
MamTrans: magnetic resonance imaging segmentation algorithm for high-grade gliomas and brain meningiomas integrating attention mechanisms and state-space models
Introduction
Gliomas and meningiomas are the two most common types of brain tumors, which pose a major threat to human health. Gliomas are highly malignant tumors originating from brain tissue, growing rapidly and exhibiting strong invasiveness, posing severe threats to patients’ quality of life and prognosis (1). Meningiomas are benign tumors that originate from the meninges, but their occupational effects can cause significant clinical symptoms. Therefore, timely detection and precise segmentation of these two clinically significant yet drastically different types of tumors are crucial for formulating individualized treatment plans and surgical planning (2). Magnetic resonance imaging (MRI), with its non-invasive nature, high resolution, and excellent soft tissue imaging capability, has been widely used for brain tumor detection and diagnosis. However, due to the extreme morphological and structural differences between gliomas and meningiomas in MRI images (3), coupled with the interference of complex surrounding environments, simultaneously achieving precise segmentation of both types using a single algorithm model is highly challenging. The existing Mamba network (4), although performing exceptionally well in processing long sequential data with inherent sequential nature and outperforming transformers, struggles to efficiently capture the complex spatial relationships between image pixels and global contextual information simultaneously due to its autoregressive modeling nature, often leading to suboptimal accuracy in segmenting medical images with blurred boundaries and diverse morphologies. Meanwhile, emerging segmentation models based on attention mechanisms, such as Transformers (5), although achieving decent performance in tumor segmentation tasks, suffer from high computational complexity caused by the self-attention mechanism, making them difficult to meet the urgent need for real-time and efficient analysis in clinical settings.
To overcome these technical bottlenecks, we propose a novel deep learning segmentation algorithm called MamTrans. This algorithm innovatively integrates attention mechanisms and Mamba models, enabling a single model to achieve high-precision segmentation of both gliomas and meningiomas, which exhibit stark morphological differences. Specifically, the proposed algorithm effectively combines self-attention modules to fully exploit the subtle differential features of the two tumor types in MRI images, achieving fine-grained segmentation. The self-attention mechanism incorporated into MamTrans for two main reasons: firstly, to leverage its global modeling and long-range dependency capturing capabilities, compensating for the shortcoming of autoregressive models like Mamba in capturing global spatial information, thereby enhancing segmentation quality; secondly, to utilize its parallel computing characteristics, meeting the demand for efficient real-time analysis. Such an ingenious design of MamTrans enables the model to better adapt to image feature recognition, while significantly improving computational efficiency while maintaining segmentation accuracy, thereby meeting the urgent demand for high-performance analysis in clinical applications.
The main innovations of this study were that: (I) proposed a new segmentation algorithm called MamTrans, which enables a single model to simultaneously segment two types of tumors with drastically different morphologies; (II) innovatively combined the attention mechanism and state-space model (SSM), allowing the module to fully utilize the subtle differentiating features between the two tumor types in images for fine-grained segmentation, achieving superior segmentation performance compared to other models.
With the introduction of the Transformer attention mechanism, Transformer has occupied a core position in various pre-trained models in the field of natural language processing, thanks to its outstanding long-range dependency modeling capability and attention mechanism, and has been widely applied to tasks such as classification, sequence generation, and machine translation. In 2023, Gu and Dao proposed the Mamba model, the first SSM-based fundamental model that can rival Transformer. It exhibits excellent computational efficiency and surpasses the bottleneck of Transformer in long sequence tasks. This breakthrough has attracted more researchers to delve into the study of fundamental models. The proposal of the Vision Mamba (VIM) model introduced SSM into the visual domain, solving the problem of SSM’s inability to directly process image data, and utilized patch embedding for visual feature extraction. Zhu et al. then proposed VIM, further enhancing the performance of SSM-based models in visual recognition tasks (6). Although pure SSM models have an advantage in long sequence tasks, their performance in visual tasks is often less than satisfactory due to the lack of ability to capture global contextual information.
To address this issue, Hatamizadeh et al. (7) ingeniously fused the advantages of SSM and self-attention mechanisms, proposing a hybrid state-space-self-attention module that combines the strengths of both, thereby enhancing the model’s ability to capture global visual features and further improving the overall performance of the model in visual tasks. We present this article in accordance with the TRIPOD+AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2180/rc).
Methods
Experimental preprocessing
Data collection and selection
This retrospective study collected cases from January 2016 to December 2023 at the Second People’s Hospital of Yibin, including a total of 181 cases of high-grade gliomas and 245 cases of meningiomas. All patients underwent surgery, and the pathological results confirmed the diagnoses. The Digital Imaging and Communications in Medicine (DICOM) format MRI data of all patients were collected. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by the Ethics Committee of the Second People’s Hospital of Yibin (No. 2023-161-01) and individual consent for this retrospective analysis was waived.
In this study, an MRI image dataset containing cases of high-grade gliomas and meningiomas was collected as the analysis subject. As shown in Figure 1, the experiment included a total of 418 cases of brain parenchymal tumors. After strict screening and manual annotation, there were 177 cases of gliomas, divided into a training set of 136 cases and a test set of 41 cases; and 241 cases of meningiomas, divided into a training set of 188 cases and a test set of 53 cases (8). All cases were solitary tumors, excluding those with a history of surgery or multiple metastases. The patients’ ages ranged from 35 to 70 years, with a roughly equal distribution of males and females. The dataset was divided using stratified random sampling to ensure a consistent distribution of each category of cases in the training and test sets.
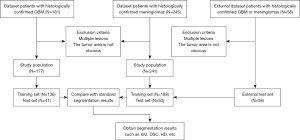
All brain tumor C1TE data were obtained from the Second People’s Hospital of Yibin and collected by professional doctors using three scanners. The MRI images of all patients were acquired using a 1.5T scanner (Erlangen, Siemens Espree, Germany). Axial T1-weighted contrast-enhanced DICOM images with a slice thickness of 1 mm were collected. The parameters for T1-weighted contrast-enhanced were as follows: slice thickness =1 mm, field of view =130 mm, repetition time =1,650 ms, echo time =3.02 ms, matrix size =512×512×176, flip angle =15°, voxel dimension =0.997×0.997×1 mm3. After obtaining the raw DICOM format MRI data, a standard data preprocessing workflow was performed.
Experimental workflow
Firstly, T1CE weighted contrast-enhanced images were collected by experienced neuroscience doctors using a scanner. The three-dimensional MRI data were reconstructed into a two-dimensional slice sequence, and three slices with distinct features from the axial position were selected to form the dataset, totaling 1,254 slice images. Subsequently, the constructed dataset underwent data augmentation, including various transformations such as brightness, contrast, and noise (9), to increase data diversity and improve the model’s generalization ability.
After data preprocessing, the data were input into the MamTrans deep learning model, and the model’s performance was evaluated. In this experiment, various mainstream convolutional neural network (CNN) segmentation models, including U-Net, U-Net++, nnU-Net, TransU-Net, and the proposed MamTrans, were compared and evaluated on the aforementioned dataset. The evaluation metrics for segmentation performance included the widely used intersection over union (IoU), Dice similarity coefficient (DSC), Hausdorff distance (HD), and F1 score, which comprehensively reflect the accuracy, completeness, and edge detail fitting of the segmentation results.
Data augmentation
To improve the segmentation accuracy of glioma and meningioma images, a series of data augmentation strategies were adopted based on the existing dataset, including transformations of the tumor subregion and background color contrast, as well as pixel value range normalization, image centering, and geometric transformations such as random rotation and translation. Through these enhancement measures, we enriched the diversity of samples in the dataset in terms of tumor type, morphology, size, position, color (10), and contrast. The data size was increased to 3,762 slices. The augmented training dataset with greater diversity helped the model learn richer tumor feature patterns, significantly improving the generalization and adaptation ability of the segmentation model in complex brain tumor scenarios (11), further enhancing the accuracy and robustness of the segmentation.
Data training
The experiment consisted of two groups: one for glioma dataset segmentation and one for meningioma segmentation. Multiple segmentation models, including U-Net, U-Net++, DeepLab, Attention U-Net, Double U-Net, and MamTrans, were used for comparison. The two datasets were separately input into different segmentation models for segmentation experiments (12), which were conducted for a total of 300 epochs. Finally, the segmentation results from each model were compared with manually annotated ground truths to obtain the accuracy metrics for each model’s segmentation experiment.
Model and architecture
This study proposes an innovative MamTrans algorithm that achieves an efficient image segmentation architecture by leveraging the advantages of attention mechanisms and SSMs. As shown in Figure 2, the decoder part of the MamTrans algorithm adopts a CNN, focusing on extracting local features from the image, while introducing the Mamba module to enable global feature recognition. The Mamba module is followed by a self-attention mechanism, further supplementing the details of global recognition. Additionally, the algorithm incorporates multilayer perceptron (MLP) layers to optimize feature extraction (13). Through this multi-stage feature extraction and fusion strategy, the MamTrans algorithm can not only accurately recover the detailed features of the image but also achieve significant improvements in computational efficiency.
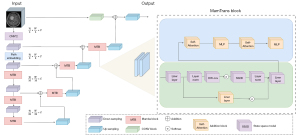
The encoder part of the MamTrans algorithm adopts the downsampling path of U-Net, progressively encoding the low-level to high-level semantic features of the image through CNNs. The Mamba module leverages a spatial state selection mechanism to model long-range dependencies on the encoded features, efficiently capturing long-range contextual dependencies in the image from a global perspective. In the decoder stage, consisting of convolutions and multiple MamTrans block (MTB) modules, the new MTB module performs feature extraction, while the MTB in the decoder part accomplishes feature fusion and recovery, progressively upsampling and employing skip connections, ultimately achieving fine-grained pixel-level segmentation prediction.
In the algorithm structure, the CNN acts as a feature extractor, generating a series of feature maps that capture low-level to high-level feature representations of the image. These feature maps undergo serialization processing, forming a serialized feature representation as input to the MTB module. The Mamba module utilizes an SSM to encode these features, extracting global contextual information. During the upsampling process, the MTB creates a rich feature representation containing both detail and global information through fusion strategies such as skip connections. The MamTrans algorithm further employs a cascaded upsampler, composed of multiple upsampling steps, to progressively refine the feature representation. Each step includes convolutional layers, activation functions, and skip connections to enhance feature fusion, achieving more precise segmentation results.
Through this innovative architectural design, the MamTrans algorithm not only achieves outstanding performance in segmentation accuracy but also exhibits significant advantages in computational efficiency. This segmentation algorithm, combining local details and global context, provides a new and efficient solution for medical image segmentation, holding important theoretical and practical value.
SSM
The SSM is a powerful mathematical tool that describes and predicts the behavior of dynamic systems by establishing mathematical relationships between system states and observations. SSMs have applications across multiple disciplines, including but not limited to control theory, signal processing, economics, and machine learning. In the field of deep learning, SSMs are particularly well-suited for processing sequential data, as they can map data sequences to a state space, effectively capturing and modeling long-term dependencies within the data.
Unlike traditional static models, SSMs can adapt to temporal changes in data through their dynamic nature, which is crucial for understanding and predicting time series, text streams, video frames, and other types of data with temporal continuity. The core advantage of SSMs lies in their ability to recursively update and predict the system through state transitions and observation equations, enabling more accurate and robust analysis when dealing with time-continuous data. In the algorithm proposed in this paper, the effective integration of SSMs with attention mechanisms will enhance the algorithm’s performance, as illustrated by Eq. [1].
MTB
As shown in Figure 3, the SSM is a key component of the MamTrans algorithm, with its core being the S4 architecture of the SSM. This architecture is similar to combining a CNN with a recurrent neural network (RNN), where the SSM can efficiently perform recursive and convolutional operations under this framework. This design endows the MamTrans algorithm with advantages in processing high-resolution images, accelerating the training process, and recognizing long-range dependencies, as described mathematically in Eq. [2].
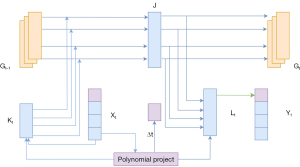
The selection of layer normalization and token mixing blocks is represented by Norm and Mixer. The Mamba module, as an application of SSM in the field of visual image processing, has evolved gradually from the fundamental SSM structure. The Visual State Space Mamba (VMamba) module inherits the characteristics of the Mamba module and further develops to adapt to the requirements of visual tasks. As the core component for image feature extraction, the MTB module consists of two main branches: the first branch contains linear and activation layers, responsible for basic feature extraction and nonlinear transformations; the second branch is more complex, enhancing the expressive power of features through a series of operations, including a linear layer, depthwise separable convolution, a special two-dimensional state space transformation (SS2D), an activation layer, and another linear layer. The block is followed by a self-attention mechanism and MLP components, with mathematical details given in Eq. [3].
Scan is a selective scanning operation, where σ is the activation function of Sigmoid using linear units (SiLU), Linear (Cin, Cout) represents linear layers with Cin and Cout as input and output embedding dimensions, and Conv and Concat represent 1D convolution and concatenation operations.
Model training
Model training was conducted using the PyTorch framework (version 1.13.0) on NVIDIA H100 GPUs equipped with CUDA 12.3. During the training process, DiceLoss was utilized as the loss function, and the Adam optimizer was employed for parameter optimization. To comprehensively evaluate the model performance, multiple benchmark architectures were selected for comparative experiments, including U-Net, U-Net++, DeepLab, Attention U-Net, Double U-Net, Swin U-Net, MISSFormer, SegTransVAE, and MamTrans. The dataset was divided into a training set and a test set, with each experimental group being trained for 300 epochs. A five-fold cross-validation method was used for validation, where the dataset was partitioned into several subsets. One subset was used as the validation set in rotation, while the remaining subsets served as the training set. This process of training and validation was repeated multiple times, and the average results were calculated. Finally, the segmentation results were compared with the ground truth annotations, and the experimental effects were demonstrated through quantitative analysis and visualization methods.
Results
Evaluation metrics
The experiment used Dice, HD, IoU, accuracy (ACC), sensitivity (SEN), specificity (SPE), and F1 score as evaluation metrics for the segmentation results. The formulas are shown below:
where true positive (TP) represents the number of pixels correctly predicted as foreground (target region) by the model. False positive (FP) represents the number of pixels incorrectly predicted as foreground by the model, while false negative (FN) represents the number of pixels incorrectly predicted as background by the model. True negative (TN) represents the number of pixels correctly predicted as background by the model (14). The experimental results will be presented in two forms: quantitative annotation and segmentation figures.
Quantitative analysis
In this study, the performance of various deep learning segmentation models was evaluated on the glioma dataset. The specific values of each model on multiple evaluation metrics are summarized in Table 1. The traditional U-Net model achieved an IoU of 74.76%, while the improved U-Net++ model had a slightly better IoU of 77.85%. The DeepLab model achieved an IoU of 75.23%, and the Attention U-Net model, which introduced an attention mechanism, significantly improved the IoU to 82.45%. The Swin U-Net model, which further integrated visual Transformers, achieved an IoU of 85.24%, The MISSFormer model demonstrated performance metrics of 86.39% for IoU and 88.08% for DSC. the SegTransVAE approach yielded marginally superior results, with an IoU of 87.45% and a DSC of 88.10%, indicating subtle improvements in segmentation accuracy. the MamTrans model proposed in this paper performed best with an IoU of 88.12%.
Table 1
| Model | Glioma data (%) | 2020 BraTS (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | IoU↑ | HD↓ | ACC↑ | DSC↑ | IoU↑ | HD↓ | ACC↑ | ||
| U-Net | 75.34 | 74.76 | 19.65 | 94.53 | 76.24 | 75.45 | 20.13 | 94.23 | |
| U-Net++ | 77.67 | 77.85 | 18.67 | 94.67 | 76.89 | 76.48 | 19.78 | 94.98 | |
| ASPP U-Net | 77.53 | 76.26 | 17.46 | 94.39 | 77.53 | 76.23 | 17.23 | 94.25 | |
| nnU-Net | 78.45 | 77.56 | 18.23 | 94.19 | 79.23 | 78.67 | 18.89 | 95.13 | |
| DeepLab | 76.11 | 75.23 | 18.93 | 95.45 | 77.24 | 79.32 | 17.25 | 95.56 | |
| Attention U-Net | 83.23 | 82.45 | 17.45 | 95.23 | 84.24 | 85.15 | 17.56 | 94.26 | |
| Swin U-Net | 87.37 | 85.24 | 14.16 | 95.87 | 88.24 | 87.27 | 13.23 | 95.39 | |
| MISSFormer | 88.08 | 86.39 | 13.85 | 95.67 | 88.37 | 87.62 | 13.36 | 95.17 | |
| SegTransVAE | 88.10 | 87.45 | 13.23 | 95.78 | 88.41 | 87.52 | 13.04 | 95.13 | |
| MamTrans | 89.23 | 88.12 | 12.67 | 95.89 | 89.56 | 89.78 | 12.78 | 95.26 | |
ACC, accuracy; DSC, Dice similarity coefficient; HD, Hausdorff distance; IoU, intersection over union.
Similarly, on the external dataset, the model demonstrated robust performance with an IoU of 90.34%, a DSC of 91.25%, and an HD of 14.17. Furthermore, on the BraTS public dataset in 2020, the proposed model achieved an IoU of 89.78% and a DSC of 89.56%, both of which surpassed the corresponding metrics of alternative segmentation approaches, thereby highlighting its superior segmentation capabilities.
In terms of another important evaluation metric, the DSC, the U-Net model achieved 75.34%, U-Net++ achieved 77.67%, and the DeepLab model improved to 76.11%. The Attention U-Net model had a DSC of 83.23%, Swin U-Net had 87.37%, and the MamTrans model again demonstrated outstanding performance with a DSC value of 89.23%.
Furthermore, on the HD metric, which reflects segmentation quality and edge detail capture capability (15), the U-Net model had an HD value of 19.65, while U-Net++ reduced it to 18.67. The DeepLab model further decreased HD to 18.93, and the Attention U-Net model significantly reduced it to 17.45. The MamTrans model also performed best on the HD metric, with a value of only 12.67, indicating its advantage in maintaining compactness and completeness of the segmentation boundary.
In the segmentation evaluation task on the meningioma dataset, we evaluated the quantitative performance of various mainstream deep learning segmentation networks, with the results shown in Table 2. The traditional U-Net model achieved an IoU of 75.24%, demonstrating a relatively excellent segmentation effect on this key metric of segmentation accuracy. The improved U-Net++ model had an IoU value of 78.57%, with a slight performance improvement. The DeepLab model’s IoU value was 79.11%, while the Attention U-Net model, which incorporated an attention mechanism, further improved the IoU to 82.34%. Notably, the MamTrans model proposed in this paper achieved the best performance on the IoU metric, reaching 90.26%, significantly outperforming the aforementioned counterpart models.
Table 2
| Model | DSC↑ (%) | IoU↑ (%) | HD↓ (%) | ACC↑ (%) |
|---|---|---|---|---|
| U-Net | 77.87 | 75.24 | 23.65 | 94.57 |
| U-Net++ | 79.25 | 78.57 | 22.33 | 94.78 |
| DeepLab | 81.11 | 79.11 | 21.16 | 95.47 |
| SCPP net | 80.24 | 79.46 | 19.17 | 95.29 |
| Attention U-Net | 83.22 | 82.34 | 18.34 | 95.78 |
| Double U-Net | 87.36 | 85.11 | 16.55 | 95.48 |
| VM-U-Net | 88.74 | 87.32 | 15.58 | 95.86 |
| MISSFormer | 89.35 | 88.25 | 15.46 | 95.78 |
| SegTransVAE | 90.15 | 89.38 | 15.39 | 95.26 |
| MamTrans | 91.27 | 90.26 | 15.14 | 96.78 |
ACC, accuracy; DSC, Dice similarity coefficient; HD, Hausdorff distance; IoU, intersection over union.
Regarding another core metric, the DSC, which evaluates the similarity between the segmentation target and the ground truth. As shown in Figure 4, the U-Net model scored 77.87% on this metric, while the U-Net++ model scored 79.25%. The MamTrans model performed exceptionally well on the DSC metric, scoring a remarkable 91.27%. This result directly reflects the high segmentation consistency between the model and the ground truth, demonstrating a clear advantage in terms of segmentation similarity.
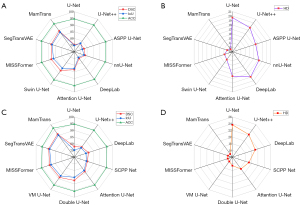
On the HD metric, which reflects segmentation quality and boundary integrity, the U-Net model had an HD value of 23.65, while the U-Net++ model reduced it to 22.33. The MamTrans model achieved the best score of 15.14 on this metric, not only significantly outperforming U-Net and U-Net++ but also directly confirming the model’s outstanding ability to maintain compactness of the segmentation boundary and capture fine details. Table 3 reflects the model’s F1, SEN, SPE, and mean absolute surface distance (MASD) metrics in the glioma segmentation experiments. Figure 5 shows the model’s receiver operating characteristic (ROC) curve, reflecting the model’s stability.
Table 3
| Model | MASD↓ | SEN↑ | SPE↑ | F1↑ (%) |
|---|---|---|---|---|
| U-Net | 4.68 | 0.8535 | 0.8745 | 75.7 |
| U-Net++ | 4.56 | 0.9054 | 0.9234 | 77.3 |
| DeepLab | 3.89 | 0.9114 | 0.9353 | 82.6 |
| Attention U-Net | 3.78 | 0.9175 | 0.9454 | 85.8 |
| Swin U-Net | 3.57 | 0.9257 | 0.9465 | 88.9 |
| MamTrans | 3.35 | 0.9367 | 0.9624 | 89.3 |
SEN, sensitivity; SPE, specificity; MASD, mean absolute scaled distance.
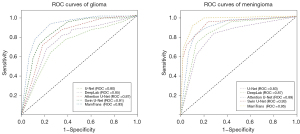
Image segmentation results
As shown in Figure 6, the results of multiple classic segmentation models on the image segmentation task are displayed. The U-Net model is able to roughly identify and segment the main regions in the image, but its segmentation performance has certain limitations, and the processing of details is not refined enough. In contrast, the Attention U-Net model demonstrates more desirable segmentation effects. Not only can it identify the main regions, but it can also better handle image details and edges (16) showing higher segmentation accuracy. Notably, the MamTrans model in Figure 6 exhibits outstanding segmentation performance. It can not only accurately restore the target region but also maintain stable and precise performance across multiple test cases. As shown in Figure 7, the segmentation results of the MamTrans model appear visually clearer, with smoother and more accurate boundaries for the target region, which is particularly important in medical image analysis as it directly affects subsequent diagnosis and treatment planning.
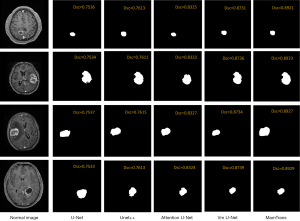
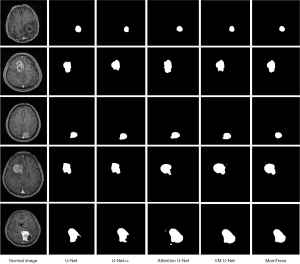
These visualization results further validate our quantitative analysis, demonstrating that the MamTrans model outperforms other compared models in terms of segmentation accuracy and stability. These findings not only provide valuable references for future research but also offer robust technical support for precise segmentation in clinical practice.
Model analysis
Ablation study
The ablation experiment results for the designed modules are presented in Table 4. Specifically, the introduction of the MTB module brought crucial feature extraction capabilities, demonstrating an outstanding ability to effectively capture medical imaging information across different types of brain tumor data. Simultaneously, the integration of the self-attention module further enhanced tumor segmentation performance, validating the effectiveness of adopting a multi-module integrated approach to predict segmentation results. Notably, the synergistic integration of the MTB and self-attention modules achieved the best performance across all evaluation metrics, with an IoU segmentation accuracy as high as 92.13%, fully demonstrating the innovative module design of the MamTrans algorithm in enhancing the model’s ability to distinguish complex tumor features and improve segmentation accuracy.
Table 4
| MTB | Self-attention | DSC↑ (%) | HD↓ (%) | IoU↑ (%) | MASD↓ |
|---|---|---|---|---|---|
| × | × | 79.35 | 21.78 | 78.45 | 5.34 |
| √ | × | 85.25 | 15.34 | 86.59 | 4.26 |
| × | √ | 87.78 | 15.67 | 87.48 | 4.23 |
| √ | √ | 90.14 | 13.47 | 91.13 | 3.38 |
×, indicates that the module has been removed from the current experimental configuration; √, indicates that the module is included in the current experimental configuration. DSC, Dice similarity coefficient; HD, Hausdorff distance; IoU, intersection over union; MASD, mean absolute scaled distance; MTB, MamTrans block.
Model complexity analysis
In Table 5, we evaluated the complexity of several segmentation models, including the number of model parameters and floating-point operation counts (FLOPs). The U-Net model performed well in terms of computational efficiency, with FLOPs of 30.9 G; however, it had certain limitations in segmentation accuracy. In contrast, models such as Attention U-Net and Swin U-Net, although achieving relatively good segmentation accuracy, had relatively lower computational efficiency. Models like Double U-Net and DeepLab had relatively low computational efficiency, which could be a limiting factor in resource-constrained environments. The MamTrans model, on the other hand, exhibited relatively low FLOPs and a smaller number of parameters compared to other models. More importantly, MamTrans not only performed well in segmentation efficiency but also demonstrated outstanding computational accuracy.
Table 5
| Model | FLOPs (G) | Param (M) | mIoU (%) |
|---|---|---|---|
| U-Net | 30.9 | 9 | 75.24 |
| U-Net++ | 239 | 9.4 | 78.57 |
| Attention U-Net | 62.5 | 49 | 82.34 |
| DeepLab | 195 | 58.8 | 81.23 |
| Double U-Net | 194.1 | 101 | 83.11 |
| Swin U-Net | 61 | 62 | 85.78 |
| MamTrans | 41.3 | 45 | 89.19 |
FLOPs, floating-point operations per second; mIoU, mean intersection over union.
These characteristics of the MamTrans model give it a significant advantage in application scenarios that require balancing computational resources and accuracy. Low FLOPs and fewer parameters mean that the model can run faster while occupying less memory, which is particularly important for real-time or resource-constrained medical image processing applications. Additionally, MamTrans’ high computational accuracy ensures the reliability of the segmentation results, which is crucial for improving the accuracy of clinical diagnoses.
Discussion
In the field of deep learning, innovative research on basic model architectures has always been a core driving force, propelling continuous progress in image segmentation techniques. From the early convolutional kernel-based segmentation networks like U-Net to the recent advanced models based on the Transformer’s self-attention mechanism, each model innovation has significantly boosted segmentation performance and accuracy. Self-attention segmentation networks, with their modular design, have demonstrated outstanding ability in capturing global dependency features in images. However, as model scales continue to expand, the computational complexity and efficiency issues of these networks have become increasingly prominent, especially in resource-constrained edge computing scenarios (17).
In contrast, segmentation algorithms based on the SSM stand out with their unique computational efficiency advantages. SSM models can selectively extract temporal context features through sequence modeling, optimizing the efficiency and performance of segmentation tasks.
The MamTrans algorithm proposed in this paper innovatively integrates CNN convolutional modules and SSM modules, achieving an organic unification of local detail feature extraction and global semantic modeling, collaboratively enhancing segmentation accuracy and model efficiency. Specifically, the CNN module focuses on extracting multi-scale local features from the input image, while the SSM module captures temporal context dependencies through sequence modeling. The complementary characteristics of these two components maximize the exploitation of local and global information in the image, ensuring segmentation accuracy while significantly improving overall algorithm efficiency (18,19). Notably, the careful design of the SSM module in the MamTrans algorithm allows it to focus only on the sequence features of the target region rather than modeling the entire global image. This efficient feature selectivity mechanism effectively reduces algorithm complexity, making a significant contribution to the algorithm’s superior computational efficiency.
In the experimental validation on large-scale glioma and meningioma image datasets, the MamTrans algorithm not only demonstrated outstanding segmentation performance (20), achieving excellent results on core evaluation metrics such as IoU, DSC, and HD, but also exhibited significantly better computational efficiency compared to self-attention models like Transformer. Its feature recognition efficiency and inference speed are superior (21), making it more suitable for clinical deployment. Overall, the MamTrans algorithm, through its innovative multi-modal fusion approach, achieves a balance between feature representation capability and computational efficiency, providing new insights for developing lightweight (22), efficient segmentation algorithms in resource-constrained scenarios. This research not only has important theoretical significance but also opens up broad prospects for the application of image segmentation in areas such as clinical diagnostic assistance (23).
Although the MamTrans model achieves excellent segmentation performance by integrating self-attention and MLP modules, this design also introduces a small amount of complexity. In practical applications, this complexity may slightly increase the computational speed (24). However, it is believed that in the critical field of medical image segmentation, accurate segmentation results are crucial for clinical diagnosis and treatment. Therefore, this complexity is acceptable to a certain extent (25). Through experiments, the MamTrans model has also demonstrated its capability in precisely identifying the characteristics of gliomas and meningiomas (26). In the future, we will continue to explore methods to simplify the model structure in order to reduce complexity while maintaining performance.
Conclusions
This study proposes an innovative MamTrans segmentation framework that innovatively integrates attention mechanisms and SSMs, enabling a single model to simultaneously achieve high-precision segmentation of gliomas and meningiomas, two tumor types with vastly different morphologies. The segmentation architecture first employs a CNN module for multi-scale feature extraction and encoding from the input images, and then utilizes an SSM module to capture temporal context dependencies. The synergy between these two modules achieves fine-grained segmentation. This innovative fusion mechanism allows MamTrans to coordinate local and global feature representations, precisely delineating the boundaries and internal details of the target regions, ultimately achieving high-precision segmentation.
In large-scale experimental evaluations on glioma and meningioma datasets, the MamTrans algorithm demonstrated outstanding segmentation performance. Quantitative results indicate that this algorithm not only efficiently identifies and segments lesion target regions but also significantly outperforms current mainstream segmentation networks in three core segmentation metrics: IoU, DSC, and HD, achieving substantial improvements in segmentation accuracy (27). Furthermore, while maintaining high segmentation accuracy, MamTrans exhibits significantly better inference speed and fewer parameters than Transformer-based segmentation models, demonstrating better potential for clinical deployment.
These experimental results objectively validate the effectiveness and superior performance of the MamTrans segmentation framework, providing compelling evidence of the algorithm’s significant application value in brain tumor segmentation tasks. This research not only provides a novel and efficient solution for medical image segmentation but also paves the way for introducing temporal modeling paradigms into the field of image analysis, holding significant theoretical and practical implications for subsequent related research and clinical applications.
Acknowledgments
The authors would like to express gratitude to all clinical doctors and technical personnel in the radiology and pathology departments, as well as researchers from Nanchang University and Henan University. We would also like to thank the Second People’s Hospital of Yibin for providing us with professional advice and guidance.
Footnote
Reporting Checklist: The authors have completed the TRIPOD+AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-2180/rc
Funding: None.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2180/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. The study was approved by Ethics Committee of the Second People’s Hospital of Yibin (No. 2023-161-01) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cao Y, Vassantachart A, Ye JC, Yu C, Ruan D, Sheng K, Lao Y, Shen ZL, Balik S, Bian S, Zada G, Shiu A, Chang EL, Yang W. Automatic detection and segmentation of multiple brain metastases on magnetic resonance image using asymmetric UNet architecture. Phys Med Biol 2021;66:015003. [Crossref] [PubMed]
- Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M. Swin-unet: Unet-like pure transformer for medical image segmentation. European Conference on Computer Vision 2022;15:205-18.
- Wang Z, Su M, Zheng JQ, Liu Y. Densely connected swin-unet for multiscale information aggregation in medical image segmentation. IEEE International Conference on Image Processing (ICIP) 2023;5:940-4.
- Wang Z, Zheng JQ, Zhang Y, Cui G, Li L. Mamba-unet: Unet-like pure visual mamba for medical image segmentation. arXiv 2024. arXiv:2402.05079.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Advances in Neural Information Processing Systems 30 (NIPS); 2017.
- Zhu L, Liao B, Zhang Q, Wang X, Liu W, Wang X. Vision Mamba: Efficient Visual Representation Learning with Bidirectional State Space Model. International Conference on Machine Learning (ICML); 2024:122-34.
- Hatamizadeh A, Kautz J. Mambavision: A hybrid mamba-transformer vision backbone. arXiv 2024. arXiv:2407.08083.
- Zhou M, Leung A, Echegaray S, Gentles A, Shrager JB, Jensen KC, Berry GJ, Plevritis SK, Rubin DL, Napel S, Gevaert O. Non-Small Cell Lung Cancer Radiogenomics Map Identifies Relationships between Molecular and Imaging Phenotypes with Prognostic Implications. Radiology 2018;286:307-15. [Crossref] [PubMed]
- Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging (Bellingham) 2019;6:014006. [Crossref] [PubMed]
- Azad R, Heidari M, Shariatnia M, Aghdam EK, Karimijafarbigloo S, Adeli E, Merhof D. Transdeeplab: Convolution-free transformer-based deeplab v3+ for medical image segmentation. International Workshop on Predictive Intelligence in Medicine 2022;25:91-102.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv 2021. arXiv:2102.04306.
- Gu A, Dao T. Mamba: Linear-time sequence modeling with selective state spaces. arXiv 2023. arXiv:2312.00752.
- Li Y, Qi Y, Hu Z, Zhang K, Jia S, Zhang L, Xu W, Shen S, Wáng YXJ, Li Z, Liang D, Liu X, Zheng H, Cheng G, Zhang N. A novel automatic segmentation method directly based on magnetic resonance imaging K-space data for auxiliary diagnosis of glioma. Quant Imaging Med Surg 2024;14:2008-20. [Crossref] [PubMed]
- Hedibi H, Beladgham M, Bouida A. A combined attention mechanism for brain tumor segmentation of lower-grade glioma in magnetic resonance images. Comput Biol Med 2025;193:110380. [Crossref] [PubMed]
- Tan M, Pang R, Le QV. EfficientDet: Scalable and Efficient Object Detection. In: Proceedings of the 2IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 2020;12:10781-90.
- Qian Z, Li Y, Wang Y, Li L, Li R, Wang K, Li S, Tang K, Zhang C, Fan X, Chen B, Li W. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 2019;451:128-35. [Crossref] [PubMed]
- Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, Cheshier SH, Napel S, Zaharchuk G, Plevritis SK. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology 2014;273:168-74. [Crossref] [PubMed]
- Takao H, Amemiya S, Kato S, Yamashita H, Sakamoto N, Abe O. Deep-learning 2.5-dimensional single-shot detector improves the performance of automated detection of brain metastases on contrast-enhanced CT. Neuroradiology 2022;64:1511-8. [Crossref] [PubMed]
- Hugo T, Matthieu C, Matthijs D, Francisco M, Alexandre S, Hervé J. Training data-efficient image transformers & distillation through attention. In: International Conference on Machine Learning 2021;16:10347-57.
- Wang WH, Xie EZ, Li X, Fan DP, Song KT, Liang D, Lu T, Luo P, Shao L. Pyramid vision transformer: A versatile backbone for dense prediction without convolutions. In: Proceedings of the IEEE/CVF International Conference on Computer Vision 2021;21:568-78.
- Xu WJ, Xu YF, Chang TL, Tu WZ. Co-scale conv-attentional image transformers. In Proceedings of the IEEE/CVF International Conference on Computer Vision 2021:9981-90.
- Hatamizadeh AL, Yin HX, Heinrich G, Kautz J, Molchanov P. Global context vision transformers. In: International Conference on Machine Learning 2023:12633-46.
- Zhou L, Wang S, Sun K, Zhou T, Yan F, Shen D. Three-dimensional affinity learning based multi-branch ensemble network for breast tumor segmentation in MRI. Pattern Recognition 2022;129:108723. [Crossref]
- Wang W, Chen C, Ding M, Yu H, Zha S, Li J, Transbts: Multimodal brain tumor segmentation using transformer. Medical Image Computing and Computer Assisted Intervention – MICCAI; 2021;12:109-19.
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993-2024. [Crossref] [PubMed]
- Zhang L, Li Y, Liang Y, Xu C, Liu T, Sun J. Dilated multi-scale residual attention (DMRA) U-Net: three-dimensional (3D) dilated multi-scale residual attention U-Net for brain tumor segmentation. Quant Imaging Med Surg 2024;14:7249-64. [Crossref] [PubMed]
- Tateishi M, Nakaura T, Kitajima M, Uetani H, Nakagawa M, Inoue T, Kuroda JI, Mukasa A, Yamashita Y. An initial experience of machine learning based on multi-sequence texture parameters in magnetic resonance imaging to differentiate glioblastoma from brain metastases. J Neurol Sci 2020;410:116514. [Crossref] [PubMed]


