conventional three-drug regimen was used consisting of methotrexate, cisplatin, and doxorubicin. This regimen was administered in 9 weeks: week 1 and week 2: methotrexate (8 mg/m2 in 6 hours for patients <14 years of age, 12 mg/m2 in 6 hours for patients ≥14 years of age); week 3: cisplatin (100 mg/m2 in 24 hours) and doxorubicin (60 mg/m2 in 8 hours); week 5 and week 6: methotrexate; week 7: cisplatin and doxorubicin; week 9: surgery. Patients who suffered severe liver dysfunction or other adverse reactions after the first week of treatment received an alternative treatment schema consisting of methotrexate, ifosfamide, cisplatin and doxorubicin in 11 weeks: week 1: methotrexate; week 2: ifosfamide (2 mg/m2/day for 5 days); week 4: cisplatin and doxorubicin; week 6: ifosfamide; week 8: ifosfamide; week 9: cisplatin and doxorubicin; week 11: surgery. Patients who suffered tumor progression or newly diagnosed lung metastasis after the first week of treatment received an alternative treatment schema consisting of methotrexate, ifosfamide, cisplatin and doxorubicin in 10 weeks: week 1 and week 2: methotrexate; week 3: cisplatin and doxorubicin; week 5 and week 7: ifosfamide; week 8: cisplatin and doxorubicin; week 10: surgery.
Histology analysis of pathological response to chemotherapy was performed by two experienced pathologists blinded to the CT images, using the criteria of Bacci et al. (18). The pGR was defined as the tumor necrosis percentages ≥90%; while non-pGR was defined as the tumor necrosis percentages <90% (19).
About three weeks after the last cycle of chemotherapy, patients underwent surgery. The type of surgery (amputation and limb salvage) was chosen depending on the comprehensive consideration involving location and extension of the tumor, and neurovascular bundle involvement. Simultaneous surgery for primary and metastatic lesions was performed for patients with metastatic lesions. Margins of the resections were defined according to the Enneking’s criteria (20).
Radiomics analysis of tumor regions
Radiomics analysis of osteosarcoma tumor regions was conducted according to the following three steps. The tumor was delineated by one experienced orthopedic surgeon using the ITK-SNAP software, reviewed and modified if necessary, by the second experienced orthopedic surgeon and an experienced radiologist (21). The two orthopedic surgeons and the radiologist were blinded to the pathological response to chemotherapy. Radiomics features were extracted from the confirmed tumor regions on CT images following image pre-processing. Subsequent classification was implemented to differentiate patients with pGR and non-pGR by using a pathological response classifier.
A total of 491 image features were extracted from each tumor region. Prior to feature extraction, image pre-processing was performed including voxel resampling and intensity normalization. All CT images were resampled to the same voxel size of 0.5×0.5×5 mm3. Voxel intensity within the tumor region was discretized to a range of 64 intensities. The feature pool was categorized as intensity statistics features, geometry features, texture features, and wavelet features. The intensity statistics features measured the voxel intensities statistical distribution within the tumor region. The geometry features described the 3D shape features of the tumor region. The texture features describing the spatial intensity correlation and distributions of the voxels within the tumor region were calculated based the gray-level co-occurrence matrix (GLCM), the gray-level size-zone matrix (GLSZM), the gray-level run-length matrix (GLRLM), and the neighborhood gray-tone-difference matrix (NGTDM). The feature extraction process was conducted based on open-source Radiomics packages in MATLAB 2017b (MathWorks, Natick, MA, USA) (22-24). Details in feature extraction were described comprehensively in the Supplementary Material (Appendix 1).
Radiomics analysis of non-tumorous regions
For non-tumorous bone feature extraction, regions of interest of 30×30×3 voxels (15×15×15 mm3, cubic area) were delineated in the region of cancellous bone structures where the tumor was located. The compact bone region was not included in the regions of interest given that the compact bone was too thin to obtain feature information. Since imaging features were extracted based on the non-tumorous bone region with a cubic size, the geometry features were not included. The feature pool for the non-tumorous bone region included three categories, involving intensity statistics features, texture features, and wavelet features. Radiomics analysis of non-tumorous regions was performed on the extracted 484 non-tumorous bone features. An example of regions of interest for the tumor region and non-tumorous bone region was shown in Figure 2.

Leave-one-out cross validation (LOOCV) and feature selection
To reduce selection bias within the training dataset, feature selection and temporary classifier construction were performed concurrently using the LOOCV method (17,25,26). The temporary classifier was constructed using features selected within each cross-validation iteration process. Note that in each iteration process, we further divided the training dataset into two separate sub-datasets: N-1 patients as the temporary training set and one patient as the temporary validation set. N was the overall number of patients in the training dataset in our study. The temporary classifier was developed using the temporary training set and validated using the temporary validation set. These training and validation procedures were repeated N times. Model performance was measured using the area under the receiver operating characteristic curve (AUC) value averaged during the LOOCV process (mean ± standard deviation).
During each LOOCV iteration process, the maximum relevance minimum redundancy (mRMR) feature selection approach was used to select a compact feature set (27,28). The mRMR method aimed to generate a feature set which had the maximum relevance to the actual label and the minimum redundancy to other features. Based on the rule of thumb, the number of predictors should remain less than 1/10 of the case number in the group (29,30). Thus, to prevent over fitting, the potential feature number was limited to 12 when deriving a temporary classifier using the multivariable logistic regression algorithm. The optimal temporary classifier was determined from 11 classifiers with feature number ranging from 2 to 12. The optimal temporary classifier was identified with the AUC value.
Furthermore, to investigate individual importance of the tumor and non-tumorous bone features, features used in the temporary classifier for more than 50% times were recorded during the LOOCV process. To investigate the additional value of non-tumorous bone features in assessing treatment response to chemotherapy, the temporary classifiers were constructed using tumor features alone, non-tumorous bone features alone, and combined features, respectively.
Classifier construction and independent validation
We developed a classifier to predict individual pathological response status using the tumor features that were selected more than 50% of the LOOCV process with multivariable logistic regression algorithm. For comparison, we further developed a pathological response classifier using the combined features selected more than 50% of the LOOCV process. Then the proposed classifiers were validated using an independent validation dataset.
Reproducibility validation of pathological response classifier
To assess the reproducibility of the region selection of the bone region and the proposed classifier, we re-delineated the non-tumorous bone ROIs of patients from the independent validation dataset. The re-delineation process was conducted by the same orthopedic surgeon following the same procedure. Furthermore, we extracted the non-tumorous bone features based on the re-delineated ROIs. Then features were normalized using values calculated based on the training dataset. We validated the proposed pathological response classifiers using re-delineated feature dataset. The performance difference between the re-delineated validation dataset and the independent validation dataset was measured using the Delong test.
Classifier construction integrating the clinical features with imaging features
We developed a clinical-imaging classifier using both imaging features and clinical features with multivariable logistic regression algorithm. The clinical features included age, sex, location of primary tumor, tumor stage, pathologic subtype, and new pulmonary metastasis. For the tumor features based clinical-imaging classifier, clinical and radiomics features were combined with those selected more than 50% of the LOOCV process. For the combined imaging features based clinical-imaging classifier, we included the clinical features and combined imaging features selected more than 50% of the LOOCV process. The proposed classifiers were validated using the independent validation dataset.
Statistical analysis
Receiver operating characteristic (ROC) analysis was used to evaluate the performance of the proposed classifiers, with AUC value to differentiate patients with pGR and non-pGR (31). The maximum AUC value was 1.0, indicating a perfect prediction. The minimum value of 0.5 indicated no predictive power. The Mann-Whitney U-test was used to assess performance difference in AUC values. The Delong test was used to access the deference in ROC curves. A threshold P value less than 0.05 was defined as the significant difference in the two-tailed analysis. The Holm-Bonferroni method was conducted to correct for multiple comparisons. All statistical analysis was performed with R software (Version 3.4.1, www.R-project.org) and MedCalc (Version 15.2.2, www.medcalc.org). The mRMR algorithm was conducted using the “mRMRe” package in R software. The ROC analysis was conducted using the “pROC” package.
Results
Radiomics analysis of tumor features
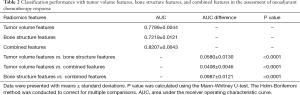
As shown in Table 2, an AUC of 0.7799±0.0044 (range, 0.7649–0.7913) was obtained for the pathological response classifier developed using the tumor features alone during the LOOCV process. ROC analysis indicated that individual tumor feature showed inferior classification performance in pathological response prediction, with an AUC of 0.5494±0.0339 (range, 0.5008–0.6304). In the LOOCV process, we found eleven tumor features were selected more than 50% times, including LZE-tumor, SRE-LLL-tumor, corrp-LLH-tumor, SZE-LLH-tumor, Complexity-LHL-tumor, corrm-LHH-tumor, SRHGE-LHH-tumor, svarh-HLH-tumor, corrp-HHL-tumor, cshad-HHH-tumor, SZHGE-HHH-tumor. The feature SRE-LLL-tumor indicated short run emphasis value extracted based on GLRLM from the wavelet decomposed image using low-pass, low-pass and low-pass filters. The full names for all other abbreviations were provided in Table S1, Supplementary Material (Appendix 1).
Full table
Radiomics analysis non-tumorous features
When we performed radiomics analysis of non-tumorous bone regions, an AUC of 0.7219±0.0121 (range, 0.6916–0.7492) was obtained using the pathological response classifier developed based on the non-tumorous bone features only during the LOOCV process. Individual non-tumorous feature showed poor classification performance in pathological response prediction with an AUC of 0.5341±0.0288 (range, 0.5002–0.6270) using ROC analysis. Two non-tumorous bone features were selected more than 50% of the time during the LOOCV process, including Entropy-bone, corrm-LLH-bone.
Radiomics analysis of combined features
The radiomics analysis of combined features showed higher AUC values (0.8207±0.0042, range, 0.8099 to 0.8347) than the analysis of tumor features alone during the LOOCV process. The AUC difference between the classifiers based on combined features and tumor features was 0.0408±0.0046, with a range from 0.0259 to 0.0538. Statistical significance was observed for the AUC difference with P value <0.0001 using Mann-Whitney U-test.
Ten image features were selected more than 50% during the LOOCV process, including seven tumor features (SRE-LLL-tumor, SZE-LLH-tumor, SRHGE-LHH-tumor, svarh-HLH-tumor, corrp-HHL-tumor, cshad-HHH-tumor, SZHGE-HHH-tumor) and three non-tumorous features (corrm-LLH-bone, SRLGE-HLH-bone, RLV-HHH-bone). Of these features, seven tumor features were all selected more than 50% times for the tumor features-based classifier; a bone feature of corrm-LLH-bone was selected more than 50% times for the non-tumorous bone features-based classifier.
Classifier construction and independent validation
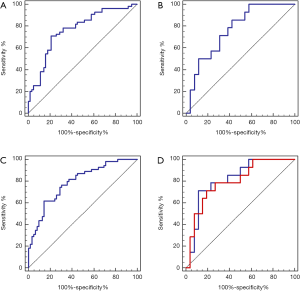
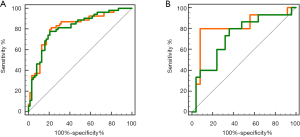
For the pathological response classifier developed using the eleven tumor features, AUC was 0.766 [95% confidence interval (CI), 0.679–0.840; significance level, P value <0.0001] for the training dataset, and 0.766 (95% CI, 0.606–0.885; significance level, P value =0.0004) for the independent validation dataset. By contrast, the combined features-based classifier showed better performance than the tumor features-based classifier in both training and validation datasets (training dataset: 0.791, 95% CI, 0.706–0.860; significance level, P value <0.0001; validation dataset: 0.816, 95% CI, 0.662–0.920; significance level, P value <0.0001). The ROC curves of both tumor features and combined features-based classifiers in the training dataset and validation dataset were showed in Figure 3.
Reproducibility validation of pathological response classifier
In the reproducibility validation test of the proposed classifiers using re-delineated ROIs, a promising performance was achieved with an AUC of 0.797 (95% CI, 0.640–0.907; significance level, P value =0.0001). We also observed a non-significant performance difference between the re-delineation dataset and the independent validation dataset (P value =0.4553). The ROC curves of the combined features-based classifier in the re-delineation validation dataset were showed in Figure 3.
Classifier construction integrating the clinical features and imaging features
Integration of clinical features with tumor feature achieved an AUC of 0.814 (95% CI, 0.731–0.880; significance level, P value <0.0001) for the training dataset and 0.728 (95% CI, 0.564–0.856; significance level, P value =0.0080) for the validation dataset. Integrating the clinical features with the combined features, we observed an AUC of 0.820 (95% CI, 0.738–0.885; significance level, P value <0.0001) for the training dataset and 0.811 (95% CI, 0.656 to 0.917; significance level, P value =0.0001) for the validation dataset. The ROC curves of these two clinical-imaging classifiers in both training dataset and validation dataset were provided in Figure 4. The difference between AUC values was 0.0059 (95% CI, −0.0311–0.0428) for the training dataset and 0.0827 (95% CI, −0.0248–0.190) for the validation dataset between the clinical/tumor classifier vs. clinical/combined classifier.
Discussion
In this study, we compared the performance of pathological response prediction to chemotherapy with the use of tumor features alone and the combination of tumor and non-tumorous bone features. Results showed statistically significant improvement in pathological response prediction performance with the combined features.
Literature showed a wide spectrum of accuracy in radiomics studies for pathological response prediction to chemotherapy in patients with osteosarcoma, probably due to the difference in data sets and model construction algorithms. Song et al. achieved a high AUC of 0.918 with a relatively small data set of thirty-five patients (10), whereas Davis et al. reported an AUC of 0.807 using a data set of thirty-four patients (9). However, both these studies only analyzed image features based on intratumoral heterogeneity. To the best of our knowledge, the present study is the first study to predict pathological response by combining tumor features and non-tumorous bone features.
For the features selected more than 50% in the LOOCV process, ten out of eleven tumor-based features, one out of two non-tumorous bone feature, and ten out of ten combined features were all wavelet features. This indicated the vital role of wavelet image features in the pathological response prediction. This observation was consistent with previous studies which also used wavelet-based features in the proposed radiomics models (32-34). The wavelet transformation splitting images into different subimages with different frequency components may further explore the spatial heterogeneity at multiple scales within the regions of interest (33).
The results of this study suggested that non-tumorous bone features could strengthen pathological response prediction to chemotherapy in osteosarcoma. Previous studies have demonstrated that the development of osteosarcoma was related to bone microenvironment and complex bone cell-tumor interactions (15,35,36). The improvement of pathological response prediction accuracy using combined features supports that tumor cell and nearby normal bone matrix interaction is important for treatment response.
For the non-tumorous bone features, only two features were selected from the LOOCV process, including Entropy-bone, corrm-LLH-bone. For the radiomics analysis of combined features, three bone features (corrm-LLH-bone, SRLGE-HLH-bone, RLV-HHH-bone) were retained. The feature of corrm-LLH-bone was selected from both processes, indicating stable predictive power in this non-tumorous bone feature. This feature measured pixel correlation to its neighbor over the whole region of interest. This indicated that the neoadjuvant chemotherapy to the osteosarcoma might cause local influence to non-tumorous regions, resulting in changes in local texture.
A few limitations to be noted: (I) the study was a retrospective study with data from single institution and small sample size, which limited the use of LOOCV method and small independent validation set. (II) Only CT images were studied, yet MR imaging might be more commonly used for patients with HOS nowadays. (III) Patients included in this study underwent three-drug regimen, whereas two regiments of three-drug regimen and two-drug regimen (cisplatin and doxorubicin) may be a preferred treatment by NCCN. The validation using patients treated by different regimens was valuable. (IV) Clinical and imaging features were studied, yet a variety of other relevant signatures, such as biological features, molecular features, immune features, etc. might affect results of the current study. Nevertheless, this is the first pilot study of using quantitative features of tumor and non-tumorous bone regions based on medical images for predicting pathological response to chemotherapy in osteosarcoma. Prospective and multicentric studies are underway involving more imaging modalities, regimens and features to further validate the findings of this study.
Conclusions
Radiomics analysis of combined tumor and non-tumorous bone features showed improved performance of pathological response prediction to chemotherapy in HOS compared to that of tumor features alone. Moreover, the proposed classifier had the potential to predict the pathological response to chemotherapy for HOS patients.
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (2018YFE0114800), Natural Science Foundation of China (NSFC Grant No. 81871351, 81950410632), Zhejiang Postdoctoral Scientific Research Preferred Project (zj2019004).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-681). The authors have no conflicts of interest to declare.
Ethical Statement: This study was approved by the Institutional Research Ethics Board of our institution. The requirement of informed consent was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Durnali A, Alkis N, Cangur S, Yukruk FA, Inal A, Tokluoglu S, Seker MM, Bal O, Akman T, Inanc M, Isikdogan A, Demirci A, Helvaci K, Oksuzoglu B. Prognostic factors for teenage and adult patients with high-grade osteosarcoma: an analysis of 240 patients. Med Oncol 2013;30:624. [Crossref] [PubMed]
- Zaikova O, Sundby Hall K, Styring E, Eriksson M, Trovik CS, Bergh P, Bjerkehagen B, Skorpil M, Weedon-Fekjaer H, Bauer HC. Referral patterns, treatment and outcome of high-grade malignant bone sarcoma in Scandinavia--SSG Central Register 25 years' experience. J Surg Oncol 2015;112:853-60. [Crossref] [PubMed]
- Bacci G, Longhi A, Fagioli F, Briccoli A, Versari M, Picci P. Adjuvant and neoadjuvant chemotherapy for osteosarcoma of the extremities: 27 year experience at Rizzoli Institute, Italy. Eur J Cancer 2005;41:2836-45. [Crossref] [PubMed]
- Bielack SS, Kempf-Bielack B, Delling G, Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M, Winkelmann W, Zoubek A, Jurgens H, Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J Clin Oncol 2002;20:776-90. [Crossref] [PubMed]
- Davis AM, Bell RS, Goodwin PJ. Prognostic factors in osteosarcoma: a critical review. J Clin Oncol 1994;12:423-31. [Crossref] [PubMed]
- Cheon GJ, Kim MS, Lee JA, Lee SY, Cho WH, Song WS, Koh JS, Yoo JY, Oh DH, Shin DS, Jeon DG. Prediction Model of Chemotherapy Response in Osteosarcoma by F-18-FDG PET and MRI. J Nucl Med 2009;50:1435-40. [Crossref] [PubMed]
- Coffin CM, Lowichik A, Zhou H. Treatment effects in pediatric soft tissue and bone tumors - Practical considerations for the pathologist. Am J Clin Pathol 2005;123:75-90. [Crossref] [PubMed]
- Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on Gadoxetic Acid-Enhanced Magnetic Resonance Imaging for Prediction of Postoperative Early and Late Recurrence of Single Hepatocellular Carcinoma. Clin Cancer Res 2019;25:3847-55. [Crossref] [PubMed]
- Davis JC, Daw NC, Navid F, Billups CA, Wu J, Bahrami A, Jenkins JJ, Snyder SE, Reddick WE, Santana VM, McCarville MB, Guo J, Shulkin BL. (18)F-FDG Uptake During Early Adjuvant Chemotherapy Predicts Histologic Response in Pediatric and Young Adult Patients with Osteosarcoma. J Nucl Med 2018;59:25-30. [Crossref] [PubMed]
- Song H, Jiao Y, Wei W, Ren X, Shen C, Qiu Z, Yang Q, Wang Q, Luo QY. Can pretreatment (18)F-FDG PET tumor texture features predict the outcomes of osteosarcoma treated by neoadjuvant chemotherapy? Eur Radiol 2019;29:3945-54. [Crossref] [PubMed]
- Wu Y, Xu L, Yang P, Lin N, Huang X, Pan W, Li H, Lin P, Li B, Bunpetch V, Luo C, Jiang Y, Yang D, Huang M, Niu T, Ye Z. Survival Prediction in High-grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine 2018;34:27-34. [Crossref] [PubMed]
- Lin P, Yang PF, Chen S, Shao YY, Xu L, Wu Y, Teng W, Zhou XZ, Li BH, Luo C, Xu LM, Huang M, Niu TY, Ye ZM. A Delta-radiomics model for preoperative evaluation of Neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imaging 2020;20:7. [Crossref] [PubMed]
- Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest 2011;121:3804-9. [Crossref] [PubMed]
- Weigelt B, Bissell MJ. Unraveling the microenvironmental influences on the normal mammary gland and breast cancer. Semin Cancer Biol 2008;18:311-21. [Crossref] [PubMed]
- Alfranca A, Martinez-Cruzado L, Tornin J, Abarrategi A, Amaral T, de Alava E, Menendez P, Garcia-Castro J, Rodriguez R. Bone microenvironment signals in osteosarcoma development. Cell Mol Life Sci 2015;72:3097-113. [Crossref] [PubMed]
- Sala E, Solomon L, Warren R, McCann J, Duffy S, Luben R, Day N. Size, node status and grade of breast tumours: association with mammographic parenchymal patterns. Eur Radiol 2000;10:157-61. [Crossref] [PubMed]
- Li H, Mendel KR, Lan L, Sheth D, Giger ML. Digital Mammography in Breast Cancer: Additive Value of Radiomics of Breast Parenchyma. Radiology 2019;291:15-20. [Crossref] [PubMed]
- Bacci G, Bertoni F, Longhi A, Ferrari S, Forni C, Biagini R, Bacchini P, Donati D, Manfrini M, Bernini G, Lari S. Neoadjuvant chemotherapy for high-grade central osteosarcoma of the extremity. Histologic response to preoperative chemotherapy correlates with histologic subtype of the tumor. Cancer 2003;97:3068-75. [Crossref] [PubMed]
- Huvos AG. Bone tumors: diagnosis, treatment and prognosis. 1987.
- Enneking WF, Spanier SS, Goodman MA. A system for the surgical staging of musculoskeletal sarcoma. Clin Orthop Relat Res 1980.106-20. [Crossref] [PubMed]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31:1116-28. [Crossref] [PubMed]
- Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol 2015;60:5471-96. [Crossref] [PubMed]
- Zhang R, Xu L, Wen X, Zhang JH, Yang PF, Zhang LX, Xue X, Wang XL, Huang Q, Guo CG, Shi YJ, Niu TY, Chen F. A nomogram based on bi-regional radiomics features from multimodal magnetic resonance imaging for preoperative prediction of microvascular invasion in hepatocellular carcinoma. Quant Imaging Med Surg 2019;9:1503-15. [Crossref] [PubMed]
- Li Y, Tan G, Vangel M, Hall J, Cai W. Influence of feature calculating parameters on the reproducibility of CT radiomic features: a thoracic phantom study. Quant Imaging Med Surg 2020;10:1775-85. [Crossref] [PubMed]
- Kohavi R. editor. A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI; 1995: Montreal, Canada.
- Wong TT. Performance evaluation of classification algorithms by k-fold and leave-one-out cross validation. Pattern Recognition 2015;48:2839-46. [Crossref]
- Yu L, Liu H. Efficient feature selection via analysis of relevance and redundancy. J Mach Learn Res 2004;5:1205-24.
- Unler A, Murat A, Chinnam RB. mr2PSO: A maximum relevance minimum redundancy feature selection method based on swarm intelligence for support vector machine classification. Inf Sci 2011;181:4625-41. [Crossref]
- Harrell FE Jr. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. Springer; 2015.
- Wei J, Yang G, Hao X, Gu D, Tan Y, Wang X, Dong D, Zhang S, Wang L, Zhang H, Tian J. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol 2019;29:877-88. [Crossref] [PubMed]
- Pesce LL, Metz CE. Reliable and computationally efficient maximum-likelihood estimation of "proper" binormal ROC curves. Acad Radiol 2007;14:814-29. [Crossref] [PubMed]
- Xu L, Yang P, Liang W, Liu W, Wang W, Luo C, Wang J, Peng Z, Xing L, Huang M, Zheng S, Niu T. A radiomics approach based on support vector machine using MR images for preoperative lymph node status evaluation in intrahepatic cholangiocarcinoma. Theranostics 2019;9:5374-85. [Crossref] [PubMed]
- Wu S, Zheng J, Li Y, Yu H, Shi S, Xie W, Liu H, Su Y, Huang J, Lin T. A Radiomics Nomogram for the Preoperative Prediction of Lymph Node Metastasis in Bladder Cancer. Clin Cancer Res 2017;23:6904-11. [Crossref] [PubMed]
- Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. [Crossref] [PubMed]
- Broadhead ML, Clark J, Myers DE, Dass CR, Choong PF. The molecular pathogenesis of osteosarcoma: a review. Sarcoma 2011;2011:959248. [Crossref] [PubMed]
- Rubio R, Abarrategi A, Garcia-Castro J, Martinez-Cruzado L, Suarez C, Tornin J, Santos L, Astudillo A, Colmenero I, Mulero F, Rosu-Myles M, Menendez P, Rodriguez R. Bone environment is essential for osteosarcoma development from transformed mesenchymal stem cells. Stem Cells 2014;32:1136-48. [Crossref] [PubMed]


