
Diagnostic interchangeability of deep convolutional neural networks reconstructed knee MR images: preliminary experience
Introduction
The long acquisition times required for magnetic resonance imaging (MRI) have limited the accessibility of this modality compared with other cross-sectional imaging modalities such as computed tomography. However, recent advances in scanner and coil technology, as well as new image reconstruction techniques such as parallel imaging (PI), simultaneous multislice (SMS) imaging, compressed sensing (CS), and machine learning reconstruction algorithms using deep learning (DL), allow for accelerated acquisition of MR images. All of these techniques reduce acquisition time by undersampling k-space. In PI, the most widely available of these techniques for clinical use, k-space is undersampled by reducing the number of phase-encoding steps, and the resulting aliased raw data are resolved using the known coil geometry and spatial sensitivities (1). PI, however, inherently results in a decrease in signal-to-noise ratio (SNR), which limits the use of high acceleration factors (AFs). This loss in SNR is avoided with SMS imaging; with this method, more than 1 slice is excited, and readout and PI techniques are then used to separate the signal from each slice. With SMS imaging, however, acceleration is usually limited to excitation of 2 slices simultaneously because of artifact and coil configurations (2). To achieve higher AFs without compromising SNR and resolution, CS has been assessed in recent studies, with promising results. To avoid aliasing associated with undersampling, k-space is randomly undersampled in CS and transformed into a sparse domain from which images are nonlinearly and iteratively reconstructed (1,3). The long image reconstruction time required for CS (sometimes ≥20 min per sequence), however, has limited its clinical adoption (4).
DL image reconstruction techniques “learn” the optimal method to reconstruct images by reconstructing images from a highly undersampled data set, comparing them to reference images from a fully sampled data set, and minimizing the difference between these images through iterative mathematical algorithms such as convolutional neural networks (CNNs) (5-13). Although this iterative method requires a slow offline training process, once training is completed, future undersampled data can subsequently be noniteratively reconstructed. Therefore, DL-based acceleration methods combine the benefits of ultrafast online reconstruction similar to conventional MRI reconstructions while achieving a high degree of acceleration without comprising SNR or resolution. The main drawback of this technique is its requirement for large amounts of training data; additionally, the reconstruction algorithm remains a “black box” due to the complexity of the deep network. As such, the diagnostic capability of such reconstructions is largely unexplored, having been assessed in only a few small studies. Additionally, the optimal balance between the amount of training data and the number of convolutional layers is unknown.
Most CNNs that have been used for MRI acceleration have used a small number of convolutional layers (6). It has been shown that increasing the number of layers can more accurately classify all of the complex features present in MR images with the same amount of training data (14). Using a model with too many layers, however, may lead to overfitting and increase reconstruction errors. In this study, we developed a new deep CNN (DCNN) reconstruction model with an optimal number of layers capable of accelerating MR image acquisition by 6-fold. The primary objective of the study was to test the interchangeability of DCNN images, as well as images from a standard 3-layer CNN, with routine nonaccelerated images for evaluation of internal derangement of the knee. As a secondary objective, the image quality of DCNN and CNN images was compared with that of the nonaccelerated images.
Methods
Data used in the preparation of this article were obtained from the Osteoarthritis Initiative (OAI) database, which is available for public access at http://www.oai.ucsf.edu/. The specific datasets used were from version 1.1. Informed consent was acquired from patients as part of the OAI, and this study did not require additional Institutional Review Board approval.
Model design
Convolutional neural network architecture
The goal of DL-based reconstruction is to obtain an image from undersampled k-space data with an image quality comparable to that of an image obtained from a fully sampled data set. Using CNN, the typical practice is to first perform the Fourier transform on the zero-filled k-space data to obtain an aliased image and to then use training data to find the CNN that maps the aliased image (input x) to the reconstructed image (output y). This nonlinear mapping can be represented as y=F(x;θ), where θ represents the parameters of the network. Figure 1 illustrates the architecture of the CNN.

In each layer, a convolution between the image from the previous layer and a set of filters was performed using the equation
During training, the objective was to learn the nonlinear relationship, or more specifically, the parameter θ, which represents all of the filter coefficients and biases. Learning was achieved through minimizing the loss function between the network prediction and the corresponding ground truth data. For MR image reconstruction, given a set of ground truth images yi and their corresponding undersampled k-space data xi, we used mean squared error (MSE) between them as the loss function as follows:
Training cohort
For training, we used 2D fat-suppressed sagittal intermediate-weighted turbo spin-echo images and 2D non-fat-suppressed coronal intermediate-weighted turbo spin-echo images from the baseline visits of 212 randomly selected patients (424 knee MRI examinations) enrolled in the OAI. This training cohort consisted of 59 men and 153 women (mean age, 64 years; range, 45–79 years). The sequence parameters for the sagittal images were as follows: repetition time (TR) =3,000 ms, echo time (TE) =30 ms, resolution =0.36 mm × 0.46 mm × 3 mm, and original image size =384×307. The sequence parameters for the coronal images were as follows: TR =3,000 ms, TE =29 ms, resolution =0.36 mm × 0.46 mm × 3 mm, and original image size =384×307.
Training models
Multiple models were trained to determine the optimal model design. All models were trained from 5,000 randomly selected images and tested on a separate 500 randomly selected images.
First, to determine the best model performance with regard to the number of convolutional layers, models were constructed with different layers (3, 10, 15, 20, and 25), and the error rates of these models were compared to select the model with the optimal number of layers (DCNN).
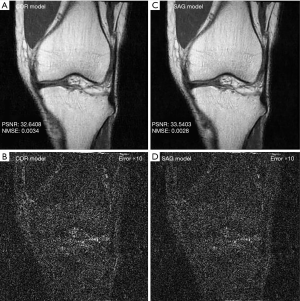
Second, to analyze whether it is necessary to train models with images in each imaging plane, coronal images were reconstructed from models trained from only coronal images (COR model), and these images were compared to coronal images reconstructed from models trained from only sagittal images (SAG model).
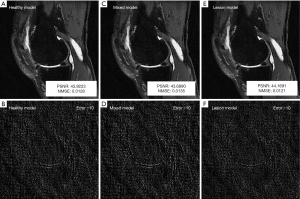
Lastly, to analyze whether it is necessary to train models with pathologic lesions, models were trained using images from patients with different degrees of osteoarthritis (OA). Specifically, 3 different models were trained using sagittal data from 2,700 patients: a “healthy” model using images from patients without OA [Kellgren-Lawrence (KL) grade =0], a “mixed” model using images from patients without OA (KL =0) and with OA (KL =2–4), and a “lesion” model using images from patients with OA (KL =2–4).
Training details
For training each model, all images were rotated 90 degrees 3 times to augment the training size. The entire dataset was normalized to a constant range based on the maximum intensity of the dataset. For k-space undersampling, a variable density sampling with an AF of 6 was used. All data were shuffled before training.
The same hyperparameters and training iterations were used for the CNNs of different layers to ensure a fair comparison between the algorithms. Specifically, a learning rate of 0.0001, momentum of 0.0, and weight decay of 0.0001 were used. It is worth noting that although smaller learning rates are preferred for deeper networks with more layers, we used a fixed learning rate because it is not trivial to determine the rate with respect to the number of layers. Padding was used with each layer during training to generate an output size that was the same as the input size. Each network was trained for 1 million iterations (training time typically 8 hours) on Caffe for Windows using a Matlab interface with two NIVIDIA GTX 1080Ti graphic processing units, each with 3,584 CUDA cores and 11 GB memory. Once fully trained, the models were able to reconstruct images from undersampled data with an average reconstruction time of 0.0001 s/image.
Clinical evaluation
Feasibility study
The feasibility of using the DCNN images (sagittal only) for clinical evaluation was initially evaluated in 10 new patients randomly sampled from the OAI cohort, including two cases for each KL grade [from KL=0 (no OA) to KL =4 (severe OA)] to ensure that cases along the entire spectrum of OA were analyzed. DCNN images and original images from these 10 cases were assessed in a blinded fashion by a musculoskeletal fellowship-trained radiologist with 30 years of experience (CW) using MOAKS (MRI OsteoArthritis Knee Score), a semiquantitative whole knee grading system. Agreement on grades between the two reconstructions was compared using kappa coefficients.
Interchangeability study
Sagittal and coronal images reconstructed using the DCNN model with an optimal number of layers and images reconstructed with a standard 3-layer model (CNN) were tested for interchangeability with the nonaccelerated (original) images in 40 additional patients. Patients were randomly proportionally sampled from the OAI cohort such that there were five patients with lesions graded as KL =0; 5, with KL =4; and 10 each, with KL =1 (questionable OA), KL =2 (mild OA), and KL =3 (moderate OA). Three musculoskeletal-trained radiologists with 13, 15, and 30 years of experience (NS, JP, CW) who were blinded to the reconstruction technique independently assessed the DCNN, CNN, and original images. For each patient, both menisci were evaluated for the presence of a definite tear, defined as increased signal reaching the articular surface on 2 slices, definite morphologic deformity, or displaced flap/fragment (DeSmet 2-touch slice rule) (17). Anterior cruciate ligaments (ACLs) were also evaluated for the presence of a tear, defined as disruption of more than 50% of the fibers, and posterior cruciate and collateral ligaments were evaluated for the presence of a partial tear (high signal and/or partial fiber disruption) or a complete tear (18). Articular cartilage was evaluated in each of six surfaces (medial and lateral femoral condyles, medial and lateral tibial plateaus, trochlea, and patella) using the International Cartilage Repair Society (ICRS) classification, a commonly used clinical grading system (19). Additionally, the visualized bones were evaluated for the presence of fractures, contusions, stress changes, and marrow-replacing lesions. Lastly, image quality was subjectively graded as nondiagnostic, poor, acceptable, or excellent.
Statistical methods
To test the primary objective of whether the accelerated images were interchangeable with the original images, the frequency of agreement for readers grading structures using the accelerated (DCNN and CNN) and original images was compared to the frequency of agreement between readers using the original images. Specifically, the proportion of cases for which two random readers agreed on the original images was calculated and compared with the proportion of cases for which two random readers agreed when one was interpreting the original image and the other was interpreting the accelerated image. The null hypothesis is that the accelerated images are not interchangeable with the original images; the alternative hypothesis is that the accelerated images are interchangeable with the original images. A noninferiority margin of 0.10 was used based on sample size calculations to ensure type I error ≤5% and power ≥80%. A logistic regression model using generalized estimating equations was used to compare the proportions, adjusting for the clustered data, and 95% confidence intervals (CIs) were constructed for the difference in the two proportions. Additionally, the percent of cases graded as nondiagnostic, poor, acceptable, or excellent for both accelerated and original images was calculated.
Results
Comparison of different model designs
Model performance as it relates to the number of convolutional layers in the network is shown in Figure 2. Reconstruction errors decreased with an increasing number of layers initially, and this decrease in reconstruction errors mathematically correlated with a visible improvement in image quality (Figure 3). This improvement, however, saturated at approximately 20 layers, after which the performance declined, and the performance of a model with 20 layers was only negligibly better than a model with 15 layers. Based on these results, a model with 15 convolutional layers (DCNN) was selected for optimal performance.


With regard to the model performance based on training with images from different imaging planes, there were negligible differences quantitatively or qualitatively based on the imaging plane(s) of the training images. The reconstruction errors were nearly identical whether models were trained using all sagittal images, all coronal images, or a mixture of sagittal and coronal images (Table 1). Similarly, there were no visible differences in the image quality of reconstructed images whether models were trained using all sagittal images, all coronal images, or a mixture of sagittal and coronal images (Figure 4).
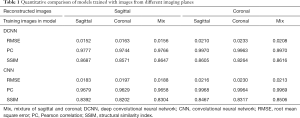
Full table
Similarly, with regard to the need to train models with pathologic lesions, models trained using only patients without OA (healthy model), patients with and without OA (mixed model), and only patients with OA (lesion model) had very similar reconstruction errors quantitatively (Table 2), and the image quality was not visibly distinguishable qualitatively (Figure 5).

Full table
Feasibility study
The 10 patients selected for the feasibility study had a mean age of 70.3 years (range, 52–79 years; four women). There was good to excellent agreement on grading between the sagittal DCNN and original images for most structures, including bone marrow lesions (kappa =0.75), menisci (kappa =0.85), ligament/tendon (kappa =1), cyst/bursa (kappa =0.71), loose bodies (kappa =1), and osteophytes (kappa =0.74). Cartilage grading agreement was also good to excellent for both thickness and size for all subcompartments (kappa range, 0.63–1) with the exception of cartilage thickness in the trochlea and posterior medial tibia thickness and cartilage size in the patella and anterior medial femoral condyle; in these structures, the agreement was moderate (kappa range, 0.46–0.58).
Interchangeability study
The 40 patients selected for the interchangeability study had a mean age of 64.6 years (range, 45–79 years; 21 women). Table 3 summarizes the frequency of pairwise agreement between readers using different reconstructions and the excess disagreement with 95% CI when one reader was using accelerated images and the other was using the original images compared to when both readers were using the original images. The agreement between readers when one reader was using either the DCNN or CNN images and the other was using the original images was very similar to the agreement when both readers were using the original images, with little to no excess disagreement. Both the DCNN and the CNN accelerated images were found to be interchangeable with the original images for assessment of medial and lateral meniscal tears, ACL tears, and cartilage defects (Figures 6,7). Interchangeability could not be tested for other structures evaluated because none of the patients had posterior cruciate ligament tears, medial collateral ligament tears, lateral collateral ligament tears, or fractures and only a few had marrow-replacing lesions.
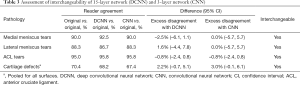
Full table


Table 4 summarizes the readers’ grading of image quality for the different reconstructions. Pooled across all three readers, image quality was graded as excellent or acceptable in 97.5% of cases (117/120) using the original images and in 95% of cases (114/120) using the DCNN images but in only 60% of cases (72/120) using the CNN images. Although the original images were graded as excellent in image quality in 87.5% of cases (105/120), the image quality of DCNN images was graded as excellent in only 20.8% of cases (25/120), and the image quality of CNN images was never graded as excellent (Figure 8). Furthermore, the CNN images were graded as poor or not diagnostic in 40% of cases (48/120). Only two cases were graded as not diagnostic, both of which were CNN reconstructions.
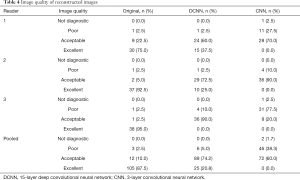
Full table

Discussion
At our institution, the acquisition time for knee MRI using our clinical protocol (five multiplanar 2D turbo spin-echo sequences and a high-resolution 3D sequence) ranges from 15 to 20 minutes. In this study, we developed a 6-fold acceleration technique using a novel 15-layer CNN (DCNN) that is capable of reducing acquisition time to 2 to 3 minutes while generating images that are diagnostically interchangeable with nonaccelerated images and have acceptable if not excellent image quality in most cases. While the diagnostic performance of DCNN was similar to that of a standard 3-layer CNN in terms of interchangeability, reconstruction errors were reduced and image quality was improved with the addition of layers in the DCNN. The image quality of the DCNN images was, however, still inferior to that of the original nonaccelerated images. Training the models using images with or without pathology did not affect performance. Similarly, training the models using images acquired in the same imaging plane or images acquired in a different imaging plane did not affect performance. These observations can be explained by the fact that the deep network is performing de-aliasing/denoising, which is rather independent of the image content.
Recently published studies have used methods such as PI, SMS imaging, CS, and DL to reduce knee MRI acquisition times and produce diagnostic images. For instance, a rapid 5-minute knee MRI protocol using PI was shown to be diagnostically interchangeable with a standard nonaccelerated protocol (20,21). As previously discussed, however, PI results in lower SNR (1), which limits acceleration beyond a factor of 2 if spatial resolution is to be maintained. Furthermore, PI is best suited for 3T imaging because of the inherently higher SNR at 3T. SMS imaging partially overcomes some of these limitations. With SMS, more than 1 slice is excited with each radiofrequency pulse and the signal from all of the slices is acquired simultaneously, thereby decreasing the acquisition time (2). The inherent aliasing of the slices that occurs at readout is then resolved by using the spatial sensitivities of the multichannel surface coils (as is done with PI). With SMS imaging, however, the acceleration does not result in the loss in SNR that occurs with PI. Additionally, SMS imaging and PI can be combined to achieve even greater AFs. In a recent study, a combined SMS + PI knee protocol with an AF of 4 was diagnostically similar to a clinically used PI protocol with an AF of 2, and the protocols demonstrated similar SNRs (22). However, because of slice cross-talk artifact and increased energy deposited into the tissues with multislice excitation, the AF is practically limited to 4 with SMS imaging. CS, on the other hand, offers the ability to accelerate images beyond a factor of 4 (3). Several recent studies have shown that CS can be used to reduce the acquisition times of 3D fast spin-echo knee MRI sequences with AFs ranging from 1.5 to 6 (23-26). CS has also been used to reduce the acquisition time of advanced metal artifact reduction sequences with an AF of 8 while maintaining artifact reduction and SNR (27). One caveat to these results is that CS is especially effective in these settings because of the ability to downsample in 2 phase-encoding directions with a 3D data set and the inherent sparsity of data with metal artifact reduction sequences; CS is not as effective with standard 2D fast spin-echo images, in which there is less data sparsity and only 1 phase-encoding direction. Additionally, although acquisition time can be reduced with CS, image reconstruction time is longer because of the iterative process, partially negating the time savings (25).
DL acceleration techniques as described in this study offer many of the same advantages as CS but without some of the limitations. Specifically, DL methods can achieve high degrees of acceleration without compromising SNR, but unlike CS, DL can be effectively applied to 2D acquisitions, as shown in this study. Furthermore, image reconstruction with DL acceleration is rapid because once the model is trained, DL image reconstruction is not an iterative process. The average reconstruction time per image in this study was 0.0001 s.
While using DL for MRI acceleration has been increasingly explored in different settings, only a few studies have evaluated DL acceleration for knee MRI (12,13,28-30). Hammernik et al. (28) was the first to explore the use of DL for accelerated MRI reconstruction, using a variational neural network (VNN) to achieve a 4-fold acceleration with image quality comparable to that of nonaccelerated images in a small cohort of 10 patients. Liu et al. (30) described a novel technique, termed Sampling-Augmented Neural neTwork with Incoherent Structure (SANTIS), which incorporated variations in undersampling while training a generative adversarial network (GAN); this technique resulted in less error in reconstructing images from undersampled knee MRI data. Recently, Knoll et al. (29) published the results of a competition that compared quantitative measurements of error and qualitative grading of image quality for 33 different DL algorithms in reconstructing undersampled knee MRI data acquired with multichannel and single-channel coil systems with AFs of 4 and 8. Of note, irrespective of coil type, AF, or reconstruction algorithm, the authors noted cases with subtle pathology evident on the fully sampled images that were not accurately depicted on the accelerated images.
Our study supports and expands on the results from these previously published studies. While these previous studies have focused on evaluation of DL accelerated images by quantitative measurements of reconstruction errors and subjective grading of image quality of accelerated images, the focus of this study was to compare the diagnostic performance of DL accelerated images. As noted above, DL accelerated images may miss important findings such as meniscal tears or cartilage defects, which are more clinically relevant than either quantitative measurements of error or qualitative measurements of image quality. As such, the primary measure in this study was the interchangeability of DL accelerated images and fully sampled images in evaluating common internal derangements in the knee, specifically meniscal tears, cartilage defects, and ACL tears. To our knowledge, this is the first study to show that accelerated images reconstructed with a DL algorithm are diagnostically interchangeability with fully sampled images. Additionally, this is the first study to show that using a deeper CNN (one with more layers) can improve image quality and produce images that are nearly always acceptable or excellent in diagnostic quality. Lastly, our results demonstrate that model performance does not require the presence of pathology in the images or the need to train on individual imaging planes or sequences.
The study design and patient population resulted in some inherent limitations. Models were trained from a homogeneous data set taken from the OAI, using images with similar contrast and resolution. Additional studies will be needed to test the generalizability of the reconstruction model with more heterogeneous data obtained using different contrast and resolution parameters. The interchangeability of images showing less common pathology such as injuries to ligaments other than the ACL, fractures, and bone lesions could not be assessed in this relatively small sample size of 40 patients. As discussed, however, the presence of pathology in the training sample did not improve reconstruction errors, suggesting that learning is likely based on other imaging features. Larger studies with more diverse pathology will be needed to validate these findings. Despite the improvement in image quality seen with a deep 15-layer CNN model compared with that of a standard 3-layer CNN model, the image quality was still inferior to the quality seen on nonaccelerated images. Further optimization of the model beyond selecting the appropriate number of layers is likely needed to improve performance. Lastly, undersampling was performed in a retrospective fashion from reconstructed images, which is inferior to prospective K space undersampling. Additional studies using prospective undersampling are needed to validate these results.
In conclusion, we have demonstrated that it is feasible to accelerate knee MRI acquisition by 6-fold through the use of a novel deep 15-layer CNN. Images obtained with this technique have acceptable image quality, and these images are diagnostically interchangeable with nonaccelerated images. Although image quality is improved with a deeper CNN model, further optimization is still needed to achieve excellent image quality. The ability to use DL to achieve highly accelerated MRI acquisitions has many important potential clinical implications, including increasing the accessibility of MRI by reducing wait times; reducing the cost of MRI by increasing efficiency; and reducing the motion artifact, patient discomfort, and claustrophobia associated with MRI as a result of long scan times.
Acknowledgments
The OAI is a public-private partnership comprising five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the National Institutes of Health, a branch of the Department of Health and Human Services, and conducted by the OAI Study Investigators. Private funding partners include Merck Research Laboratories; Novartis Pharmaceuticals Corporation; GlaxoSmithKline; and Pfizer, Inc. Private sector funding for the OAI is managed by the Foundation for the National Institutes of Health. This manuscript was prepared using an OAI public use data set and does not necessarily reflect the opinions or views of the OAI investigators, the NIH, or the private funding partners.
Funding: This work was supported in part by the National Institutes of Health R21EB020861. This study was also partially supported by a Society of Skeletal Radiology (SSR) Research Seed Grant (“Highly Accelerated Knee MRI using a Novel Deep Convolutional Neural Network Algorithm: A Multi-Reader Comparison Study”).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-20-664). DL serves as an unpaid editorial board member of Quantitative Imaging in Medicine and Surgery. NS reports grants from Society of Skeletal Radiology, during the conduct of the study; CW reports grants from Society of Skeletal Radiology, during the conduct of the study; NO reports other from Quantitative Imaging Biomarker Alliance (QIBA), outside the submitted work. The other authors have no conflicts of interest to declare.
Ethical Statement: Data used in the preparation of this article were obtained from the Osteoarthritis Initiative (OAI) database, which is available for public access at http://www.oai.ucsf.edu/. The specific datasets used were from version 1.1. Informed consent was acquired from patients as part of the OAI, and this study did not require additional Institutional Review Board approval.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Glockner JF, Hu HH, Stanley DW, Angelos L, King K. Parallel MR imaging: a user's guide. Radiographics 2005;25:1279-97. [Crossref] [PubMed]
- Barth M, Breuer F, Koopmans PJ, Norris DG, Poser BA. Simultaneous multislice (SMS) imaging techniques. Magn Reson Med 2016;75:63-81. [Crossref] [PubMed]
- Lustig M, Donoho D, Pauly JM. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med 2007;58:1182-95. [Crossref] [PubMed]
- Hollingsworth KG. Reducing acquisition time in clinical MRI by data undersampling and compressed sensing reconstruction. Phys Med Biol 2015;60:R297-322. [Crossref] [PubMed]
- Sandino CM, Dixit N, Cheng JY, Vasanawala SS. Deep convolutional neural networks for accelerated dynamic magnetic resonance imaging. Long Beach: 31st Conference on Neural Information Processing Systems, 2017.
- Wang S, Su Z, Ying L, Peng X, Zhu S, Liang F, Feng D, Liang D. Accelerating magnetic resonance imaging via deep learning. Proc IEEE Int Symp Biomed Imaging 2016;2016:514-7. [PubMed]
- Lee D, Yoo J, Ye JC. Deep residual learning for compressed sensing MRI. Melbourne: 2017 IEEE 14th International Symposium on Biomedical Imaging, 2017.
- Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A deep cascade of convolutional neural networks for dynamic MR image reconstruction. IEEE Trans Med Imaging 2018;37:491-503. [Crossref] [PubMed]
- Wang S, Huang N, Zhao T, Yang Y, Ying L, Liang D. 1D Partial Fourier Parallel MR imaging with deep convolutional neural network. Honolulu: ISMRM 25th Annual Meeting & Exhibition, 2017.
- Wang S, Xiao T, Tan S, Liu Y, Ying L, Liang D. Undersampling trajectory design for fast MRI with super-resolution convolutional neural network. Honolulu: ISMRM 25th Annual Meeting & Exhibition, 2017.
- Jin KH, McCann MT, Froustey E, Unser M. Deep convolutional neural network for inverse problems in imaging. IEEE Trans Image Process 2017;26:4509-22. [Crossref] [PubMed]
- Johnson PM, Recht MP, Knoll F. Improving the speed of MRI with artificial intelligence. Semin Musculoskelet Radiol 2020;24:12-20. [Crossref] [PubMed]
- Lin DJ, Johnson PM, Knoll F, Lui YW. Artificial intelligence for MR image reconstruction: an overview for clinicians. J Magn Reson Imaging 2020. Epub ahead of print. [Crossref] [PubMed]
- Kim J, Lee JK, Lee KM. Accurate image super-resolution using very deep convolutional networks. Las Vegas: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 Las Vegas: IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2016.
- Krogh A, Hertz JA. A simple weight decay can improve generalization. In: Moody JE, Hanson SJ, Lippmann RP, editors. Advances in neural information processing systems 4. San Mateo: Morgan Kauffmann Publishers, 1992:950-7.
- De Smet AA, Tuite MJ. Use of the "two-slice-touch" rule for the MRI diagnosis of meniscal tears. AJR Am J Roentgenol 2006;187:911-4. [Crossref] [PubMed]
- Hong SH, Choi JY, Lee GK, Choi JA, Chung HW, Kang HS. Grading of anterior cruciate ligament injury. Diagnostic efficacy of oblique coronal magnetic resonance imaging of the knee. J Comput Assist Tomogr 2003;27:814-9. [Crossref] [PubMed]
- Brittberg M, Peterson L. Introduction of an articular cartilage classification. ICRS Newsletter 1998;1:58.
- Subhas N, Benedick A, Obuchowski NA, Polster JM, Beltran LS, Schils J, Ciavarra GA, Gyftopoulos S. Comparison of a fast 5-minute shoulder MRI protocol with a standard shoulder MRI protocol: a multiinstitutional multireader study. AJR Am J Roentgenol 2017;208:W146-54. [Crossref] [PubMed]
- Alaia EF, Benedick A, Obuchowski NA, Polster JM, Beltran LS, Schils J, Garwood E, Burke CJ, Chang IJ, Gyftopoulos S, Subhas N. Comparison of a fast 5-min knee MRI protocol with a standard knee MRI protocol: a multi-institutional multi-reader study. Skeletal Radiol 2018;47:107-16. [Crossref] [PubMed]
- Fritz J, Fritz B, Zhang J, Thawait GK, Joshi DH, Pan L, Wang D. Simultaneous multislice accelerated turbo spin echo magnetic resonance imaging: comparison and combination with in-plane parallel imaging acceleration for high-resolution magnetic resonance imaging of the knee. Invest Radiol 2017;52:529-37. [Crossref] [PubMed]
- Altahawi FF, Blount KJ, Morley NP, Raithel E, Omar IM. Comparing an accelerated 3D fast spin-echo sequence (CS-SPACE) for knee 3-T magnetic resonance imaging with traditional 3D fast spin-echo (SPACE) and routine 2D sequences. Skeletal Radiol 2017;46:7-15. [Crossref] [PubMed]
- Fritz J, Fritz B, Thawait GK, Raithel E, Gilson WD, Nittka M, Mont MA. Advanced metal artifact reduction MRI of metal-on-metal hip resurfacing arthroplasty implants: compressed sensing acceleration enables the time-neutral use of SEMAC. Skeletal Radiol 2016;45:1345-56. [Crossref] [PubMed]
- Fritz J, Raithel E, Thawait GK, Gilson W, Papp DF. Six-fold acceleration of high-spatial resolution 3D SPACE MRI of the knee through incoherent k-space undersampling and iterative reconstruction-first experience. Invest Radiol 2016;51:400-9. [Crossref] [PubMed]
- Kijowski R, Rosas H, Samsonov A, King K, Peters R, Liu F. Knee imaging: rapid three-dimensional fast spin-echo using compressed sensing. J Magn Reson Imaging 2017;45:1712-22. [Crossref] [PubMed]
- Fritz J, Ahlawat S, Demehri S, Thawait GK, Raithel E, Gilson WD, Nittka M. Compressed sensing SEMAC: 8-fold accelerated high resolution metal artifact reduction MRI of cobalt-chromium knee arthroplasty implants. Invest Radiol 2016;51:666-76. [Crossref] [PubMed]
- Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, Knoll F. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med 2018;79:3055-71. [Crossref] [PubMed]
- Knoll F, Murrell T, Sriram A, Yakubova N, Zbontar J, Rabbat M, Defazio A, Muckley MJ, Sodickson DK, Zitnick CL. Advancing machine learning for MR image reconstruction with an open competition: overview of the 2019 fastMRI challenge. Magn Reson Med 2020. Epub ahead of print. [Crossref] [PubMed]
- Liu F, Samsonov A, Chen L, Kijowski R, Feng L. SANTIS: Sampling-augmented neural network with incoherent structure for MR image reconstruction. Magn Reson Med 2019;82:1890-904. [Crossref] [PubMed]


