
Combination of generative adversarial network and convolutional neural network for automatic subcentimeter pulmonary adenocarcinoma classification
Introduction
Lung cancer is the leading cause of cancer-related death worldwide. Non-small cell lung cancer (NSCLC), including squamous carcinoma, pulmonary adenocarcinoma, and large cell carcinoma, accounts for about 85% of all lung cancers (1). However, due to the lack of routine and low-cost biomarkers, the diagnosis and classification of early-stage NSCLC is challenging; this is especially true for subcentimeter pulmonary adenocarcinoma nodules (2,3). Different subtypes of pulmonary adenocarcinoma, including adenocarcinoma in situ (AIS), minimally invasive adenocarcinoma (MIA), and invasive adenocarcinoma (IAC), vary in invasiveness rates and risks of recurrence, which correspond to different treatment regimens (4,5). Misdiagnosis of pulmonary adenocarcinoma can lead to many serious consequences, such as adverse treatment and even medical malpractice. Computed tomography (CT), a widely used imaging technique in clinic, can provide internal lung information and facilitate the diagnosis of pulmonary adenocarcinoma. However, as there is a limited resolution of mini-nodules and a large number of images require interpretation, it is hard for radiologists to distinguish between different subtypes of pulmonary mini-nodules (6). Therefore, bronchoscopy-guided biopsy or CT-guided biopsy should be conducted if the CT examinations indicate invasiveness. However, this process not only causes pain and injury in patients but is also associated with complications such as pneumothorax and intrapulmonary hemorrhage (7,8). Furthermore, the low sensitivity of the biopsy makes it difficult to accurately obtain the target tissue and cause the misdiagnosis by pathologists. Ever since the Luna challenge 16 (9) and the 2017 Kaggle Data Science Bowl were held, many studies have focused on the classification of benign and malignant nodules, and have achieved good results (10,11) based on the public The Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) dataset (12). However, the benign and malignant labels in the LIDC-IDRI database are only based on the judgment of the radiologists and lack pathological evaluations (a golden standard in clinic) after surgical resection. These limitations reduce the clinical value of the research. Some studies on pulmonary adenocarcinoma classification have instead focused on the modeling of radiomics features and other manual labeling characteristics (13-16). These methods are labor-intensive and place more burden on doctors. To clarify mini-nodule classification and to assist in the daily work of radiologists, we collected the nodule dataset with pathological labels after surgical resection and proposed a computer-assisted automatic pulmonary adenocarcinoma diagnosis system based on unenhanced chest CT scan images.
One critical problem in the medical imaging field is the small number of samples. Compared with the millions of labeled data in natural image datasets [e.g., ImageNet (17)], medical image collection is limited by ethical privacy and relies on experts with related experience. Despite the recent release of large datasets like CheXpert (18), most medical image datasets are still small in general. For example, one of the most popular datasets of the human brain contains just 373 MRI images, and another nucleus segmentation dataset contains only 30 cropped digital microscopic tissue images (19,20). These insufficient sample sizes hinder the application of deep learning methods, and researchers have put forward many solutions, such as transfer learning, multi-task learning, and data augmentation (21-23). Data augmentation is the most direct and universal method. Common data augmentation techniques, such as cropping, flipping, and rotation, are widely used in the field of image processing to alleviate the data shortage problem (24). However, these techniques can only produce highly correlated and relatively small datasets and have a lack of variation. The generative adversarial network (GAN), one of the most attractive generative models, can be used for data augmentation (25). It has the advantage of simulating data distribution without the explicit modeling of potential probability density functions. The samples generated by a GAN can offer more variability and thus enrich the dataset. GANs are composed of two components: a generator network and a discriminator network. The generator aims to produce fake images and the discriminator tries to distinguish between real and fake images. After iterative and alternate training of the generator and discriminator, the generator can fit the distribution of images, producing high-quality images that can deceive the discriminator and even people. A large number of studies on the GAN data augmentation theory, training techniques, and natural/medical image augmentation applications have been published (26-29). However, to our knowledge, few studies have tried to use GANs to augment pulmonary nodules. Currently, the literature on GAN synthetic pulmonary nodules includes only a few conference papers (30), reporting on radiologists being fooled by images generated by DC-GAN (31-33) and integrating GAN into detection/segmentation/super-resolution tasks. Other reports have focused on inpainting the erased nodules based on some variants of pix2pix (34-36). However, little is known about how to combine different GAN techniques to generate nodules from scratch and how the classification performance can be improved by directly adding GAN synthetic data. Therefore, there is an urgent need to provide more information for different GAN techniques and to better understand the potential of GAN augmentation in the medical imaging field.
In this study, we first compared and integrated the different GAN techniques. Wasserstein distance loss, gradient penalty [from wGAN-GP (26)], pixel-level condition [from pix2pix (27)], progressive growing [from pgGAN (37)], and pixel-wise normalization (38) were sequentially implemented and compared. Secondly, we designed a convolutional neural network (CNN) for the subcentimeter pulmonary adenocarcinoma classification. We not only compared the performance of CNN under different augmentation methods, but we also evaluated the state-of-the-art methods of our datasets. The results suggested that GAN has the potential to alleviate the data insufficiency problem and to improve the classification performance of the CNN. Our lightweight CNN model is also convenient for implementation in hospital diagnostic systems, which can assist in reducing radiologists’ daily work and can promote the development of precision medical care.
The highlights of this study are the following:
- The deep learning pulmonary adenocarcinoma classification method obtained state-of-the-art results;
- Good guidance for building a patch-based algorithm on a small dataset was achieved;
- A thorough evaluation of several GAN techniques was conducted.
Methods
Data collection
The CT image datasets were collected from the Shanghai Public Health Clinical Center and Zhongshan Hospital Affiliated to Fudan University. The classification of the pulmonary adenocarcinoma was confirmed through the pathological analysis of surgical specimens. The CT images were obtained by the following four CT scanners: Brilliance (Philips Medical Systems Inc., Netherlands), SOMATOM Definition AS (Siemens AG, Munich, Germany), SCENARIA (64 channels/128 slices, Hitachi Ltd., Tokyo, Japan), or Aquilion One (Canon Medical Supply Co., Ltd, Tokyo, Japan). An unenhanced chest CT examination was performed to obtain the whole lung scan of each patient, and the thickness of all the CT images was 1 mm. Only ground-glass nodules measuring 5–10 mm on pathologic examination and without severe respiratory motion artifacts were included.
The dataset was composed of 206 nodules with postoperative pathological labels, with 30 adenocarcinomas in situ (AISs), 119 MIAs, and 57 IACs. The typical examples of each class are presented in Figure 1. Compared with AISs and MIAs, IACs were always larger in morphology, with a blurry tumor-lung interface, higher density, and bubble-like shape. However, even with many morphological descriptions on these three subtypes of nodules, it is still challenging to make an accurate diagnosis of the nodule based on CT images. The difficulty is partially due to the significant variations within the category and the limited perception of the human eye for the tiny pixel differences of the mini-nodules.
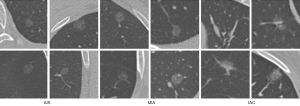
The contours and locations of the nodules were labeled by a junior radiologist (SYY, with 4 years of experience in chest radiology) using an in-house annotation tool based on a region growing algorithm (39). Then, the binary masks of these nodules were reviewed by a senior radiologist (FS, with more than 15 years of experience in chest radiology). Different from the benign/malignant label judged by radiologists in the public LIDC dataset, the classification labels in our dataset were determined by pathological evaluation after surgical resection of the tumors. Pathological evaluation is the clinical gold standard and was used for our dataset. All specimens were classified according to the consensus of the two experienced pathologists using the 2011 International Multidisciplinary Classification Standard for Lung Adenocarcinoma (5). Because of different sources in our CT scans, we processed each nodule into several two-dimensional (2D) slices, instead of a three-dimensional (3D) cube, and fed them into the 2D-CNN in turn.
Data processing
In this step, the images of the nodule itself were obtained. The area of the nodule in every CT scan was calculated based on the radiologists’ annotation. Three CT images with the largest nodule area were selected. A few of them were not consecutive because our selection criteria were only based on the nodule area. Three 64×64×1 pixel images centered on the nodule were cut, and one nodule could generate three images in this step. These images were named as the raw dataset.
Having sufficient data is important because even a small CNN containing hundreds of parameters is easy to over-fit. To avoid this problem, the common strategy of data augmentation was employed. Common augmentation techniques include translation, rotation, scaling, and flipping. We first translated the images randomly by [1,20%×64] pixels. Then rotations were performed with angles at 30°,45°,60°,90°,120°,135°,150°. Afterwards, the images were rescaled with a stochastic ratio ranging from 80% to 120%. Finally, the nodule patches were flipped up-down and left-right. The images generated by the above operations were 64×64 pixels, consistent with the raw dataset created previously. These techniques not only amplified the dataset but also acquired image information near the nodule. The dataset generated in this step was named as the common augmented dataset.
GAN-based image synthesis
Generative methods are widely used in image synthesis. Currently, the most advanced generation models include autoregressive models (40), variational autoencoders (41), and GAN (25). A GAN has the strength of producing sharp images and the weakness of an unstable training process. The three most well-known GAN techniques were tested in order to synthesize nodule images. Firstly, the original wGAN-GP (26) and pix2pix (27) model with a deep convolutional structure were implemented. However, these synthesized images were blurry, and the quality was not satisfactory. Inspired by the idea of pgGAN, we implemented a progressive growing wGAN, with sliced Wasserstein distance loss, progressive growing, and pixel-wise normalization. It stabilized the training process and generated high-quality images. The well-trained models and related code for this training can be found at https://github.com/wangyunpengbio/nodule_generation_with_progressive_growing_wGAN. The GAN was implemented using Python 3.6 based on TensorFlow 1.12.0 deep learning library, and the training process was accelerated by four graphics processing units (GPU type: Nvidia Corporation TITAN Xp 12G). The details of this model are described below.
Wasserstein GAN
The Wasserstein distance, also known as the Earth mover’s distance, measures the probability distance between PX and PY. It is defined in detail below.
|
| [1] |
Progressive growing
The generator and discriminator networks were symmetric with each other and trained at the same time. Whenever a new layer was added, we faded it smoothly to prevent a large impact on the already well-trained, smaller-resolution network. Firstly, the network was trained to fit a low-resolution image. Then, we continuously and incrementally increased the resolution so that the model could gradually adapt to the high-dimensional distribution. This effectively solved the instability problem in GAN training and reduced the training time. Previous work (37) demonstrates that this technique can speed up the training by 2–6 times.
Pixel-wise normalization
Escalation of signal magnitudes, caused by unbalanced training of the two networks, often occurred in the GAN network. The feature map was thus normalized in the pixel level after every convolution layer. The normalization formula was as follows:
|
| [2] |
where ax,y and bx,y denotes the original and normalized feature map in pixel (x, y), and N represents the total number of feature maps. This normalization method was originally proposed in 2012 (38) to alleviate signal magnitudes problems, and has been widely used in academia and industry.
GAN architecture
The architecture of the GAN used for pulmonary adenocarcinoma generation is shown in Figure 2A. The generator consists of nine convolution layers. Firstly, 512-dimensional random noise was fed into a fully connected layer, and then the 4×4 pixels feature map was generated by the first convolution layer. Afterward, the feature map was passed through four blocks and each block was composed of two convolution layers. The detailed structure of the block is shown in Figure 2B. These blocks constantly doubled the height and width of the feature map, finally generating a 64×64 pixel image.
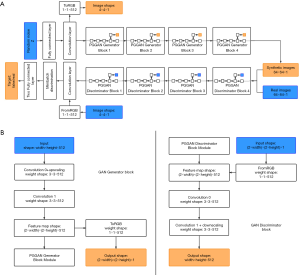
The discriminator also consisted of nine convolution layers. Mirrored with the generator, it started with a 64×64 pixel image, and passed through four blocks, which were composed of two convolution layers, and ended with a 4×4 pixel feature map. Mini-batch discrimination was added into the final convolution layer to make training more stable. Finally, this feature map was passed through two, small, fully connected layers to get the last true or false target.
Both the generator and discriminator contained 22 million parameters, with a 3×3 kernel of the convolution layer. Adam was used as the optimization function, and the loss function was the same as wGAN-GP. The batch size was 128 when the network was trained on the images with a resolution lower than 16×16. To avoid exceeding the memory limit, the batch size was set to 64 and 32 for images with the resolution of 32×32 and 64×64, respectively. The common augmented dataset mentioned above was chosen as the input of the GAN. The model was trained until a total of 5,000,000 images were fed into the network. The nodule patches generated in this step were called the GAN synthetic dataset.
CNN classification
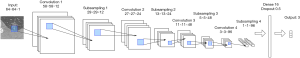
We tested and compared different network structures and finally designed the most suitable network architecture of the CNN for pulmonary adenocarcinoma classification (Figure 3). Because of the small image size and limited dataset, many prominent CNN architectures like VGGNet, ResNet, MobileNet were not suitable for this task. This specially designed small-scale CNN contained fewer parameters and was thus less prone to overfitting. The input images were fixed to 64×64 pixels, and the intensity was normalized to 0 and 1. The architecture was composed of four convolution layers, four max-pooling layers, and one fully connected layer. Relu was chosen as an activation function. The network ended with a soft-max layer, predicting the classes of the image. Furthermore, the dropout method was employed after each max-pooling layer (dropout rate =0.25) to prevent over-fitting. The entire model contained only 55,000 parameters, which was lightweight and easy to train. The small size of the model also meant it could be more conveniently deployed by in-hospital diagnostic systems. Adam was used as the optimization function, and cross-entropy was chosen as the loss function. The model was trained for 500 epochs, with a batch size of 128. The implementation of the CNN was based on the Keras framework, and the training process was accelerated by four graphics processing units (GPU type: Nvidia Corporation TITAN Xp 12G).
Workflow
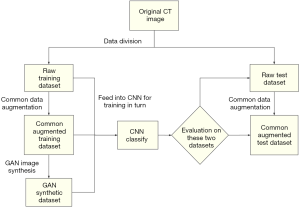
The flowchart for the experiments conducted to evaluate augmented data and CNN classification is presented in Figure 4. For each experiment, all data were divided into training sets and test sets, with data split at the patient level. Due to class imbalance (AIS:MIA:IAC =30:119:57), the IACs and MIAs were first down-sampled to match the number of AISs. Only 30 of 119 MIAs, 30 of 57 IACs, and all 30 AISs were randomly chosen in each experiment. After setting up the dataset with the 90 patients’ images, we used three-fold cross-validation. Each fold contained 30 patient images, including 10 AISs, 10 MIAs, and 10 IACs. Three images with the largest nodule area were cut from every patient’s nodule and named as a raw dataset. After common augmentation, every image in the raw dataset was augmented into 70 images, and the whole common augmentation dataset contained 18,900 (90×3×70) images. The GAN models were trained for each nodule class on the previously divided training set. Each GAN synthesized 10,000 images of nodule patches for each nodule class, and a total of 30,000 images from three categories were generated. Finally, the common augmentation dataset and the GAN synthetic dataset were fed into the CNN for training and evaluation according to the flowchart shown in Figure 4.
Results
Images generated by GAN
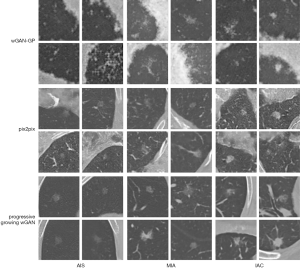
Examples of nodule patches generated by the different types of GAN are presented in Figure 5. In every experiment, three GAN models were trained for each nodule class. The nodule patches generated by the wGAN-GP were of very low quality. Pix2pix captured the vascular characteristics but failed to mimic the lung wall. Our progressive-growing wGAN synthesized images with high quality and clarity, containing more specific nodule features such as blood vessels and the chest wall. More importantly, it also captured the unique characteristics of each nodule class. The synthesized IACs had a bubble-like shape, and the synthesized AISs-MIAs contained a clear tumor-lung interface.
CNN classification
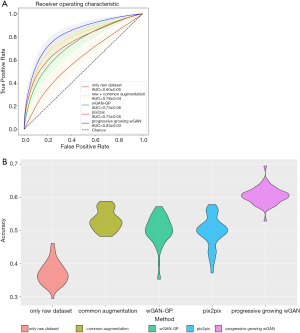
To reasonably analyze the GAN-generated images and our three-category classification CNN, we considered this clinically important subtask: binary classification of IA nodules (IAC) and non-IA nodules (AIS and MIA). The disease-free survival rate for the patients with MIA is close to 100% when treated with complete resection (5). However, the disease-free survival rate for those with IAC only ranges from 60% to 70% (43,44), indicating the need for more aggressive treatments (e.g., chemotherapy). Many previous studies have also merged AIS and MIA into one category (45,46). The data extraction process was random, so we repeated 50 experiments, and finally calculated the average receiver operating characteristic curve (ROC). The three-category classification accuracy was also recorded. The raw dataset, common augmented dataset, and GAN synthetic dataset were added into CNN for training in turn. The changes in ROC curves and CNN classification accuracy are presented in Figure 6. First, the performance of the CNN was unstable and inadequate when only using the raw dataset (binary area under the curve (AUC): 0.60±0.05; three-category accuracy: 37%±3.4%). Second, an additional dataset significantly improved the performance of the CNN (binary area under the curve (AUC) improvement: 0.13–0.23; three-category accuracy improvement: 16.2–23.5%). Lastly, the CNN performed best with the dataset generated by the progressive-growing wGAN (binary AUC: 0.83±0.03; three-category accuracy: 60.5%±2.6%).
t-SNE (47) was employed for dimension reduction and visualization to analyze the result further. Features were extracted from the last layer of the well-trained CNN and then fed into t-SNE for dimension reduction. Afterward, the images were finally visualized into 2D scatterplots (Figure 7). We extracted the features from the test set images to illustrate whether the CNN network was improved by adding synthetic data. Furthermore, we only took 200 cases at random for drawing to avoid too much point density on the scatter plot. Three classes of nodules represented by three colors were gradually dispersed from left to right. Figure 7 shows the improvements of the CNN’s feature extraction ability under additional datasets.
Comparison with transfer learning
Transfer learning is also a common solution for a small dataset. To present a comprehensive comparison, several well-known networks (VGG16, ResNet50, MobileNet) were transferred to our classification task. The fully connected layers of the original network were replaced by a global average pooling layer and a fully connected layer with three outputs. Adam was used for optimization, and an early stop with the patience set to 10 was used to avoid overfitting (training was halted when the validation loss did not decrease after 10 epochs). The classification accuracy of the different finetuned networks under different datasets are shown in Table 1. First, adding data, especially the progressive-growing wGAN-synthesized dataset, could significantly contribute to the fine-tuning process, (highest accuracy under the GAN-synthesized dataset). These results revealed the potential of transfer learning combined with a GAN. However, the proposed CNN (Number of parameters: 55,000) was still much lighter than the other massive pretrained networks and achieved the best performance (accuracy: 60.5%±2.6%).

Full table
Expert assessment
The quality of the synthetic images was assessed by radiologists. We designed a visual Turing test involving two experts, a primary radiologist, and a senior radiologist. A dataset that was mixed up with 100 real samples and 100 samples generated by our progressive growing wGAN was created. Radiologists were asked to classify every image in that dataset as being either real or fake. The radiologists were told some tricks to identify the GAN synthetic images to allow the radiologists to be more experienced in distinguishing between true and fake images, such as strange lung walls and checkerboard artifacts. They were also told the total number of true and fake images. The result of this test is summarized in Table 2. The true-positive ratio (how many real images have been identified as real), the false-positive ratio (how many synthetic images have been classified as real), and the accuracy were calculated. The average accuracy of distinguishing fake nodules for radiologists was only 60.5% (little higher than random), suggesting there to be a high-quality of GAN synthetic images.

Full table
Comparison with other classification methods
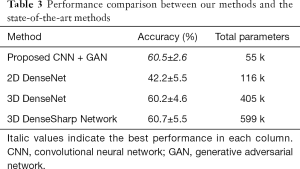
In our final experiment, the performance of the GAN plus CNN–based system proposed in this work was compared with a recently published state-of-the-art subcentimeter pulmonary adenocarcinoma classification method, called the 3D DenseSharp Network (48). 2D DenseNet and 3D DenseNet (49) were also within our range of comparison. 3D DenseSharp Network is an enhanced 3D DenseNet model created through the introduction of segmentation loss. To compare these approaches, we ran 2D DenseNet, 3D DenseNet, and 3D DenseSharp on the dataset of the current work. The optimized hyperparameters for the subcentimeter pulmonary adenocarcinoma classification task were the same as the public code found in their original article: patch size =32×32×3; λ=0.2. Following their experimental setup, the weights of the DenseNet networks were randomly initialized, rather than pretrained on ImageNet. The ground truth masks used in DenseSharp Network were collected as described in the above methods section. We trained in four-fold cross-validations 12 times. Table 3 summarizes the accuracy and the number of total parameters across the different methods. Primarily, the proposed GAN plus CNN-based system achieved high and stable accuracy (60.5%±2.6%) that was comparable to the state-of-the-art methods (60.7%±5.5%). Additionally, our methods (number of parameters: 55,000) were much more lightweight than the state-of-the-art methods (number of parameters: 599,000).
Full table
Discussion
The purpose of this study was to generate synthetic medical images with a GAN to augment datasets and to improve the performance on CNN classification tasks. We tried and compared several GAN models and finally designed the progressive-growing wGAN. By adding synthetic images, the CNN model showed much better performance, comparable to the state-of-the-art methods. The CNN model also contained fewer parameters, which is important for applications in hospital computer-aided design (CAD) systems.
The GAN was the main point of our work and achieved good results. Some researchers have studied the use of a GAN for pulmonary nodule generation (30). Many challengers of the Kaggle competition have also tried to use a GAN for data augmentation. Comprehensive experiments were conducted (comparison with pix2pix, wGAN-GP), and the GAN proposed in this article can synthesize images with higher quality.
The CNN classification was also an important part of our work. Experiments were conducted to determine what extent the augmentation data could improve the classification performance of the CNN model. From Figure 6, we could see that adding a common augmented dataset improved the accuracy of CNN significantly, either in binary or three-category classification tasks. These results verified the idea that common augmentation methods could stabilize training and improve the generalization of the model. Interestingly, the GAN did not always outperform common augmentation methods (e.g., wGAN-GP and pix2pix). These results can be attributed to the lower image quality generated by an inappropriate GAN model. After using the data generated by our progressive-growing wGAN, the performance of the CNN reached the highest level (an accuracy increase of 23.5% from the raw dataset and 7.3% from the common augmentation dataset). These results demonstrated that our progressive-growing wGAN did have the ability to enlarge small datasets and improve CNN classification task performance.
Visualization can intuitively show how the augmented data improve the CNN model. We found that the three classes of nodules represented by three colors could be gradually dispersed from left to right (Figure 7). These findings showed that the performance of the model would continue to improve as more synthetic data were added to the training set. From the transition from the first to the second subgraph, we noted that among the three kinds of mixed samples, the points represented by IACs would be the first to be separated. These results were closely related to the unique features of the IACs compared to the MIAs or AISs. In clinical treatment, IACs are also significantly different from MIAs and AISs. Some confusing features between MIAs and AISs still exist, as shown in the second subgraph; further, they are not completely separated. By adding the GAN-synthetic dataset, the MIAs and AISs were slightly more separate, indicating a gradual improvement in the classifier.
Transfer learning was also compared with the proposed methods, but there were many limitations. One limitation was that these networks were pretrained on ImageNet with massive datasets and contained a huge amount of parameters (Table 1). These large sizes of networks have caused efficiency problems in practical applications. Moreover, the performance of the fine-tuned big networks was still not as good as the proposed CNN training from scratch. The best performance of transfer learning was VGG16 (Accuracy: 60.2%±4%). However, its performance was still worse than the proposed methods, even after adding a dataset synthesized by the progressive-growing wGAN. These results could be attributed to the oversized model capacity, making it difficult to fine-tune with such a small dataset.
Expert assessment was conducted for the GAN synthesis evaluation. Table 2 summarizes the experts’ results. It was notable that the accuracy for distinguishing between true and fake was 56% and 65%, respectively, which is only slightly better than chance. Consistent with common sense, the primary radiologist had a slightly lower accuracy due to limited experience. Overall, the radiologists correctly found 81% real samples, but 60% of the synthetic samples were mistaken as real. All in all, radiologists could not reliably distinguish between true and fake samples. These interesting results have led to the conclusion that the generated samples were of high quality and authentic.
Discerning the details between different images has been the advantage of deep learning. Zhao et al. (48) showed that it was even difficult for radiologists to make a classification on pulmonary adenocarcinoma from CT images (accuracy: 52.35% junior radiologists, 55.85% senior radiologists). In the final experiment, we compared our methods with state-of-the-art methods (Table 3). After training with the GAN-augmented data, our classifiers achieved an accuracy of 60.5% on average, which was comparable to the state-of-the-art methods. Also, the performance of the classifier obtained by our training method was more stable, with a smaller variance in the accuracy during repeated experiments. Moreover, our classifier contained fewer parameters than state-of-the-art methods. All of these showed the potential for a great value in practical applications.
Despite many advantages of our generative and classification models, there was still room for improvement. First, for the GAN model, the generated images in our experiments had a lower resolution of 64×64 pixels compared to the original 512×512-pixel CT images. Although numerous studies have documented (50,51) the synthesizing of high-resolution images, they have not addressed the problem of the huge cost of computing resources. Therefore, how to compress the network with novel theories in computer vision will be the focus of our future work. Additionally, the input volume of our CNN is 2D. One extension could be transforming the models into 3D and introducing some more advanced techniques and network architectures. Investigation of the GAN architectures that generate 3D images would also be worthwhile. Another plan was to incorporate more features into the CNN. However, diagnosis by radiologists is not only based on CT images but also include a series of comprehensive judgments about patient’s age, medical history, smoking, and so on. We will integrate this kind of contextual information into our model in future work to further improve the performance of our model.
Conclusions
Automatic pulmonary adenocarcinoma classification from a CT scan can be an essential part of the diagnosis system. In this article, we proposed a classification approach based on deep learning that had an excellent performance comparable to the state-of-the-art methods and that was more lightweight. Also, a comprehensive comparison between different GAN techniques was conducted, revealing how and to what extent the GAN improved the performance of classifiers. Through these experiments, we can provide guidance for how to combine different GAN techniques to synthesize images at unprecedented levels of realism on small datasets. The proposed framework has the potential to be generalized to synthesize other objects of interest in medical images. This article can inspire the building of a robust artificial intelligence auxiliary diagnosis system.
Acknowledgments
Funding: This work is supported by funding from the National Key Research and Development Program of China (No. 2018YFC0910700), the Shanghai Committee of Science and Technology (No. 18511102704), and the Intelligent Medical Special Research Foundation of the Shanghai Health and Family Planning Commission (No. 2018ZHYL0104).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/qims-19-982). The authors have no conflicts of interest to declare.
Ethical Statement: The study was approved by the ethics committee of the Shanghai Public Health Clinical Center. The need for individual consent was waived by the committee.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- He J, Hu Y, Hu M, Li B. Development of PD-1/PD-L1 Pathway in Tumor Immune Microenvironment and Treatment for Non-Small Cell Lung Cancer. Sci Rep 2015;5:13110. [Crossref] [PubMed]
- Aoki MN, Amarante MK, de Oliveira CEC, Watanabe MAE. Biomarkers in Non-Small Cell Lung Cancer: Perspectives of Individualized Targeted Therapy. Anticancer Agents Med Chem 2018;18:2070-7. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- Donahue JM, Morse CR, Wigle DA, Allen MS, Nichols FC, Shen KR, Deschamps C, Cassivi SD. Oncologic Efficacy of Anatomic Segmentectomy in Stage IA Lung Cancer Patients With T1a Tumors. Ann Thorac Surg 2012;93:381-7. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE, Garg K, Austin JH, Asamura H, Rusch VW, Hirsch FR, Scagliotti G, Mitsudomi T, Huber RM, Ishikawa Y, Jett J, Sanchez-Cespedes M, Sculier JP, Takahashi T, Tsuboi M, Vansteenkiste J, Wistuba I, Yang PC, Aberle D, Brambilla C, Flieder D, Franklin W, Gazdar A, Gould M, Hasleton P, Henderson D, Johnson B, Johnson D, Kerr K, Kuriyama K, Lee JS, Miller VA, Petersen I, Roggli V, Rosell R, Saijo N, Thunnissen E, Tsao M, Yankelewitz D. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Sun Z, Ng KH, Sarji SA. Is utilisation of computed tomography justified in clinical practice? Part IV: applications of paediatric computed tomography. Singapore Med J 2010;51:457-63. [PubMed]
- Becker H, Herth F, Ernst A, Schwarz Y. Bronchoscopic Biopsy of Peripheral Lung Lesions Under Electromagnetic Guidance: A Pilot Study. Journal of Bronchology & Interventional Pulmonology 2005;12:9-13.
- Eberhardt R, Anantham D, Ernst A, Feller-Kopman D, Herth F. Multimodality Bronchoscopic Diagnosis of Peripheral Lung Lesions. Am J Respir Crit Care Med 2007;176:36-41. [Crossref] [PubMed]
- Setio AAA, Traverso A, de Bel T, Berens MSN. Bogaard Cvd, Cerello P, Chen H, Dou Q, Fantacci ME, Geurts B, Gugten Rvd, Heng PA, Jansen B, de Kaste MMJ, Kotov V, Lin JYH, Manders JTMC, Sóñora-Mengana A, García-Naranjo JC, Papavasileiou E, Prokop M, Saletta M, Schaefer-Prokop CM, Scholten ET, Scholten L, Snoeren MM, Torres EL, Vandemeulebroucke J, Walasek N, Zuidhof GCA, Ginneken Bv, Jacobs C. Validation, comparison, and combination of algorithms for automatic detection of pulmonary nodules in computed tomography images: The LUNA16 challenge. Medical Image Analysis 2017;42:1-13. [Crossref] [PubMed]
- Jung H, Kim B, Lee I, Lee J, Kang J. Classification of lung nodules in CT scans using three-dimensional deep convolutional neural networks with a checkpoint ensemble method. BMC Medical Imaging 2018;18:48. [Crossref] [PubMed]
- Dai Y, Yan S, Zheng B, Song C. Incorporating automatically learned pulmonary nodule attributes into a convolutional neural network to improve accuracy of benign-malignant nodule classification. Phys Med Biol 2018;63:245004. [Crossref] [PubMed]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J Digit Imaging 2013;26:1045-57. [Crossref] [PubMed]
- Li M, Narayan V, Gill RR, Jagannathan JP, Barile MF, Gao F, Bueno R, Jayender J. Computer-Aided Diagnosis of Ground-Glass Opacity Nodules Using Open-Source Software for Quantifying Tumor Heterogeneity. AJR Am J Roentgenol 2017;209:1216-27. [Crossref] [PubMed]
- Teramoto A, Tsujimoto M, Inoue T, Tsukamoto T, Imaizumi K, Toyama H, Saito K, Fujita H. Automated Classification of Pulmonary Nodules through a Retrospective Analysis of Conventional CT and Two-phase PET Images in Patients Undergoing Biopsy. Asia Ocean J Nucl Med Biol 2019;7:29-37. [PubMed]
- Shi Z, Deng J, She Y, Zhang L, Ren Y, Sun W, Su H, Dai C, Jiang G, Sun X, Xie D, Chen C. Quantitative features can predict further growth of persistent pure ground-glass nodule. Quant Imaging Med Surg 2019;9:283-91. [Crossref] [PubMed]
- Mao L, Chen H, Liang M, Li K, Gao J, Qin P, Ding X, Li X, Liu X. Quantitative radiomic model for predicting malignancy of small solid pulmonary nodules detected by low-dose CT screening. Quant Imaging Med Surg 2019;9:263-72. [Crossref] [PubMed]
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. ImageNet Large Scale Visual Recognition Challenge. International Journal of Computer Vision 2015;115:211-52. [Crossref]
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpanskaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, Ng AY. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. arXiv e-prints 2019. arXiv:1901.07031.
- Kumar N, Verma R, Sharma S, Bhargava S, Vahadane A, Sethi A. A Dataset and a Technique for Generalized Nuclear Segmentation for Computational Pathology. IEEE Transactions on Medical Imaging 2017;36:1550-60. [Crossref] [PubMed]
- Marcus DS, Fotenos AF, Csernansky JG, Morris JC, Buckner RL. Open Access Series of Imaging Studies: Longitudinal MRI Data in Nondemented and Demented Older Adults. J Cogn Neurosci 2010;22:2677-84. [Crossref] [PubMed]
- Gurovich Y, Hanani Y, Bar O, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz P, Kamphausen SB, Zenker M, Bird LM, Gripp KW. DeepGestalt - Identifying Rare Genetic Syndromes Using Deep Learning. arXiv e-prints 2018. arXiv:1801.07637.
- Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, McKeown A, Yang G, Wu X, Yan F, Dong J, Prasadha MK, Pei J, Ting MYL, Zhu J, Li C, Hewett S, Dong J, Ziyar I, Shi A, Zhang R, Zheng L, Hou R, Shi W, Fu X, Duan Y, Huu VAN, Wen C, Zhang ED, Zhang CL, Li O, Wang X, Singer MA, Sun X, Xu J, Tafreshi A, Lewis MA, Xia H, Zhang K. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-1131.e9. [Crossref] [PubMed]
- Nishio M, Sugiyama O, Yakami M, Ueno S, Kubo T, Kuroda T, Togashi K. Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS One 2018;13:e0200721. [Crossref] [PubMed]
- Stephen RC, Wiles LJ, Robert HE, Gregory SM, Gale HD, James CA. Modeling With Limited Data: The Influence of Crop Rotation and Management on Weed Communities and Crop Yield Loss. Weed Science 2009;57:175-86. [Crossref]
- Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative Adversarial Networks. arXiv e-prints 2014. arXiv: 1406.2661.
- Gulrajani I, Ahmed F, Arjovsky M, Dumoulin V, Courville A. Improved Training of Wasserstein GANs. arXiv e-prints 2017. arXiv:1704.00028.
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-Image Translation with Conditional Adversarial Networks. arXiv e-prints 2016. arXiv: 1611.07004.
- Shin HC, Tenenholtz NA, Rogers JK, Schwarz CG, Senjem ML, Gunter JL, Andriole K, Michalski M. Medical Image Synthesis for Data Augmentation and Anonymization using Generative Adversarial Networks. arXiv e-prints 2018. arXiv: 1807.10225.
- Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing 2018;321:321-31. [Crossref]
- Chuquicusma MJM, Hussein S, Burt J, Bagci U. How to Fool Radiologists with Generative Adversarial Networks? A Visual Turing Test for Lung Cancer Diagnosis. arXiv e-prints 2017. arXiv:1710.09762.
- Tang Y, Cai J, Lu L, Harrison AP, Yan K, Xiao J, Yang L, Summers RM. CT Image Enhancement Using Stacked Generative Adversarial Networks and Transfer Learning for Lesion Segmentation Improvement. arXiv e-prints 2018. arXiv:1807.07144.
- Teramoto A, Fujita H, Yamamuro O, Tamaki T. Automated detection of pulmonary nodules in PET/CT images: Ensemble false-positive reduction using a convolutional neural network technique. Med Phys 2016;43:2821-7. [Crossref] [PubMed]
- Yi X, Babyn P. Sharpness-Aware Low-Dose CT Denoising Using Conditional Generative Adversarial Network. J Digit Imaging 2018;31:655-69. [Crossref] [PubMed]
- Jin D, Xu Z, Tang Y, Harrison AP, Mollura DJ. CT-Realistic Lung Nodule Simulation from 3D Conditional Generative Adversarial Networks for Robust Lung Segmentation. arXiv e-prints 2018. arXiv:1806.04051.
- Yang J, Liu S, Grbic S, Arindra Adiyoso Setio A, Xu Z, Gibson E, Chabin G, Georgescu B, Laine AF, Comaniciu D. Class-Aware Adversarial Lung Nodule Synthesis in CT Images. arXiv e-prints 2018. arXiv:1812.11204.
- Han C, Kitamura Y, Kudo A, Ichinose A, Rundo L, Furukawa Y, Umemoto K, Li Y, Nakayama H. Synthesizing Diverse Lung Nodules Wherever Massively: 3D Multi-Conditional GAN-based CT Image Augmentation for Object Detection. arXiv e-prints 2019.
- Karras T, Aila T, Laine S, Lehtinen J. Progressive Growing of GANs for Improved Quality, Stability, and Variation. arXiv e-prints 2017. arXiv: 1710.10196.
- Krizhevsky A, Sutskever I, Hinton G. ImageNet Classification with Deep Convolutional Neural Networks. Neural Information Processing Systems 2012;25.
- Ren H, Zhou L, Liu G, Peng X, Shi W, Xu H, Shan F, Liu L. An unsupervised semi-automated pulmonary nodule segmentation method based on enhanced region growing. Quant Imaging Med Surg 2020;10:233-42. [Crossref] [PubMed]
- van den Oord A, Kalchbrenner N, Kavukcuoglu K. Pixel Recurrent Neural Networks. arXiv e-prints 2016.
- Kingma DP, Welling M. Auto-Encoding Variational Bayes. arXiv e-prints 2013. arXiv:1312.6114.
- Arjovsky M, Chintala S, Bottou L. Wasserstein GAN. arXiv e-prints 2017. arXiv: 1701.07875.
- Yim J, Zhu LC, Chiriboga L, Watson HN, Goldberg JD, Moreira AL. Histologic features are important prognostic indicators in early stages lung adenocarcinomas. Mod Pathol 2007;20:233-41. [Crossref] [PubMed]
- Borczuk AC, Qian F, Kazeros A, Eleazar J, Assaad A, Sonett JR, Ginsburg M, Gorenstein L, Powell CA. Invasive size is an independent predictor of survival in pulmonary adenocarcinoma. Am J Surg Pathol 2009;33:462-9. [Crossref] [PubMed]
- Lim HJ, Ahn S, Lee KS, Han J, Shim YM, Woo S, Kim JH, Yie M, Lee HY, Yi CA. Persistent Pure Ground-Glass Opacity Lung Nodules ≥ 10 mm in Diameter at CT Scan: Histopathologic Comparisons and Prognostic Implications. Chest 2013;144:1291-9. [Crossref] [PubMed]
- Son JY, Lee HY, Kim JH, Han J, Jeong JY, Lee KS, Kwon OJ, Shim YM. Quantitative CT analysis of pulmonary ground-glass opacity nodules for distinguishing invasive adenocarcinoma from non-invasive or minimally invasive adenocarcinoma: the added value of using iodine mapping. Eur Radiol 2016;26:43-54. [Crossref] [PubMed]
- van der Maaten L, Hinton G. Viualizing data using t-SNE. J Mach Learn Res 2008;9:2579-605.
- Zhao W, Yang J, Sun Y, Li C, Wu W, Jin L, Yang Z, Ni B, Gao P, Wang P, Hua Y, Li M. 3D Deep Learning from CT Scans Predicts Tumor Invasiveness of Subcentimeter Pulmonary Adenocarcinomas. Cancer Res 2018;78:6881-9. [Crossref] [PubMed]
- Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. arXiv e-prints 2016. arXiv:1608.06993.
- Brock A, Donahue J, Simonyan K. Large Scale GAN Training for High Fidelity Natural Image Synthesis. arXiv e-prints 2018. arXiv:1809.11096.
- Donahue J, Simonyan K. Large Scale Adversarial Representation Learning. arXiv e-prints 2019. arXiv:1907.02544.



