In vivo fluorescence imaging of hepatocellular carcinoma using a novel GPC3-specific aptamer probe
Introduction
Liver cancer is a leading cause of cancer-related death worldwide. The most common form of liver cancer is hepatocellular carcinoma (HCC), one of the most frequent malignant neoplasms worldwide (1,2). To prolong survival or to even cure HCC, early detection and accurate diagnosis are of significant values to the patients.
Tumor targeted imaging plays an increasingly important role in the early diagnosis and evaluation of cancers (3). Aptamer, a single-stranded RNA or DNA (ssDNA) oligonucleotide with secondary and tertiary structures, has high specificity to certain targets. It can be served as a promising targeting agent with some unique advantages compared with the monoclonal antibody, such as the lack of immunogenicity, higher stability and faster tissue penetration. Recently, the aptamers have been experimentally explored in various in vivo cancer imaging targeting colorectal cancers, leukemia, glioma, breast and prostate cancer (4-9). Glypican-3 (GPC3), a membrane-bound heparin sulfate proteoglycan, is highly expressed in most of HCCs, whereas negative in normal liver tissue or benign hepatocellular nodules (10-14). It was found that the frequency of GPC3 mRNA expression was significantly higher than that of AFP in small HCCs (15). Therefore, GPC3 may be used as a reliable and early biomarker for HCC with high sensitivity and specificity (10,16).
Optical imaging is one of the most commonly used imaging tools, especially in the preclinical cancer studies. Near-infrared (NIR) fluorescence imaging has many advantages including the improved tissue penetration and the minimized effect of tissue autofluorescence when compared with other fluorescence imaging techniques (17), which are expected to be popularized in clinical research. Lymphoscintigraphy with NIR fluorescence imaging has been employed on breast cancer patients (18,19). Fluorescence-guided surgery, which provides valuable information for more precise tumor dissection is becoming the standard of care for some tumor resections (20).
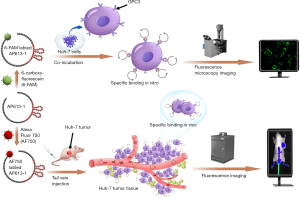
This study is to introduce a novel GPC3 specific aptamer (AP613-1) screened by a systematic evolution of ligands by exponential enrichment (SELEX) strategy based on capillary electrophoresis (CE), to verify its specific binding property in vitro, and to evaluate its targeting efficiency in vivo by performing NIR fluorescence imaging on an HCC xenograft model (Figure 1).
Methods
Chemicals and materials
The purified GPC3 protein (100 µg/mL) was purchased from Fitzgerald (Fitzgerald, MA, USA). Dulbecco’s phosphate buffered saline (PBS) and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (Saint Louis, MO, USA). All the chemicals and reagents used were of analytical grade and prepared with deionized water.
Cell culture
A human liver cancer cell line (Huh-7, GPC3 positive) and a human lung cancer line (A549, GPC3 negative) purchased from Shanghai Cell Bank (Chinese Academy of Sciences, Shanghai, China) were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) plus 100 units/mL penicillin/streptomycin and incubated at 37 °C in 5% CO2.
Selection of GPC3-bound aptamer by CE
The selection and identification of GPC3-bound aptamers were done on the P/ACE MDQ CE system (Beckman Coulter, Inc., Fullerton, CA, USA) installed with 32 Karat software according to a previous report (21). Briefly, CE of the purified GPC3 protein mixed with the ssDNA library was performed. GPC3-bound aptamers were collected and amplified by routine PCR to generate ssDNA sub-library that would be used in next round of separation. After six rounds of selection, nineteen ssDNA were obtained and the binding affinities of them to GPC3 were validated by flow cytometry (Becton Dickson, FACScan®, Mansfield, MA, USA). Then the ssDNA with the minimum equilibrium dissociation constants (Kd) to GPC3 was selected and then truncated (AP613-1, 5’-TAACGCTGACCTTAGCTGCATGGCTTTACATGTTCCA-3’, Kd =59.85±15.39 nM).
Flow cytometry assay
Generally, about 1×105 Huh-7 cells were incubated with 6-carboxyfluorescein (6-FAM)-labeled AP613-1 (1 µM) in 200 µL binding buffer at 37 °C for 60 min in the dark and washed twice with 0.8 mL PBS buffer. The samples were then resuspended in 0.5 mL PBS buffer and detected with a flow cytometer (Becton Dickson, FACScan®, Mansfield, MA, USA) by counting 10,000 events. The initial ssDNA library was used as a negative aptamer control. A549 cells were used as a negative cell control.
Aptamer-based in vitro immunofluorescence imaging
AP613-1 was labeled with 6-FAM at 5’ end. Huh-7 cells and A549 cells were seeded into 24-well plates (Corning, NY, USA) at a density of 2×104 cells/well for 24 h at 37 °C. Subsequently, the cells were incubated with 500 nM (10 µg/mL) 6-FAM-labeled AP613-1 in 1% BSA for 60 min in the dark, washed with PBS three times and then imaged by a fluorescence microscope (Olympus, Tokyo, Japan). 6-FAM-labeled initial ssDNA library was also used instead of 6-FAM-labeled AP613-1 above as a negative control. Meanwhile, immunofluorescence staining using anti-GPC3 antibody (mouse anti-human; 10 µg/L; Abcam, Cambridge, MA, USA) was performed as a positive control.
Xenograft model
This experiment was approved by the institutional Ethics and Animal Care Committee of Zhongshan Hospital, Fudan University. Male athymic BALB/c (Balb/C-nu) mice (4 weeks old, 15–20 g weight) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. Sixteen four-week-old male BALB/c nude mice were inoculated subcutaneously with 5×106 tumor cells into the right upper flank (Huh-7, n=12) or bilateral upper flanks (right Huh-7, left A549, n=4). The inoculation was done with the mice in a prone position and at the flank area where the fluorescence signal of the tumors would not be affected by the surrounding tissues to the greatest extent. The tumors were allowed to grow for 3 weeks to reach 1.0–1.5 cm in diameter before fluorescence imaging.
In vivo NIR fluorescence imaging
The nude mice were first anesthetized with an intraperitoneal injection of pentobarbital sodium solution (80 mg/kg). Those with unilateral tumors (n=12) were randomly divided into experimental and control groups with 6 mice per group. Each mouse of the experimental group was injected with 200 µL physiological saline containing 0.65 nmol of Alexa Fluor 750 (AF750, Ex/Em =749 nm/775 nm) labeled AP613-1 (Thermo Fisher Scientific, Shanghai, China) via its tail vein whereas each control mouse received the same volume of labeled initial ssDNA library as a negative control. In addition, each mouse with bilateral different tumors (n=4) was injected with 200 µL physiological saline containing 0.65 nmol of labeled aptamer to demonstrate its selectivity to HCC. According to previous reports and our own experience, an imaging time point of 2 hours after injection was chosen (5,22,23). Surface fluorescence images of the dorsal side of mice were taken by the IVIS® Lumina K III (PerkinElmer, USA). A 750 nm excitation filter and a 780 nm emission filter were selected. All the fluorescence images were presented with the manufacturer’s software (Living Image® software, version 4.4).
Ex vivo NIR fluorescence imaging
After in vivo fluorescence imaging, the mice were sacrificed immediately by cervical dislocation. Tumors and vital organs (heart, liver, spleen, lung and kidney) of the mice were removed and washed with deionized water three times quickly in the dark. Ex vivo fluorescence images of the tumors and tissues were obtained on the IVIS Spectrum® under the same parameters stated above.
Analysis of the fluorescence image
On the fluorescence images, freehand regions of interest (ROIs) were drawn carefully along the margins of tumors in vivo or ex vivo, and along the removed vital organs. The mean fluorescence intensities [average radiance efficiency, (p/s/cm2/sr)/(µW/cm2)] were generated automatically by Living Image® 4.4 software and recorded.
Statistical analysis
The statistical analyses were performed using the SPSS software package (v. 16.0.1, Chicago, IL) and P values <0.05 were considered statistically significant. The difference between the fluorescence intensities from all the tumors in vivo and ex vivo was analyzed by paired t-test. Spearman rank correlation analysis was performed to assess the correlation between the tumor fluorescence intensities in vivo and ex vivo and the correlation coefficient rho (r, 0.00–0.20, poor correlation; 0.21–0.40, fair; 0.41–0.60, moderate; 0.61–0.80, good; and 0.81–1.00, excellent) was also calculated to define the degree of correlation. Differences in fluorescence intensities of the tumors in vivo and ex vivo, and the vital organs ex vivo between the experimental group and the control group were analyzed by independent t-test. Paired t-test was used to compare the difference in fluorescence intensities between the Huh-7 tumors and the A549 tumors in vivo and ex vivo from the mice with bilateral tumors.
Results
In vitro targeting efficiency
Compared to the initial ssDNA library, there was a significant difference between the binding strength of AP613-1 to Huh-7 and to A549 cells. When AP613-1 was used, marked binding to Huh-7 cells was observed by flow cytometry while no significant binding to A549 cells was seen (Figure 2A).
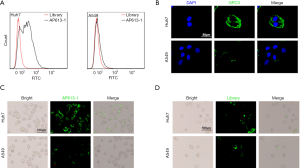
To verify the positive expression of GPC3 in Huh-7 cell line and the negative expression in A549 cell line, a GPC-3 antibody-based immunofluorescence staining was performed. It showed that Huh-7 was positively stained while A549 was not, demonstrating that Huh-7 cells displayed GPC3 expression whereas A549 did not (Figure 2B). Huh-7 and A549 cells were stained by 6-FAM-labeled AP613-1 and fluorescence microscopy subsequently showed much greater fluorescence signals from Huh-7 cells than from A549 cells (Figure 2C). Moreover, no specific staining was found in both Huh-7 and A549 when using 6-FAM-labeled initial ssDNA library (Figure 2D). All these results above demonstrated that AP613-1 could specifically target GPC3 protein in vitro.
Correlation between the fluorescence intensities in vivo and ex vivo
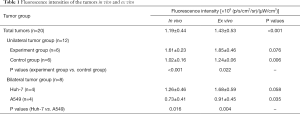
The paired t-test showed that fluorescence intensities of all the tumors (n=20) in vivo were significantly lower than ex vivo (P<0.001, Table 1). Lower fluorescence intensities of the tumors in vivo than ex vivo were also found in each group (Table 1). However, the correlation coefficient rho (r) was 0.968 (P<0.001), which demonstrates an excellent correlation between the tumor fluorescence intensities from in vivo and ex vivo. Therefore, fluorescence intensities of the tumor in vivo could reflect the intensities from ex vivo.
Full table
In vivo NIR fluorescence imaging
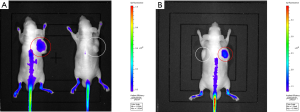
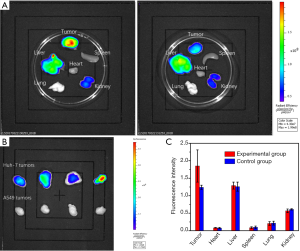
Fluorescence intensities of the tumors in each group were listed in Table 1. As shown in Table 1, fluorescence intensities of the tumors in the experimental group were significantly higher than tumors in the control group (Figure 3A) in vivo (P<0.001). In the mice with bilateral tumors, the Huh-7 tumors showed significantly higher fluorescence intensities than the A549 tumors (Figure 3B) in vivo (P=0.016).
Ex vivo NIR fluorescence imaging and biodistribution experiments
Similar to the results from in vivo, fluorescence intensities of the tumors in the experimental group were significantly higher than tumors in the control group (Figure 4A) ex vivo (P=0.022, Table 1). Significantly higher ex vivo fluorescence intensities in the Huh-7 tumors than the A549 tumors were also found (Figure 4B) in the mice with bilateral tumors (P=0.004, Table 1). Based on the results of in vivo and ex vivo, specific targeting of AP613-1 to Huh-7 cells could be validated. To observe the biodistribution of AF750 labeled AP613-1 and AF750 labeled initial ssDNA library, ex vivo fluorescence intensities of the dissected Huh-7 tumors and the vital organs including hearts, livers, spleens, lungs and kidneys in both groups were measured and analyzed (Figure 4A,C). No significant difference was found between the experimental and the control groups for the fluorescence intensities of the hearts, livers, spleens, lungs and kidneys (P=0.340, 0.345, 0.552, 0.824 and 0.112, respectively). Both the experimental and the control mice displayed the same distribution of the imaging agents with high fluorescence intensities in livers and kidneys followed by the lungs, spleens and hearts. In the experimental group, the fluorescence intensities of the Huh-7 tumors were higher than all the other organs, whereas in the control group they were nearly equal to those of livers.
Discussion
Up to now, it is still challenging to detect an early or small HCC. Compared with conventional imaging methods such as CT, MRI and ultrasound, targeted molecular imaging may be important in the early diagnosis of this tumor with higher sensitivity and specificity. GPC3 may be served as a targeted biomarker for this purpose as it is highly expressed in most HCCs (14,24). Due to its high expression of GPC3, Huh-7 cell line was chosen as the targeted cell line in the present study (25). Meanwhile, according to a previous research, GPC3 is minimally expressed in lung adenocarcinoma (26). Therefore, A549, a lung adenocarcinoma cell line was chosen as a negative control. Immunofluorescence staining results in our study further confirmed the previous reports.
In recent years, a few studies have focused on the potential role of GPC3 as a biomarker for HCC-targeted imaging. A nanoparticle which could target GPC3-expressing HCC cells was used for both MR and NIR fluorescence imaging in 2011 (27). After that, an MRI-specific superparamagnetic iron oxide anti-GPC3 probe was introduced to image early HCC cells with improved accuracy compared to anti-AFP probe (28). However, these were in vitro studies and the targeting moiety used was the monoclonal antibody, which might result in untoward effects when administered in vivo owing to its immunogenicity and low penetrability. These weaknesses may be overcome when an aptamer is used instead of the monoclonal antibody. With much smaller size than the antibody, an aptamer can permeate through the tumor vessel walls more easily and attach to the targets, which facilitate the detection of early HCC even with small amount of neovasculature. In the present study, a GPC3 specific aptamer was selected first and then used as the targeting ligand and the aptamer-based fluorescence imaging was performed both in vitro and in vivo.
Usually, the aptamer is obtained from the SELEX process. However, conventional SELEX is expensive and time-consuming. At least 8–12 rounds in 4–6 weeks are required by a conventional SELEX due to lack of filtration techniques with higher efficiency (29). Moreover, some minor deviations in screening may negatively affect the quality of aptamer. Therefore, CE-SELEX, an improved method with high-efficiency, simplified-process and shortened-period was adopted in this study for screening aptamer (21,30,31).
Similar to the previous studies on other tumors (5,32), we established two mice groups with the control group injected with the random oligonucleotide to verify the specific affinity of an aptamer to Huh-7 tumors. Moreover, an additional group of mice with both Huh-7 and A549 tumors on either side of the body was established to further confirm the specific affinity of AP613-1 to GPC3 without having to consider numerous individual differences between mice such as weight and metabolism. Our study demonstrated that AP613-1 could specifically target the GPC3 positive HCC and potentially be used as an imaging probe. When analyzing the distribution of AF750 labeled AP613-1, we found the fluorescence intensities of the Huh-7 tumors were higher than other vital organs, further demonstrating the targeting ability of this agent to HCC.
Due to its long wavelength emission, enhanced photostability and pH-insensitivity over a wide molar range, AF750 was chosen as the fluorescent dye in the in vivo NIR fluorescence imaging to obtain stable images in this study (33,34). In order to avoid the overlap between the tumor and the surrounding tissues in the direction of fluorescence imaging, which could impair the accurate measurement of the tumor fluorescence intensity, the tumor cells were inoculated subcutaneously at the lateral flank of the mice to minimize this superimposition effect. Fluorescence strengths of the tumors in vivo were found lower than ex vivo in the present study. This phenomenon was attributed to the fluorescence attenuation caused by the skin covering the tumor. However, the fluorescence intensities from the tumors in vivo and ex vivo were in excellent correlation, which allowed us to directly measure the fluorescence intensities from the tumors in vivo to represent the binding strength of aptamer to HCC. Quantitative analysis of the fluorescence intensities in vivo is important because subtle differences could be discovered without executing the experimental animals to remove the tumors. The quantitative approach in this study may not only verify the targeting capability more precisely but also hold the promise to monitor the therapeutic effects on the tumor in vivo after certain treatments.
The major limitation of this study is that subcutaneous rather than orthotopic Huh-7 xenografts were employed for the examination because of the poor penetration of fluorescent signals from deep tissues. Although its specific and strong binding affinity was verified in this study, AP613-1 labeled with other agents such as magnetic nanoparticles or 18F, would be more pragmatic in the clinical setting when used for in vivo orthotopic HCC imaging on an MRI or PET-CT platform.
Conclusions
In conclusion, by means of in vitro and in vivo studies, we found a novel aptamer AP613-1 which may be used as a probe targeting GPC3 positive HCC with high binding affinity. Significantly higher fluorescence intensities from subcutaneous Huh-7 tumors in nude mice administered with AF750 labeled AP613-1 could be revealed by the NIR fluorescence imaging. With the specific binding affinity and inherent advantages of aptamer over antibodies, aptamer-mediated NIR fluorescence imaging may play an important role in the clinical and experimental HCC researches such as intraoperative HCC visualization, delivery and monitoring of preclinical tumor-targeted therapies via the quantification of the fluorescence strength.
Acknowledgements
We thank Prof. Jingwu Kang from Shanghai Institute of Organic Chemistry of Chinese Academy of Sciences for his technical assistance in CE-SELEX. We also thank Huiyan Li and Xiao Guo from School of Basic Medical Sciences of Fudan University for their technical assistance in animal imaging.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: This experiment was approved by the institutional Ethics and Animal Care Committee of Zhongshan Hospital, Fudan University.
References
- Venook AP, Papandreou C, Furuse J, de Guevara LL. The incidence and epidemiology of hepatocellular carcinoma: a global and regional perspective. Oncologist 2010;15 Suppl 4:5-13. [Crossref] [PubMed]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [Crossref] [PubMed]
- McDermott S, Kilcoyne A. Molecular imaging-its current role in cancer. QJM 2016;109:295-9. [Crossref] [PubMed]
- Jacobson O, Weiss ID, Wang L, Wang Z, Yang X, Dewhurst A, Ma Y, Zhu G, Niu G, Kiesewetter DO, Vasdev N, Liang SH, Chen X. 18F-Labeled Single-Stranded DNA Aptamer for PET Imaging of Protein Tyrosine Kinase-7 Expression. J Nucl Med 2015;56:1780-5. [Crossref] [PubMed]
- Shi H, He X, Wang K, Wu X, Ye X, Guo Q, Tan W, Qing Z, Yang X, Zhou B. Activatable aptamer probe for contrast-enhanced in vivo cancer imaging based on cell membrane protein-triggered conformation alteration. Proc Natl Acad Sci U S A 2011;108:3900-5. [Crossref] [PubMed]
- Hwang DW, Ko HY, Lee JH, Kang H, Ryu SH, Song IC, Lee DS, Kim S. A nucleolin-targeted multimodal nanoparticle imaging probe for tracking cancer cells using an aptamer. J Nucl Med 2010;51:98-105. [Crossref] [PubMed]
- Roy K, Kanwar RK, Kanwar JR. LNA aptamer based multi-modal, Fe3O4-saturated lactoferrin (Fe3O4-bLf) nanocarriers for triple positive (EpCAM, CD133, CD44) colon tumor targeting and NIR, MRI and CT imaging. Biomaterials 2015;71:84-99. [Crossref] [PubMed]
- Kang WJ, Lee J, Lee YS, Cho S, Ali BA, Al-Khedhairy AA, Heo H, Kim S. Multimodal imaging probe for targeting cancer cells using uMUC-1 aptamer. Colloids Surf B Biointerfaces 2015;136:134-40. [Crossref] [PubMed]
- Yu MK, Kim D, Lee IH, So JS, Jeong YY, Jon S. Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles. Small 2011;7:2241-9. [Crossref] [PubMed]
- Wang XY, Degos F, Dubois S, Tessiore S, Allegretta M, Guttmann RD, Jothy S, Belghiti J, Bedossa P, Paradis V. Glypican-3 expression in hepatocellular tumors: diagnostic value for preneoplastic lesions and hepatocellular carcinomas. Hum Pathol 2006;37:1435-41. [Crossref] [PubMed]
- Libbrecht L, Severi T, Cassiman D, Vander Borght S, Pirenne J, Nevens F, Verslype C, van Pelt J, Roskams T. Glypican-3 expression distinguishes small hepatocellular carcinomas from cirrhosis, dysplastic nodules, and focal nodular hyperplasia-like nodules. Am J Surg Pathol 2006;30:1405-11. [Crossref] [PubMed]
- Yamauchi N, Watanabe A, Hishinuma M, Ohashi K, Midorikawa Y, Morishita Y, Niki T, Shibahara J, Mori M, Makuuchi M, Hippo Y, Kodama T, Iwanari H, Aburatani H, Fukayama M. The glypican 3 oncofetal protein is a promising diagnostic marker for hepatocellular carcinoma. Mod Pathol 2005;18:1591-8. [Crossref] [PubMed]
- Mast AE, Higuchi DA, Huang ZF, Warshawsky I, Schwartz AL, Broze GJ Jr. Glypican-3 is a binding protein on the HepG2 cell surface for tissue factor pathway inhibitor. Biochem J 1997;327:577-83. [Crossref] [PubMed]
- Kandil D, Leiman G, Allegretta M, Trotman W, Pantanowitz L, Goulart R, Evans M. Glypican-3 immunocytochemistry in liver fine-needle aspirates: a novel stain to assist in the differentiation of benign and malignant liver lesions. Cancer 2007;111:316-22. [Crossref] [PubMed]
- Hsu HC, Cheng W, Lai PL. Cloning and expression of a developmentally regulated transcript MXR7 in hepatocellular carcinoma: biological significance and temporospatial distribution. Cancer Res 1997;57:5179-84. [PubMed]
- Kandil DH, Cooper K. Glypican-3: a novel diagnostic marker for hepatocellular carcinoma and more. Adv Anat Pathol 2009;16:125-9. [Crossref] [PubMed]
- Hilderbrand SA, Weissleder R. Near-infrared fluorescence: application to in vivo molecular imaging. Curr Opin Chem Biol 2010;14:71-9. [Crossref] [PubMed]
- Sevick-Muraca EM, Sharma R, Rasmussen JC, Marshall MV, Wendt JA, Pham HQ, Bonefas E, Houston JP, Sampath L, Adams KE, Blanchard DK, Fisher RE, Chiang SB, Elledge R, Mawad ME. Imaging of lymph flow in breast cancer patients after microdose administration of a near-infrared fluorophore: feasibility study. Radiology 2008;246:734-41. [Crossref] [PubMed]
- Ogasawara Y, Ikeda H, Takahashi M, Kawasaki K, Doihara H. Evaluation of breast lymphatic pathways with indocyanine green fluorescence imaging in patients with breast cancer. World J Surg 2008;32:1924-9. [Crossref] [PubMed]
- Elliott JT, Dsouza AV, Davis SC, Olson JD, Paulsen KD, Roberts DW, Pogue BW. Review of fluorescence guided surgery visualization and overlay techniques. Biomed Opt Express 2015;6:3765-82. [Crossref] [PubMed]
- Dong L, Tan Q, Ye W, Liu D, Chen H, Hu H, Wen D, Liu Y, Cao Y, Kang J, Fan J, Guo W, Wu W. Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX. Sci Rep 2015;5:15552. [Crossref] [PubMed]
- Park JY, Lee TS, Song IH, Cho YL, Chae JR, Yun M, Kang H, Lee JH, Lim JH, Cho WG, Kang WJ. Hybridization-based aptamer labeling using complementary oligonucleotide platform for PET and optical imaging. Biomaterials 2016;100:143-51. [Crossref] [PubMed]
- Kryza D, Debordeaux F, Azema L, Hassan A, Paurelle O, Schulz J, Savona-Baron C, Charignon E, Bonazza P, Taleb J, Fernandez P, Janier M, Toulme JJ. Ex Vivo and In Vivo Imaging and Biodistribution of Aptamers Targeting the Human Matrix MetalloProtease-9 in Melanomas. PLoS One 2016;11:e0149387. [Crossref] [PubMed]
- Saffroy R, Pham P, Reffas M, Takka M, Lemoine A, Debuire B. New perspectives and strategy research biomarkers for hepatocellular carcinoma. Clin Chem Lab Med 2007;45:1169-79. [Crossref] [PubMed]
- Lei CJ, Yao C, Pan QY, Long HC, Li L, Zheng SP, Zeng C, Huang JB. Lentivirus vectors construction of SiRNA targeting interference GPC3 gene and its biological effects on liver cancer cell lines Huh-7. Asian Pac J Trop Med 2014;7:780-6. [Crossref] [PubMed]
- Aviel-Ronen S, Lau SK, Pintilie M, Lau D, Liu N, Tsao MS, Jothy S. Glypican-3 is overexpressed in lung squamous cell carcinoma, but not in adenocarcinoma. Mod Pathol 2008;21:817-25. [Crossref] [PubMed]
- Park JO, Stephen Z, Sun C, Veiseh O, Kievit FM, Fang C, Leung M, Mok H, Zhang M. Glypican-3 targeting of liver cancer cells using multifunctional nanoparticles. Mol Imaging 2011;10:69-77. [Crossref] [PubMed]
- Li Y, Chen Z, Li F, Wang J, Zhang Z. Preparation and in vitro studies of MRI-specific superparamagnetic iron oxide antiGPC3 probe for hepatocellular carcinoma. Int J Nanomedicine 2012;7:4593-611. [PubMed]
- Cox JC, Hayhurst A, Hesselberth J, Bayer TS, Georgiou G, Ellington AD. Automated selection of aptamers against protein targets translated in vitro: from gene to aptamer. Nucleic Acids Res 2002;30:e108. [Crossref] [PubMed]
- Mendonsa SD, Bowser MT. In vitro evolution of functional DNA using capillary electrophoresis. J Am Chem Soc 2004;126:20-1. [Crossref] [PubMed]
- Mendonsa SD, Bowser MT. In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis. Anal Chem 2004;76:5387-92. [Crossref] [PubMed]
- Shi H, Tang Z, Kim Y, Nie H, Huang YF, He X, Deng K, Wang K, Tan W. In vivo fluorescence imaging of tumors using molecular aptamers generated by cell-SELEX. Chem Asian J 2010;5:2209-13. [Crossref] [PubMed]
- Mahadevan K, Patthipati VS, Han S, Swanson RJ, Whelan EC, Osgood C, Balasubramanian R. Highly fluorescent resorcinarene cavitand nanocapsules with efficient renal clearance. Nanotechnology 2016;27:335101. [Crossref] [PubMed]
- Hilderbrand SA, Kelly KA, Niedre M, Weissleder R. Near infrared fluorescence-based bacteriophage particles for ratiometric pH imaging. Bioconjug Chem 2008;19:1635-9. [Crossref] [PubMed]


