
Slice-stacking T2-weighted MRI for fast determination of internal target volume for liver tumor
Introduction
Primary liver cancer and liver metastases are the main causes of worldwide cancer morbidity and mortality. Surgical resection is the primary choice for liver cancer treatment. However, nearly 70–90% of liver metastases cannot be fully removed through surgeries (1). Therefore, modern high precision radiation therapy has become an emerging approach for liver cancer treatment. However, radiotherapy for liver tumors has been confronted with two challenges: (I) low tolerance of whole liver to radiation dose (2) and (II) mobility of both tumor and surrounding organs at risk (OARs) (3). Severe radiation-induced liver disease (RILD) may occur after radiation therapy and can be classified into two types: classical and non-classical. Patients without underlying liver disease may have classical RILD such as presence of fatigue, abdominal pain, hepatomegaly and with underlying liver disease may have non-classical RILD such as jaundice and notable evaluation of serum transaminase (2). Thus, caution must be taken to deliver a desired dose to the tumor while sparing the remaining liver from overdosing. Stereotactic body radiation therapy (SBRT), delivering a few fractions of high dose radiation treatments, can provide a sharp dose gradient outside the tumor and into the surrounding normal tissue (4,5). To reduce the impact of respiratory motion on treatment accuracy, breath-hold or abdominal compression are commonly used (6-9). However, these techniques are not comfortable for patients and not compatible for patients who have expiratory dyspnea. To manage tumor motion and deformation due to respiration, four-dimensional computed tomography (4D-CT) is the current clinical standard in radiation therapy. However, 4D-CT is known to have certain limitations. A major limitation is that it has a lower soft tissue contrast, causing inaccurate OARs delineation. In addition, due to a longer scan acquisition time to acquire anatomy information for the breathing cycle period, an extra dose is delivered to patient (10). The different types of artifacts caused by data acquisition and postprocessing also limits the application of 4D-CT in radiotherapy (10).
These limitations of 4D-CT can be potentially overcome by four-dimensional magnetic resonance imaging (4D-MRI) (11,12). 4D-MRI is more desirable for target volume delineation in treatment planning since it provides both respiratory motion information and better soft tissue contrast especially for abdominal cancer (10,13,14). In free-breathing liver SBRT, 4D-MRI can be used to generate AIP for OARs contouring and maximum intensity projection (MIP) for internal target volume (ITV) determination. Two main approaches have been developed in 4D-MRI acquisition: (I) fast 3D MR sequence for real-time volumetric images acquisition (15-17), and (II) fast 2D MR sequence for continuous image acquisition (18-20). The first approach can achieve real-time 4D-MRI without additional post-processing. However, it requires fast imaging technique to acquire images with high frame rates and the image quality is low due to the lack of spatial resolution and motion artifacts (21). The second approach sorts acquisition data retrospectively into different phase, based on the breathing surrogate’s trajectory information acquired during the scan. Several works have been investigated to generate 4D-MRI with different surrogate. Paganelli et al. computed mutual information to identify the respiratory phases without using external surrogate for liver cancer (22). Cai et al. used body area as a respiratory surrogate to reconstruct 4D-MRI for lung and abdominal cancer (23). van de Lindt et al. sorted multi-slice 2D images by computing correlation coefficients between exhale image and all other acquired images to determine the respiratory phases for liver cancer (24). 4D-MRI generated from retrospective sorting technique can improve the image quality. However, the acquisition time is long, typically requiring 15–20 repeated scans (10–15 minutes) to acquire anatomic information for a breathing cycle period. Besides the limitations from currently available hardware and software, 4D-MRI is also limited to the patient breathing regularity and reconstruction method, leading to incomplete ITV in MIP (14).
A novel method called Slice-stacking technique can potentially overcome the limitations of MIP generated from 4D-MRI. It applies a multi-slice MRI acquisition to generate a 3D-MIP (MIPSS-MRI) for ITV contouring. Using slice-stacking technique, MIPSS-MRI is generated by determining the maximum pixel intensity throughout all scanning 2D images (at same slice location) instead of images from all 4D phases. Adamson et al. verified that it is feasible to use slice-stacking technique to generate MIPSS-MRI for multiple 2D cine-MRI (25). In their study, MIPSS-MRI is generated using slice-stacking technique and the volume measurements between SS-MRI and cine-MRI reaches a good agreement.
In our study, the feasibility of using slice-stacking technique to generate MIPSS-MRI for a sequential Half-Fourier-Acquired Single-shot Turbo spin Echo (HASTE) 2D sequence MR images were investigated. 4D digital extended cardiac-torso (XCAT) phantom (26) was used to simulate sequential HASTE 2D sequence MR images with a regular breathing curve and a realistic patient breathing curve, and the ITV volume agreement for liver cancer using slice-stacking technique was investigated.
Methods
Image acquisition
The images with respiratory motion was simulated on 4D XCAT phantom (27), which is controlled by two-time curves: changes in the diaphragm height in superior posterior (SP) direction and changes in the amount of anterior posterior (AP) expansion of the chest. These two inputs are selected as 1 cm in SI direction and 0.5 cm in AP direction in this study. Given a time point in the breathing curve, organs including heart, liver, stomach, spleen and kidneys all move and deform with scaled down motions from the defined diaphragm inputs. A breathing curve was inserted as an input for the 4D XCAT phantom to simulate sequential 2D axial images with respiratory motion.
For the regular breathing motion, a sinusoidal wave with a period of 3.1 s was simulated and the sampling period for image generation was 0.77 s. The tumor diameter was designed to be 10, 30 and 50 mm to cover normal liver tumor size, with a volume of 1.5 cm3, 23 cm3 and 90 cm3 respectively. The resolution of images was 1 mm × 1 mm and the slice thickness was 5 mm. The total slice number was 50 and the repetition number of scans was 20.
For the irregular breathing motion, five patients’ breathing curves were selected for simulation. Figure 1 shows the process of patient’s breathing curve before inserting into 4D XCAT phantom. Original patient breathing curves (Figure 1A) were sampled with a sample frequency of 0.3 s. To acquire adequate breathing curve length to generate images, the breathing curve was extended by connecting a symmetrical curve about the y-axis (Figure 1B). In addition, extremely irregular breathing motion, like sudden cough or deep breath, were removed before simulating images by detecting peaks and valleys calculating their frequencies in histogram (Figure 1C). The extremely irregular breathing motion, occurring at the far left and the far right of the histogram, were removed based on a threshold of a sudden increase (set to be 50% from far left and right to center). From the histogram after reduction in Figure 1D, the pre-processed breathing curve in Figure 1E was completed and could be applied in 4D XCAT digital phantom for image simulation. Tumor size and image parameters were set the same as in simulation with regular breathing motion.

For patient study, three patients with hepatic carcinoma were involved to demonstrate the feasibility. The study was approved by institutional ethics board of Duke University Medical Center and informed consent was taken from all the patients. Images were acquired with a 1.5T Siemens MRI scanner and acquired in an axial half-Fourier acquisition single-shot turbo spin-echo (HASTE/SSFSE) MR sequence, which is a T2-weighted and high-speed sequence. The following imaging parameters were used: repetition time (TR): 550–562 ms; echo time (TE): 57–92 ms; flip angle: 60–123°; slice thickness: 5 mm; pixel resolution: 1.72 mm ×1.72 mm. The repetition number of scans is 15 or 20. SS-MRI was generated using random 5 scans and total 15 or 20 scans, and the ITV relative volume difference was analyzed.
Slice-stacking method
For 4D-MRI, the MIP images are generated by determining the maximum pixel intensity throughout all 4D phases. In this study, SS-MRI is generated by determining the maximum pixel intensity throughout scanning sequential 2D images instead of images from all 4D phases.
All scanning images were used to generate a MIP as a reference, MIPref. MIPi, generated using i repetition number of scans, was used to analyze the relationship between Dice’s similarity coefficient (DSC) and the number of repetitions, and the relationship between the relative ITV volume difference and the number of repetitions. DSC and relative volume difference can be calculated as:
[1]
[2]
where, ITVi is the tumor voxel number of ITV in MIP generated with i repetition number of scans and ITVref is the tumor voxel number of ITV in MIP generated with all scanning images. For the simulation study, the total repetition number of scans is 20. To test the reproducibility of slice-stacking method, MIPi were generated in multiple sets by randomly selecting the order of scans n (n = 1, 2, …, 20). MIPi can be reproductively generated in multiple sets Rr,i,n, where r is the number of multiple sets, i is the repetition number of scans and n is the randomly selected order of scans. Rr,i,n can be randomly chosen such as { Rr,i,n: (R1,i,n: n = 1, 2, 3, 4, 5), (R2,,i,n: n = 1, 3, 5, 7, 9), …, (R30,i,n: n = 1, 6, 13, 17, 20)}. For the patient study, we compared the ITV relative volume difference and DSC generated from random 5 scans and total 15 or 20 scans. The ITV was manually contoured using ITK-SNAP software (28). The ITK-SNAP is a software for structure segmentation in 3D medical images. To demonstrate the reproducibility, for one patient, we repeated selecting 5 scans randomly from total scans to generate MIP and contour ITV to quantify the variance of ITV relative volume difference and DSC from different 5 scans data. MIPi was also reproductively generated in multiple sets to test the reproductivity.
Results
Figure 2 shows the relationship between the DSC of ITV and number of scans, and the relationship between the ITV relative difference and number of scans for MRI images generated with regular breathing motion. The red lines inside the blue box represent the median value of DSC or relative volume difference. The black spots inside the blue box represent the average value of DSC or relative volume difference. In each box, the central mark indicates the median, and the bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers are lines extending above and below each box. Whiskers are drawn from the ends of the interquartile ranges to the furthest observations within the whisker length (the adjacent values). Observations beyond the whisker length are marked as outliers. By default, an outlier is a value that is more than 1.5 times the interquartile range away from the top or bottom of the box. Outliers are indicated with a red + sign.
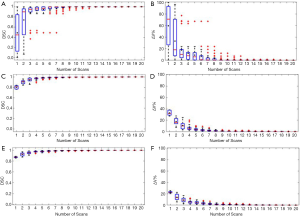
As shown in Figure 2A and Figure 2B, for liver tumor with 10 mm diameter, the mean DSC of ITV is larger than 0.94 and the mean relative ITV volume difference is less than 10% (approximately 0.15 cm3) when using 5 repeated scanning images to reconstruct MIP. Figure 2C and Figure 2D shows that for liver tumor with 30 mm diameter, the mean DSC of ITV is larger than 0.97 and the mean relative ITV volume difference is less than 4.1% (approximately 0.93 cm3) when using 5 repeated scanning images to reconstruct MIP. Figure 2E and Figure 2F indicates that for liver tumor with 50 mm diameter, the mean DSC of ITV is larger than 0.97 and the mean relative ITV volume difference is less than 4.4% (approximately 4 cm3) when using 5 repeated scanning images to reconstruct MIP.
Figure 3 shows the relationship between the DSC of ITV and number of scans, and the relationship between the ITV relative difference and number of scans for MR images generated with irregular breathing motion. Figure 3A and Figure 3B show that for liver tumor with 10 mm diameter, the mean DSC of ITV is larger than 0.94 and the mean relative ITV volume difference is approximately 10% (approximately 0.14 cm3) when using 5 repeated scanning images to reconstruct MIP. Figure 3C and Figure 3D indicate that for liver tumor with 30 mm diameter, the mean DSC of ITV is larger than 0.97 and the mean relative ITV volume difference is approximately 5.5% (approximately 1.13 cm3) when using 5 repeated scanning images to reconstruct MIP. Figure 3E and Figure 3F demonstrate that for liver tumor with 50 mm diameter, the mean DSC of ITV is approximately 0.98 and the mean relative ITV volume difference is less than 4.1% (approximately 3.34 cm3) when using 5 repeated scanning images to reconstruct MIP.
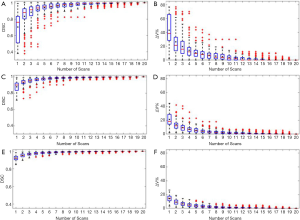
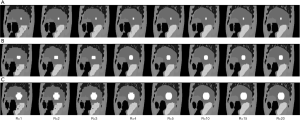
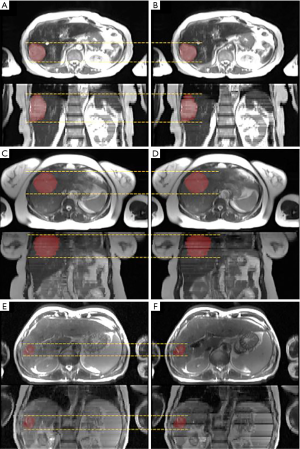
Figure 4 shows the ITV volume variation in the center sagittal image when using 1, 2, 3, 4, 5, 10, 15 and 20 scans to generate SSMRI with regular breathing curve. After 5 repeated scans with regular motion, the shape and position of ITV is generally consistent. The ITV volume information and evaluation are summarized in Table 1. From the MIP generated form 5 scans, the mean ITV relative volume difference is 2.66% and the mean DSC is 0.97. Figure 5 shows the ITV volume in axial view and sagittal view from three patient studies. From axial view and sagittal view, the shape and position of ITV structure is generally consistent. For the reproducibility, the mean ITV relative volume difference for patient 1 is 2.70%±1.57% and the mean DSC is 0.96±0.01, which shows a consistency of ITV volume generated from random 5 scans.
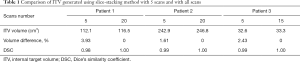
Full table
Discussion
For abdominal motion, an adequate knowledge of organ range of motion is important to provide a better radiation treatment. In abdominal tumor radiotherapy, 4D imaging technique has been commonly used to manage the motion of tumor and surrounding organs and to deliver a precise dose coverage. Currently, 4D-MRI retrospective reconstruction requires high image quality and advanced post-image processing. Current investigations on 4D-MRI retrospective reconstruction requires different surrogate (24-27), which is dependent on the precision of information from surrogate. However, for 4D-MRI retrospect sorting methods, large slice thickness may not quantify motion of a small target (less than 5 mm), and the reconstructed volume may be discontinuous on slice direction. In addition, stitch motion artifacts may occur on reconstructed 4D images (11,12,29).
The proposed technique in this study is a simple and quick method to reconstruct MIP and to generate clinically acceptable ITV in radiation therapy, addressing limitations of 4D-MRI retrospective reconstruction. In this study, we used all acquisition images instead of 4D-MRI to generate MIPref and ITVref. Using all acquisition images as reference can provide a more precise ITV, minimizing the error and lost information from 4D-MRI technique and maximally evaluating the DSC and volume error using SS-MRI technique. We used the same repetition number of scans (15-20) as required to generate 4D-MRI, therefore, the scanning period or repetition scan is the same. Our result shows that the repetition number of scans required to generate tumor ITV from MIPSS-MRI is 3–4 times less than that of MIPref (15–20 repetitions), as the same number of repetitions of MIP from 4D-MRI. Therefore, it is feasible to include that the total acquisition time of our proposed method is 3–4 times less than that of 4D-MRI.
In our study, we demonstrated that the number of repeated scans required to generate tumor ITV for slice-stacking method less than that of 4D-MRI. A longer scan time can capture more respiratory variability and make the 4D motion envelop more robust. Under a same acquisition time, our proposed technique can provide a more precise ITV compared to that of 4D-MRI since we would include every scan to generate MIP. Our results showed that this method could be implemented to liver tumor with diameter not smaller than 10 mm. In addition, our proposed method is not limited to different imaging protocols. In our study, we demonstrated the application on HASTE 2D axial MR images. This method can also be applied to other 2D or 3D MR sequences and different slice orientation.
There are several limitations in our study. First, we did not directly compare the ITV generated form slice-stacking method to the ITV from 4D-MRI, thus lacking an intuitive comparison between slice stacking technique and 4D-MRI retrospective method. In our study, we selected MIP generated from all scanning images MIPref as a gold standard to investigate the feasibility of slice stacking since the MIPref includes complete motion information, which might be missing after 4D-MRI retrospective reconstruction. Second, for slice-stacking technique, DSC and relative ITV volume difference depends on the tumor size and patents’ breathing pattern. If the tumor size is small, it is expected that the DSC would be smaller and relative volume difference would be larger, when using 5 repeated scanning volumes to reconstruct MIP. More outlier points were seen for the small tumor, which indicates that this technique might not be valid for small tumor or more repetition number of scans might be needed to generate ITV for small tumor. In addition, more tumor size simulation should be conducted to investigate the relationship between DSC and tumor size, and between relative volume difference and tumor size. In our simulation study with irregular breathing motion, more outlier points were found compared to regular breathing motion. It indicates that a more normal breathing motion would have a better construction of ITV. Third, a quantification of irregular breathing motion magnitude was not included. In irregular motion simulation study, we processed patients’ breathing motion and excluded the extremely abnormal motion based on a threshold. However, we did not quantify the threshold and investigate the difference if using these abnormal breathing motion. To address this limitation, for patients with an abnormal breathing motion, a breath gating technique might be used to exclude the scanning images with abnormal breathing magnitude. Finally, for patient studies, the ITV contouring in ITK-SNAP varies based on the physicians and image quality, and the number of cases is limited. More patient studies should be included in future study.
Conclusions
Our study showed the feasibility of quickly generating liver tumor ITV using slice-stacking method on T2-w sequential MR images. The number of scans required to generate tumor ITV for slice-stacking method (5–7 repetition) is 3–4 times less than that of 4D-MRI (15–20 repetitions), indicating the image acquisition time for slice-stacking based ITV determination (3–4 minutes on average) is significantly reduced as compared to that required by 4D-MRI.
Acknowledgments
Funding: This project was partly supported by NIH R21 CA165384, NIH R01 CA226899, GRF 151021/18M, GRF 151022/19M and HMRF 06173276.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-41). JC reports grants from NIH, grants from GRF, grants from HMRF, during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by institutional ethics board of Duke University Medical Center and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Scorsetti M, Clerici E, Comito T. Stereotactic body radiation therapy for liver metastases. J Gastrointest Oncol 2014;5:190-7. [PubMed]
- Benson R, Madan R, Kilambi R, Chander S. Radiation induced liver disease: A clinical update. J Egypt Natl Canc Inst 2016;28:7-11. [Crossref] [PubMed]
- Yoganathan SA, Maria Das KJ, Agarwal A, Kumar S. Magnitude, impact, and management of respiration-induced target motion in radiotherapy treatment: A comprehensive review. J Med Phys 2017;42:101-15. [Crossref] [PubMed]
- Michel R, Françoise I, Laure P, Anouchka M, Guillaume P, Sylvain K. Dose to organ at risk and dose prescription in liver SBRT. Rep Pract Oncol Radiother 2017;22:96-102. [Crossref] [PubMed]
- Tétreau R, Llacer C, Riou O, Deshayes E. Evaluation of response after SBRT for liver tumors. Rep Pract Oncol Radiother 2017;22:170-5. [Crossref] [PubMed]
- Eccles CL, Patel R, Simeonov AK, Lockwood G, Haider M, Dawson LA. Comparison of Liver Tumor Motion With and Without Abdominal Compression Using Cine-Magnetic Resonance Imaging. Int J Radiat Oncol Biol Phys 2011;79:602-8. [Crossref] [PubMed]
- Keall PJ, Mageras GS, Balter JM, Emery RS, Forster KM, Jiang SB, Kapatoes JM, Low DA, Murphy MJ, Murray BR, Ramsey CR, Van Herk MB, Vedam SS, Wong JW, Yorke E. The management of respiratory motion in radiation oncology report of AAPM Task Group 76a). Med Phys 2006;33:3874-900. [Crossref] [PubMed]
- Langen KM, Jones D. Organ motion and its management. Int J Radiat Oncol Biol Phys 2001;50:265-78. [Crossref] [PubMed]
- Ozhasoglu C, Murphy MJ. Issues in respiratory motion compensation during external-beam radiotherapy. Int J Radiat Oncol Biol Phys 2002;52:1389-99. [Crossref] [PubMed]
- Hugo GD, Rosu M. Advances in 4D radiation therapy for managing respiration: Part I - 4D imaging. Z Med Phys 2012;22:258-71. [Crossref] [PubMed]
- Stemkens B, Paulson E, Tijssen R. Nuts and bolts of 4D-MRI for radiotherapy. Phys Med Biol 2018;63:21TR01. [Crossref] [PubMed]
- Paganelli C, Whelan B, Peroni M, Summers P, Fast M, van de Lindt T, McClelland J, Eiben B, Keall P, Lomax T, Riboldi M, Baroni G. MRI-guidance for motion management in external beam radiotherapy: current status and future challenges. Phys Med Biol 2018;63:22TR03. [Crossref] [PubMed]
- Yang J, Cai J, Wang H, Chang Z, Czito BG, Bashir MR, Palta M, Yin FF. Is diaphragm motion a good surrogate for liver tumor motion? Int J Radiat Oncol Biol Phys 2014;90:952-8. [Crossref] [PubMed]
- von Siebenthal M, Székely G, Gamper U, Boesiger P, Lomax A, Cattin P. 4D MR imaging of respiratory organ motion and its variability. Phys Med Biol 2007;52:1547-64. [Crossref] [PubMed]
- D Dinkel J. Hintze C, Tetzlaff R, Huber PE, Herfarth K, Debus J, Kauczor HU, Thieke C. 4D-MRI analysis of lung tumor motion in patients with hemidiaphragmatic paralysis. Radiother Oncol 2009;91:449-54. [Crossref]
- Blackall JM, Ahmad S, Miquel ME, Mcclelland JR, Landau DB, Hawkes DJ. MRI-based measurements of respiratory motion variability and assessment of imaging strategies for radiotherapy planning. Phys Med Biol 2006;51:4147-69. [Crossref] [PubMed]
- Cusumano D, Dhont J, Boldrini L, Chiloiro G, Teodoli S, Massaccesi M, Fionda B, Cellini F, Azario L, Vandemeulebroucke J, De Spirito M. Valentini Vi, Verellen Dirk. Predicting tumour motion during the whole radiotherapy treatment: a systematic approach for thoracic and abdominal lesions based on real time MR. Radiother Oncol 2018;129:456-62. [Crossref] [PubMed]
- Yang J, Cai J, Wang H, Chang Z, Czito BG, Bashir MR, Yin FF. Four-Dimensional Magnetic Resonance Imaging Using Axial Body Area as Respiratory Surrogate: Initial Patient Results. Int J Radiat Oncol Biol Phys 2014;88:907-12. [Crossref] [PubMed]
- Liu Y, Yin FF, Chen NK, Chu ML, Cai J. Four-dimensional magnetic resonance imaging with retrospective k-space reordering: a feasibility study. Med Phys 2015;42:534-41. [Crossref] [PubMed]
- Liu Y, Yin FF, Czito BG, Bashir MR, Cai J. T2-weighted four-dimensional magnetic resonance imaging with result-driven phase sorting. Med Phys 2015;42:4460-71. [Crossref] [PubMed]
- Tryggestad E, Flammang A, Han-Oh S, Hales R, Herman J, McNutt T, Roland T, Shea S, Wong J. Respiration-based sorting of dynamic MRI to derive representative 4D-MRI for radiotherapy planning. Med Phys 2013;40:051909. [Crossref] [PubMed]
- Paganelli C, Summers P, Bellomi M, Baroni G, Riboldi M. Liver 4DMRI: A retrospective image-based sorting method. Med Phys 2015;42:4814-21. [Crossref] [PubMed]
- Cai J, Chang Z, Wang Z, Segars WP, Yin FF. Four-dimensional magnetic resonance imaging (4D-MRI) using image-based respiratory surrogate: A feasibility study. Med Phys 2011;38:6384-94. [Crossref] [PubMed]
- van de Lindt TN, Fast MF, van der Heide UA, Sonke JJ. Retrospective self-sorted 4D-MRI for the liver. Radiother Oncol 2018;127:474-80. [Crossref] [PubMed]
- Adamson J, Chang Z, Wang Z, Yin FF, Cai J. Maximum Intensity Projection (MIP) Imaging Using Multi-Slice Cine MRI. Med Phys 2010;37:5914-20. [Crossref] [PubMed]
- Segars WP, Sturgeon G, Mendonca S, Grimes J, Tsui BMW. 4D XCAT phantom for multimodality imaging research. Med Phys 2010;37:4902-15. [Crossref] [PubMed]
- Segars WP, Tsui BMW, Cai J, Yin FF, Fung GSK, Samei E. Application of the 4-D XCAT Phantoms in Biomedical Imaging and Beyond. IEEE Trans Med Imaging 2018;37:680-92. [Crossref] [PubMed]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: Significantly improved efficiency and reliability. NeuroImage 2006;31:1116-28. [Crossref] [PubMed]
- Wang C, Subashi E, Yin FF, Chang A, Cai J. A Spatiotemporal-Constrained Sorting Method for Motion-robust 4D-MRI: A Feasibility Study. Int J Radiat Oncol Biol Phys 2019;103:758-66. [Crossref] [PubMed]


