3D convolutional neural network for differentiating pre-invasive lesions from invasive adenocarcinomas appearing as ground-glass nodules with diameters ≤3 cm using HRCT
Introduction
Lung cancer is the leading cause of cancer-related death worldwide. With the development of multi-slice spiral computed tomography (CT) technology and the popularization of low-dose spiral CT screening, an increasing number of lung cancer cases are likely to be detected in the early stage, which significantly reduces the number of deaths due to lung cancer. In high-resolution CT (HRCT), the nodules showed solid, partially solid, or non-solid lesions, and most of the partially solid and non-solid lesions were identified as malignant (1,2).
To solve the problem of the accurate classification of pulmonary nodules, in 2011, the international association for lung cancer research (IASLC), the American Thoracic Society (ATS) and the European Respiratory Society (ERS) proposed a new classification of lung adenocarcinoma. Ground-glass nodules (GGNs) were classified as atypical adenomatous hyperplasias (AAHs), in situ adenocarcinomas (AISs), minimally invasive adenocarcinomas (MIAs), or invasive adenocarcinomas (IACs) depending on the size of the lesion and the presence of solid components in the pathological analysis (3). The new classification had an important effect on the choice of treatment and follow-up of patients because the prognosis of different pathological subtypes varies greatly. Studies have shown that disease-free survival in early stage AIS and MIA patients is close to 100% (4,5), while disease-free survival in IACs patients is 60–70% (6-8). Therefore, it is very important to accurately evaluate the risk of malignant lesions on diagnostic CT for early intervention.
Lobectomy is still the standard surgical treatment for early non-small cell lung cancer (NSCLC); however, some studies (9-12) have shown that selecting the appropriate sublobar resection can achieve a prognosis comparable to lobectomy for peripheral small cell lung cancer. Sublobar resection has the advantages of preserving lung function, low perioperative morbidity and mortality, and the opportunity to resect the subsequent primary lung tumor (13). Patients with AIS or MIA may receive sublobar resection. Although invasive features of small-sized NSCLC have been identified by retrospective studies based on imaging features, these features are still difficult to apply to guide surgical treatment due to their unascertained accuracy. For these reasons, imaging techniques need to be further improved to guide sublobar resection.
Traditional computer-aided diagnosis (CAD) methods (14,15) utilize various feature extraction protocols to describe the characteristics of nodules. Machine learning algorithms have been employed to classify lesion properties. However, choosing appropriate features for nodule detection is difficult due to the variety of nodule locations, sizes, shapes, and densities. Instead of manual intervention in traditional feature engineering, deep learning techniques (16) automatically acquire features for nodule detection and classification. The cascade convolution neural network (CNN) framework was here used to detect nodules and to determine whether a nodule is benign or malignant.
Ever since the IASLC/ATS/ERS proposed a new classification of lung adenocarcinomas in 2011, preoperative HRCT has been used not only to diagnose pulmonary nodules such as malignant tumors but also to distinguish pre-invasive lesions (PILs) (AAH, AIS, or MIA) and IACs. In our hospital, the classification of GGNs by preoperative HRCT has become one basis for guiding resections of lung cancer.
The purpose of this study was to develop and validate a 3D CNN using quantitative imaging biomarkers obtained from HRCT scans to classify GGNs as PILs (AAH, AIS, or MIA) or IACs.
Methods
Patient selection
Ethical approval was obtained for this retrospective analysis, and informed consent requirement was waived. Our study recruited consecutive lung adenocarcinoma patients with pulmonary lesions (F/M, 1075/470; mean age 55.8±10.6 years; age range, 32–84 years) between January 2010 and March 2017. Within our institution’s PACS database, we selected patients with the words “GGN”, “GGO”, “ground glass opacity”, “ground glass nodule”, “non-solid nodule”, “sub-solid nodule” or “mixed nodule”. All patients underwent routine chest CT scans before surgery on their lung lesions. The inclusion criteria consisted of: (I) histopathologically confirmed PILs (AAH, AIS, MIA) and IACs; (II) the presence of solitary pulmonary nodule (with lesion measuring >5 and <30 mm in the longest diameter). Patients who met one of the following criteria were excluded: (I) previous systemic treatment (chemotherapy or radiotherapy), which could produce changes in texture features; (II) incomplete CT datasets at our institution; and (III) two or more lesions that had been resected.
CT acquisition and reconstruction
CT examinations including the chest and the upper abdomen were performed as part of the routine CT imaging protocol for the selected patients at our institution by using the Somatom Definition AS (Siemens Healthcare, Germany), Sensation 64 (Siemens Healthcare, Germany) and Brilliance (Philips Healthcare, the Netherlands), with a tube voltage of 120 kV and a current of 200 mA. The target lesion (the largest one if there were multiple nodules) was selected for reconstruction. All imaging data were reconstructed using a standard reconstruction algorithm, and the reconstruction parameters were as follows: slice thickness, 0.625 mm; increment, 1 mm; pitch, 1.078; field of view, 15 cm; and matrix, 512×512.
Preprocessing
First, all raw data were converted to Hounsfield units (HUs), a standard measure of radio-density in CT images. The preprocessing procedure consisted of two steps: lung mask extraction and the 3D reconstruction. Lung mask extraction removes tissues that are not in the lung but are similar to nodules in their spherical shapes, which reduces false positive signals in the detected lesions. The 3D reconstruction enables the developed algorithm to preprocess the data collected from different scanners or imaging protocols. For lung mask extraction, morphological operations (erosion and dilation) were utilized to calculate all connected components, and strategies (e.g., convex hull and dilation) were employed to generate the left and right lungs (17). Then, 3D reconstruction was performed by resizing the data to 1 mm × 1 mm × 1 mm resolution, and the luminance was clipped between −1,200 and 600, scaled to 0–255 and convert to unsigned 8 bits integer.
Automatic lung cancer diagnosis
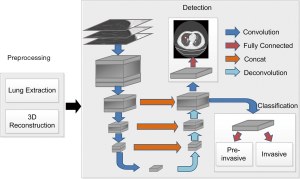
Automatic diagnosis of lung cancer involves the detection of all possible nodules and classification of GGNs as PILs or invasive lung adenocarcinoma. A 3D CNN framework was designed for nodule detection and invasive lung adenocarcinoma probability evaluation. The flow of the automatic diagnosis framework is shown in Figure 1.
For better nodule detection performance, multi-resolution data was used to train the nodule detection network. For high resolution CT (HRCT) data (thickness = 0.625 mm), raw resolution helps to retain initial information. For thicker data (e.g., thickness ≥1.5 mm), resizing to unified resolution (e.g., 1 mm × 1 mm × 1 mm) improves nodule detection, particularly on CT data with collected with small thickness. Thus, the data with raw resolution and 1 mm × 1 mm × 1 mm resized resolution were used during the training process, and raw resolution was used to predict nodules on HRCT data. One problem that should be considered during the training process is that high-resolution CT data cause GPU memory exploration. Thus, small 3D patches were cropped from the preprocessed data. The size of each cropped patch is 128×128×128×1 (height × length × width × channel) in space. If the cropped patch goes beyond the lung scan, the excess of cropped patch over the scanned data was padded with the value of 170, which corresponding to the intensity of normal tissue in CT. The cropped patch of 3D CT data will be entered into the network during network training. The amount of the training set, the validation set, and the test set were 1,075, 270, and 200, respectively.
An encoder-decoder structure-based framework was designed to detect nodules in the input data. The encoder-decoder structure (18) enables the network to conveniently combine multi-scale resolution. A region proposal network proposed in Faster-R-CNN (19) was employed to first roughly locate the nodules and then to further regress the position and the diameter with the trained model. When training detection network, the learning rate was initialized as 0.01, and the learning rate decreased a factor of 10 every 50 epochs. Three different scales of square anchor boxes were used and the corresponding length with respect to the anchor boxes were 2, 10, and 30 mm, respectively. In the process of network design, key techniques, such as 3D convolution and hard-example mining, were used to promote the recall and accuracy of the nodule detection.
Based on the nodule detection results, the probability of one subject suffering invasive lung adenocarcinoma was evaluated with a shallow neural network. The classification network consisted of a 64-neuron, fully connected layer and a one-neuron output layer. The classification network was added as a branch in the decoder structure in the detection network. Accurate nodule detection enables convincing cancer classification; thus, detection and classification networks were alternately trained after the first 100 iterations. In this way, the information obtained from detection presented compelling evidence in making the final classification verdict.
Performance of radiologists with time constraints
An experienced radiologist (SPW, 14 years of medical imaging experience and 10 years of specialized chest imaging) labeled lung nodules on the test dataset and provided a list of nodule locations. Three radiologists volunteered to participate in the study. They understood and agreed with the basic principles and objectives of this study. As determined by the research ethics committee, the radiologists participating in the review panel were not required to provide written informed consent. Radiologists 1, 2, and 3 had 3, 7, and 15 years of experience, respectively. The average age of the three radiologists was 47.7 years (range, 32–45 years) and the average number of years of practice was 7.3 years (range, 3–15 years). Radiologist 3 had experience in chest imaging as a special interest area.
Three radiologists with time constraints (radiologists WTC) independently evaluated the corresponding CT slices of these nodules using our hospital’s daily diagnostic display (E-3620, MP Single-head Nio Grayscale Display, BRACO, Italy). The three radiologists were asked to express their confidence in the interpretation of each nodule according to five levels: definitely PIL, probably PIL, equivocal, probably IAC, and definitely IAC. To obtain an estimate of the sensitivity and specificity from each radiologist, the five confidence levels were split in two by considering the definitely PIL and probably PIL confidence levels as negative findings and all the other levels as positive findings. To obtain the ROC curve, the threshold was changed to cover the entire range of possible ratings by the radiologist, and the sensitivity was plotted as a function of false positive score (1-specificity). A performance comparison was conducted between the 3D CNN model and the three radiologists in the classification between PILs and IACs in CT using the same test data.
Statistical analysis
The results of our automatic lung cancer diagnosis algorithm were evaluated by comparing them to those of three radiologists with different years of diagnostic experience. The indicators of sensitivity, specificity, and accuracy were used to quantitatively assess the performance of the automatic algorithm. In addition, for clear comparisons with the results obtained by the radiologists, ROC curves and the AUC of the ROC were plotted (20). A higher AUC indicated a better invasive lung adenocarcinoma diagnosis result.
Results
Patient characteristics
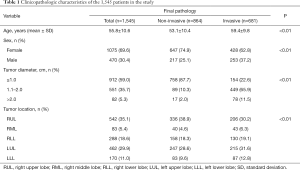
Upon histological examination, 864 (55.9%) cases were PILs, and 681 (44.1%) cases were IACs. The mean age in the IAC group was significantly greater than that in the PIL group (P<0.01). The end-to-end network predicted the nodule position and diameter, together with the probability, with respect to the IACs. The statistical results listed in Table 1 indicate that 87.4% of the nodules classified as IACs had a diameter larger than 10 mm. In contrast, 87.7% of the nodules diagnosed as PILs had a diameter smaller than 10 mm. It is rather remarkable that the female patients made up a higher proportion of the subjects suffering from both PILs and IACs (P<0.01).
Full table
CNN performance
We used the AUC values of the overall classification accuracy, sensitivity, and specificity and the area under the ROC curve to evaluate the performance and discrimination of the deep learning model (CNN). Their quantitative measurements were calculated using accuracy, sensitivity, and specificity.
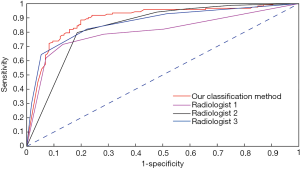
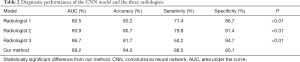
The ROC analysis results from the CNN model and from the three radiologists are shown in Figure 2. The CNN model had the largest AUC value of 89.2%, which was significantly different from the AUC of the three radiologists (radiologist 1: 80.5%; radiologist 2: 83.9%; and radiologist 3: 86.7%). Discriminant analysis showed that the CNN model was the best for the differential diagnosis of PILs and IACs. The total accuracy rates of radiologist 1, radiologist 2, radiologist 3, and the CNN model were 80.2%, 80.7%, 81.7%, and 84.0%, respectively. The sensitivity and accuracy of the CNN model were significantly higher than those of the three radiologists (P<0.05).
Discussion
This study demonstrated the deep learning algorithm was able to achieve a better AUC than a panel of three pathologists WTC were able to when participating in a simulation exercise involving the identification of PILs and IACs in stage IA lung adenocarcinoma. To our knowledge, this is the largest study based on HRCT and a deep learning algorithm to predict the invasive status of early lung adenocarcinoma so far. The sensitivity and accuracy of our GGNs classification model were 88.5% and 84.0%, respectively, which would be helpful in identifying IACs before operation. The results of our automatic classification method are competitive compared with other traditional methods (15), such as logistic regression (accuracy =81.5%), random forest (accuracy =83.0%), and adaboosting (82.1%). Automatic interpretation of radiological image features of pulmonary nodules has potential benefits, such as improved efficiency, reproducibility, and improved prognosis, by providing early detection and treatment.
PILs included AAHs, AISs, and MIAs, and the disease-free survival rate after sublobectomy was 100% (4,12). However, lobectomy plus systemic lymph node dissection or sampling is the standard surgical approach for invasive adenocarcinoma (12). Given that intraoperative frozen sections are difficult to use to accurately determine the invasiveness of adenocarcinoma; preoperative identification of nodular invasiveness is helpful for preoperative planning (21). The CNN model predicted the invasiveness of GGNs with higher accuracy, sensitivity, and AUC values than those of the three radiologists.
A number of studies have reported the use of HRCT (22-26) or radiomics (27-29) in predicting early lung adenocarcinoma invasiveness. Methods such as HRCT and radiomics are limited by the significant pretreatment required to extract imaging biomarkers, reducing the reproducibility. Deep learning simplifies multistage pipelines by learning predictive features directly from the image, allowing for greater reproducibility. In this study, our accuracy (84.0%) was similar to or higher than that reported above, demonstrating that accurate predictions can be achieved in PIL and IAC patients without predesigned features. The sensitivity of our method is higher than those of the three radiologists. In fact, during the clinical diagnosis, the doctors tend to put more attention on patients suffered invasive lung cancer, which influences the value of sensitivity. Thus, our classification method is working on producing a high accuracy and high sensitivity. The results of the radiologists may be influenced by their experience. Some radiologists may prone to classify lung cancer as pre-invasive type, which produce a low sensitivity and a high specificity.
The algorithm was superior to the three time-limited radiologists in distinguishing the invasive status of lung adenocarcinomas. However, given sufficient time, radiologists may outperform the CNN algorithm in distinguishing PILs from IACs. In this study, the three radiologists were given only 3 hours to evaluate the CT images of all 230 nodules, each less than 1 minute. It is a remarkable fact that the locations of all nodules were provided for the three radiologists before reading. Thus, the radiologists only need to draw their attention on distinguishing whether a nodule is pre-invasive or invasive. There were two problems with this approach: one was that this was an impractical assessment of short-term CT images, and the other was that, in routine clinical practice, radiologists are unlikely to review 230 consecutive pulmonary nodules. In most hospitals, radiologists require additional image post-processing and windowing techniques, which are factors that affect diagnostic performance. In addition, the simulation exercise invited the radiologists to review 230 nodular CT sections within approximately 3 hours to distinguish between PILs and IACs. Although feasible in this simulation environment, this does not represent the progression of work in other settings. Reducing the time limit for task completion can improve the accuracy of GGN diagnostic review. Although there is no perfect control, it is clear that there were limitations on this study.
In fact, the 3D CNN-based automatic method made an advance in lung cancer diagnosis in comparison with human diagnosis. First, on the basis of the 3D convolution technique, the CNN algorithm makes full use of information from sagittal, coronal, and axial views. In this way, the automatic diagnosis algorithm obtains more comprehensive and accurate information with respect to nodule size, shape, and location, which improves the reliability of the diagnostic results. Second, as in human beings, CNN works as memory network by learning critical features in cancer classification. In this process, CNNs are more flexible in adjusting learning strategies and are able to determine the critical clinical features even for confusing cases. In contrast, radiologists may be limited by experiences, and the results are likely to be tendentious at times. For example, the results in Table 2 indicate some radiologist tendencies to classify IACs as PILs.
Full table
A limitation of our developed automatic lung cancer diagnosis method is the effect of an insufficient data size on final performance. To compensate for the limitation, various data enhancement strategies, such as rotation, enhancement, and scaling, were employed during the training process. In the future, data collection should be continued for further enhancement of classification sensitivity and specificity with respect to IACs and PILs, respectively.
The focus on this work in the future will be to use a larger network in the hope of achieving more accurate results. Deeper networks can potentially improve the model’s results and generalization capabilities; however, more multicenter data will be needed to minimize the effects of overfitting. A larger patient population will further improve the performance of the algorithm, especially considering the heterogeneity of the image acquisition parameters. The rates of IACs in our study varied among different patient groups. Ideally, all patient CT scans will be acquired using consistent acquisition parameters (kV, mAs, resolution, slice thickness, and contrast agent), and the rates of IACs will be the same. However, this will be a challenge in practice because the CT scanner model and acquisition parameters, as well as the captured patient’s demographic data, vary widely among different institutions. Our research validated the algorithm by using data from a single institution, and we hope to use data from multiple institutions in the future to enhance the model’s generalization capability.
Conclusions
In this paper, we have shown that the performance of a deep CNN trained on large datasets to classify IACs from PILs is superior to that of experienced radiologists. Our model may be used as a non-invasive tool to supplement invasive tissue sampling, to guide patient surgery protocols and to manage the disease early and during follow-up. We believe that deep CNNs or similar deep learning methods show great advantages, and these algorithms can ultimately help doctors identify PILs and IACs.
Acknowledgements
Funding: This research is supported by, Shanghai Science and Technology Funds (No. 20130424), NSFC (No. 81371550). We thank engineers from Tencent Youtu Lab for debugging automatic diagnosis algorithm.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: Ethical approval was obtained for this retrospective analysis, and informed consent requirement was waived.
References
- Park CM, Goo JM, Lee HJ, Lee CH, Chun EJ, Im JG. Nodular ground-glass opacity at thin-section CT: histologic correlation and evaluation of change at follow-up. Radiographics 2007;27:391-408. [Crossref] [PubMed]
- Lee HY, Choi YL, Lee KS, Han J, Zo JI, Shim YM, Moon JW. Pure ground-glass opacity neoplastic lung nodules: histopathology, imaging, and management. AJR Am J Roentgenol 2014;202:W224-W33. [Crossref] [PubMed]
- Travis WD, Brambilla E, Noguchi M, Nicholson AG, Geisinger KR, Yatabe Y, Beer DG, Powell CA, Riely GJ, Van Schil PE. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011;6:244-85. [Crossref] [PubMed]
- Liu S, Wang R, Zhang Y, Li Y, Cheng C, Pan Y, Xiang J, Zhang Y, Chen H, Sun Y. Precise diagnosis of intraoperative frozen section is an effective method to guide resection strategy for peripheral small-sized lung adenocarcinoma. J Clin Oncol 2016;34:307-13. [Crossref] [PubMed]
- Moon Y, Lee KY, Park JK. The prognosis of invasive adenocarcinoma presenting as ground-glass opacity on chest computed tomography after sublobar resection. J Thorac Dis 2017;9:3782. [Crossref] [PubMed]
- Yim J, Zhu LC, Chiriboga L, Watson HN, Goldberg JD, Moreira AL. Histologic features are important prognostic indicators in early stages lung adenocarcinomas. Mod Pathol 2007;20:233. [Crossref] [PubMed]
- Yoshizawa A, Motoi N, Riely GJ, Sima CS, Gerald WL, Kris MG, Park BJ, Rusch VW, Travis WD. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011;24:653. [Crossref] [PubMed]
- Zhang J, Wu J, Tan Q, Zhu L, Gao W. Why do pathological stage IA lung adenocarcinomas vary from prognosis?: a clinicopathologic study of 176 patients with pathological stage IA lung adenocarcinoma based on the IASLC/ATS/ERS classification. J Thorac Oncol 2013;8:1196-202. [Crossref] [PubMed]
- Tsutani Y, Miyata Y, Nakayama H, Okumura S, Adachi S, Yoshimura M, Okada M. Oncologic outcomes of segmentectomy compared with lobectomy for clinical stage IA lung adenocarcinoma: propensity score–matched analysis in a multicenter study. J Thorac Cardiovasc Surg 2013;146:358-64. [Crossref] [PubMed]
- Kodama K, Higashiyama M, Okami J, Tokunaga T, Imamura F, Nakayama T, Inoue A, Kuriyama K. Oncologic outcomes of segmentectomy versus lobectomy for clinical T1a N0 M0 non-small cell lung cancer. Ann Thorac Surg 2016;101:504-11. [Crossref] [PubMed]
- Cao C, Gupta S, Chandrakumar D, Tian DH, Black D, Yan TD. Meta-analysis of intentional sublobar resections versus lobectomy for early stage non-small cell lung cancer. Ann Cardiothorac Surg 2014;3:134. [PubMed]
- Dembitzer FR, Flores RM, Parides MK, Beasley MB. Impact of histologic subtyping on outcome in lobar vs sublobar resections for lung cancer: a pilot study. Chest 2014;146:175-81. [Crossref] [PubMed]
- Cheng X, Onaitis MW, D’amico TA, Chen H. Minimally Invasive Thoracic Surgery 3.0: Lessons Learned From the History of Lung Cancer Surgery. Ann Surg 2018;267:37-8. [Crossref] [PubMed]
- Statnikov A, Wang L, Aliferis CF. A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics 2008;9:319. [Crossref] [PubMed]
- Li M, Narayan V, Gill RR, Jagannathan JP, Barile MF, Gao F, Bueno R, Jayender J. Computer-Aided Diagnosis of Ground-Glass Opacity Nodules Using Open-Source Software for Quantifying Tumor Heterogeneity. AJR Am J Roentgenol 2017;209:1216-27. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JA, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Kuhnigk JM, Dicken V, Bornemann L, Bakai A, Wormanns D, Krass S, Peitgen HO. Morphological segmentation and partial volume analysis for volumetry of solid pulmonary lesions in thoracic CT scans. IEEE Trans Med Imaging 2006;25:417-34. [Crossref] [PubMed]
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, 2016:424-32.
- Ren S, He K, Girshick R, Sun J. Faster R-CNN: towards real-time object detection with region proposal networks. IEEE Trans Pattern Anal Mach Intell 2017;39:1137-49. [Crossref] [PubMed]
- Avila-Flores R, Medellin RA. Ecological, taxonomic, and physiological correlates of cave use by mexican bats. J Mammal 2004;85:675-87. [Crossref]
- Walts AE, Marchevsky AM. Root cause analysis of problems in the frozen section diagnosis of in situ, minimally invasive, and invasive adenocarcinoma of the lung. Arch Pathol Lab Med 2012;136:1515-21. [Crossref] [PubMed]
- Zhang Y, Shen Y, Qiang JW, Ye JD, Zhang J, Zhao RY. HRCT features distinguishing pre-invasive from invasive pulmonary adenocarcinomas appearing as ground-glass nodules. Eur Radiol 2016;26:2921-8. [Crossref] [PubMed]
- Gao JW, Rizzo S, Ma LH, Qiu XY, Warth A, Seki N, Hasegawa M, Zou JW, Li Q, Femia M. Pulmonary ground-glass opacity: computed tomography features, histopathology and molecular pathology. Transl Lung Cancer Res 2017;6:68. [Crossref] [PubMed]
- Lee SM, Park CM, Goo JM, Lee HJ, Wi JY, Kang CH. Invasive pulmonary adenocarcinomas versus preinvasive lesions appearing as ground-glass nodules: differentiation by using CT features. Radiology 2013;268:265-73. [Crossref] [PubMed]
- Yue X, Liu S, Yang G, Li Z, Wang B, Zhou Q. HRCT morphological characteristics distinguishing minimally invasive pulmonary adenocarcinoma from invasive pulmonary adenocarcinoma appearing as subsolid nodules with a diameter of ≤3 cm. Clin Radiol 2018;73:411.e7-411.e15. [Crossref] [PubMed]
- Ichinose J, Kohno T, Fujimori S, Harano T, Suzuki S, Fujii T. Invasiveness and malignant potential of pulmonary lesions presenting as pure ground-glass opacities. Ann Thorac Cardiovasc Surg 2014;20:347-52. [Crossref] [PubMed]
- Alpert JB, Rusinek H, Ko JP, Dane B, Pass HI, Crawford BK, Rapkiewicz A, Naidich DP. Lepidic Predominant Pulmonary Lesions (LPL): CT-based Distinction From More Invasive Adenocarcinomas Using 3D Volumetric Density and First-order CT Texture Analysis. Acad Radiol 2017;24:1604-11. [Crossref] [PubMed]
- Hwang IP, Park CM, Park SJ, Lee SM, McAdams HP, Jeon YK, Goo JM. Persistent pure ground-glass nodules larger than 5 mm: differentiation of invasive pulmonary adenocarcinomas from preinvasive lesions or minimally invasive adenocarcinomas using texture analysis. Invest Radiol 2015;50:798-804. [Crossref] [PubMed]
- Son JY, Lee HY, Lee KS, Kim JH, Han J, Jeong JY, Kwon OJ, Shim YM. Quantitative CT analysis of pulmonary ground-glass opacity nodules for the distinction of invasive adenocarcinoma from pre-invasive or minimally invasive adenocarcinoma. PLoS One 2014;9. [Crossref] [PubMed]