
Random forest with preoperative core biopsy categories: a novel method for refining ultrasonic Breast Imaging Reporting and Data System evaluation
Introduction
Breast lesions, a highly prevalent condition across the world, and histologically categorized into benign, malignant, and borderline types. These lesions can be managed through various approaches, including imaging follow-up, biopsy, or surgical intervention (1,2).
Imaging modalities can serve as a preliminary method for assessing the histological characteristics of breast lesions, including initial determination of their benign or malignant status. For benign lesions, continued follow-up with persistent monitoring of their progression is recommended through the use of techniques such as magnetic resonance imaging (MRI), mammography, and ultrasonography. However, MRI is frequently associated with a high false-positive rate in identifying malignant tumors and involves substantial cost (3); meanwhile, mammography is limited in its ability to detect tumors within dense breast tissue (4). On the other hand, ultrasound is distinguished by being a nonradioactive, cost-effective, and readily accessible diagnostic tool. Given that the majority of women in Asian countries have dense breast tissue, ultrasound screening has emerged as the diagnostic modality of choice in these regions (5).
The ultrasound lexicon of the American College of Radiology Breast Imaging Reporting and Data System (ACR BI-RADS) is extensively applied to estimate the likelihood of malignancy. However, based on our clinical experience, lesions classified as BI-RADS category 3 and 4A continue to represent a critical but challenging diagnostic group. Chae et al. demonstrated that lesions classified as category 3 have a low malignancy rate (6), while in a study by Barr et al., a mere 0.1% of lesions exhibited suspicious malignant changes during a 6-month follow-up period (7). In clinical practice, many benign lesions are often classified as category 4A due to the diagnostic uncertainty experienced by physicians, leading to a high number of unnecessary biopsies. If these lesions could be accurately classified as BI-RADS category 3, patients could potentially forego a biopsy. Attempts have been made to enhance the diagnostic accuracy of BIRADS. For example, Weng et al. used contrast-enhanced ultrasound (8), and Zhao et al. employed strain elastography (9). Nonetheless, there remains a scarcity of appropriate diagnostic techniques to complement BI-RADS, particularly for lesions categorized as 3 and 4A.
Percutaneous imaging-guided core needle biopsy (CNB) can provide a definitive pathological classification for breast tumors, which may differ from that of the BI-RADS category. CNB categories range from B1 to B5 (10), offering valuable insights into the characteristics of breast lesions. These categories enable clinicians to make precise decisions related to clinical management. Leveraging these CNB categories, we developed a novel approach to enhance the accuracy of the BI-RADS classification system.
Machine learning has demonstrated considerable potential in the management of malignant tumors, particularly in the imaging assessment of breast cancer (11-14). By automatically analyzing and extracting patterns from existing data, machine learning can use these patterns to predict outcomes for unknown data. Its ability to perform classification tasks with high precision has been widely recognized across various medical fields, including radiology, critical care medicine, and cardiology (13,15-18). For instance, Panourgias et al. employed an MRI-based inductive decision tree to classify B3 lesions within BI-RADS 4 and 5 categories, achieving high accuracy (88.7%) and an excellent area under the receiver operating characteristic (ROC) curve (AUC =0.992) in the training set. However, the model’s performance appeared to decline in the test set (AUC =0.5), likely due to the limited sample size (19). In another study, Bahl et al. developed a mammogram-based random forest (RF) model to predict the risk of pathologic upgrade of high-risk breast lesions (B3 lesions) to cancer. Their model demonstrated the ability to reduce the number unnecessary surgeries by nearly one-third (20). These studies highlight the effectiveness of machine learning in analyzing B3 lesions from different perspectives and attest to its robust performance (19,20). However, despite these advancements, the application of machine learning to the classification of core needle biopsy category (CBC) based on imaging remains underexplored. To our knowledge, few ultrasound studies have used machine learning to investigate the relationship between CBC and BI-RADS or to refine BI-RADS classification via CBC. This presents a promising opportunity for future research to leverage machine learning in improving the precision of breast lesion classification and clinical decision-making.
Our study aimed to leverage machine learning to predict CBC by assessing ultrasound and clinical characteristics, with the ultimate goal of refining the BI-RADS classification, particularly for category 3 and 4A lesions. This approach may be able to reduce the number unnecessary biopsies by improving the accuracy of lesion characterization. We present this article in accordance with the TRIPOD+AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2070/rc).
Methods
Participants
This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Ethical approval for this study was granted by the institutional review board of Guangdong Provincial People’s Hospital (No. KY2023-1069-01), who waived the requirement for informed consent due to the retrospective nature of the analysis. Histological characteristics of breast nodules were extracted from pathology reports. We included 1082 consecutive female patients aged 12–96 years (mean age 42.22±13.37 years) who attended Guangdong Provincial People’s Hospital between March 1 and December 31, 2019. A total of 1,185 nodules (815 benign and 370 malignant) satisfied the inclusion criteria, with all ultrasound images archived in the medical system.
The inclusion criteria for patients were as follows: (I) nodules clinically suspicious for breast cancer and recommended for biopsy; (II) nodules classified as B1, B2, B3, or B5 based on CNB results; and (III) nodules categorized as BI-RADS 3 or higher. Meanwhile, the exclusion criteria were (I) lesions identified as metastatic tumors and (II) patients who had undergone systemic hormone therapy or adjuvant chemotherapy.
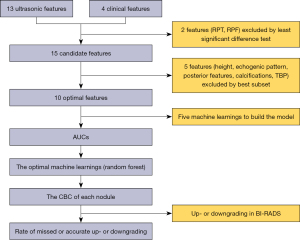
The workflow of the study is illustrated in Figure 1.
Clinical features and ultrasonic image acquisition
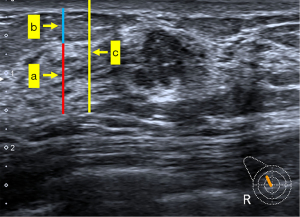
The clinical features examined in this study included height, weight, body mass index (BMI), and age. We used a 14-MHz linear transducer (Toshiba Aplio 500, Tokyo, Japan) to capture ultrasonic images. Images of the nodules were acquired in a standard manner, and contained at least two orthogonal planes (radial and antiradial or transverse and longitudinal). According to the ACR BI-RADS fifth edition classification criteria and a previous study (21), all images were analyzed retrospectively by two breast radiologists (Reader 1 with 10 years of experience and Reader 2 with 5 years of experience). The radiologists maintained strict records of 13 ultrasonic features: shape, orientation, margin, echogenic pattern, posterior features, calcifications, vascularity distribution, vascularity grade, tumor size, BI-RADS category, anteroposterior thickness of the breast parenchyma (TBP), anteroposterior thickness ratio of breast parenchyma to mammary fat (RPF), and anteroposterior thickness ratio of breast parenchyma to tissue before pectoralis fascia (RPT). RPF and RPT were the adjusted parameters of TBP and were obtained after TBP was corrected according to the thickness of tissue but before pectoralis fascia and thickness of fat, respectively (Figure 2). Detailed feature descriptions are provided in Appendix 1 (22). Both Reader 1 and Reader 2 were blinded to histological results but had access to patient age. We assessed the inter- and intraobserver agreement for all 13 sonographic features. In cases of discrepancy between readers, final determinations were reached through consensus discussion.
Core biopsy reporting categories
To facilitate the analysis of the clinical and ultrasound characteristics of the lesions, all lesions were categorized into four groups based on histological examination (10). (I) The B1 group included normal tissue. (II) The B2 group included fibroadenoma, fibrocystic changes, sclerosing adenosis, duct ectasia, and other nonparenchymal lesions such as abscesses and fat necrosis. (III) The B3 group included lesions with uncertain malignant potential. These lesions may exhibit benign histology on core biopsy but are known to display heterogeneity or carry an increased risk of associated malignancy. This category encompassed atypical intraductal epithelial proliferation, flat epithelial atypia, lobular neoplasia, phyllodes tumors, papillary lesions, radial scars, mucocele-like lesions, and other rare lesions. Due to their uncertain malignant potential, B3 lesions are recommended for expanded excision during surgeries. (IV) Finally, the B5 group included malignant nodules.
It should be noted that the B4 classification includes suspicious nodules. Typically, this classification is applied when errors occur in pathological section preparation, such as crushed deformation or poorly fixed core tissue samples, leading to suspicion of cancerous tissue within the sample. In such cases, remaking the pathological sections is necessary to confirm the tumor characteristics. Consequently, B4 lesions were excluded from this study.
Statistical analysis
Statistical analyses were conducted with SPSS version 22.0 (IBM Corp., Armonk, NY, USA). A two-sided significance threshold of P<0.05 was applied. Continuous variables were compared using the least significant difference (LSD) test, whereas categorical variables were assessed with the Bonferroni correction. The best subset method was used to select the optimal predictive features for model development.
Machine learning in characteristics analysis
SPSS Modeler 18.0 software (IBM Corp.) was used to implement the machine learning workflow, which directly predicted the probability of CBC for each nodule. The procedure consisted of the following steps. First, the optimal features were selected using the best subset method. Second, during implementation, the dataset was randomly split into training and validation cohorts using the partition node at a ratio of 7:3 (training: validation). Subsequently, the balance node was applied to address class imbalance issues. Finally, given the variety of available machine learning algorithms, we employed several widely used models to perform the classification task. These models included RF, support vector machine (SVM), k-nearest neighbor (KNN), multilayer perceptron (MLP), and logistic regression (LR).
Performance of the machine learning models
The diagnostic performance of each algorithm was evaluated using ROC curve analysis, with the AUC calculated for comparison. The algorithm demonstrating the highest AUC was selected. We then applied this optimal algorithm to perform CBC prediction of contralateral breast cancer for each nodule.
Up- and downgrading in BI-RADS
If the CBC prediction was B1 or B2, the BI-RADS category was downgraded by one level. Conversely, if the CBC prediction was B3 or B5, the BI-RADS category was upgraded by one level. Lesions classified as BI-RADS 4 or 5 were recommended for biopsy. Specifically, for BI-RADS 4B or 4C lesions, regardless of whether they were upgraded or downgraded, biopsy remained necessary. Therefore, the focus of this study was on BI-RADS-US category 3 and 4A lesions, and we calculated the number and rate of accurate or missed upgrades and downgrades.
Results
Clinical and ultrasonic characteristics
A total of 1,185 lesions were examined in this study, and the distribution of BI-RADS classifications was as follows: 42 were category 3 (3.5%), 167 were category 4A (14%), 399 were category 4B (34%), 296 were category 4C (25%), and 281 were category 5 (24%). Meanwhile, the distribution of CBC was follows: 44 were category B1 (4%), 714 were category B2 (60%), 57 were category B3 (5%), and 370 were category B5 (31%). The baseline ultrasonic and clinical features are summarized in Table 1. Significant differences (P<0.05) were observed in 15 features, including age, height, weight, BMI, echo pattern, shape, margin, orientation, posterior features, calcification, vascularity distribution, vascularity grade, BI-RADS category, tumor size, and TBP. However, no significant differences were found for RPT or RPF (P>0.05).
Table 1
| Feature | BI-RADS | P | ||||
|---|---|---|---|---|---|---|
| 3 (n=42) | 4A (n=167) | 4B (n=399) | 4C (n=296) | 5 (n=281) | ||
| Age (years) | 36.95±13.0 | 37.66±11.54 | 38.41±11.87 | 43.47±13.54 | 49.81±12.7 | <0.001 |
| Height (cm) | 158.76±4.95 | 158.68±4.79 | 159.18±4.76 | 158.14±5.12 | 157.9±5.04 | 0.009 |
| Weight (kg) | 53.86±7.39 | 53.68±7.27 | 54.33±7.78 | 55.94±8.17 | 57.46±8.36 | <0.001 |
| BMI, kg/m2 | 21.39±3.03 | 21.35±2.97 | 21.45±2.96 | 22.36±3.19 | 23.04±8.36 | <0.001 |
| Echo pattern | <0.001 | |||||
| Hyperechoic | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Complex cystic and solid | 3 (7.1) | 1 (0.6) | 3 (0.8) | 1 (0.3) | 0 (0) | |
| Hypoechoic | 34 (81.0) | 109 (65.3) | 163 (40.9) | 107 (36.1) | 117 (41.6) | |
| Isoechoic | 5 (11.9) | 8 (4.8) | 3 (0.8) | 1 (0.3) | 11 (3.9) | |
| Heterogeneous | 0 (0) | 49 (29.3) | 230 (57.6) | 187 (63.2) | 153 (54.4) | |
| Shape | <0.001 | |||||
| Oval | 27 (64.3) | 120 (71.9) | 69 (17.3) | 8 (2.7) | 5 (1.8) | |
| Round | 1 (2.4) | 1 (0.6) | 8 (2.0) | 1 (0.3) | 3 (1.1) | |
| Irregular | 14 (33.3) | 46 (27.5) | 322 (80.7) | 287 (97.0) | 273 (97.2) | |
| Margin | <0.001 | |||||
| Circumscribed | 29 (69.0) | 128 (76.6) | 85 (21.3) | 13 (4.4) | 6 (2.1) | |
| Indistinct | 4 (9.5) | 19 (11.4) | 103 (25.8) | 44 (14.9) | 23 (8.2) | |
| Angular | 9 (21.4) | 16 (9.6) | 203 (50.9) | 208 (70.3) | 125 (44.5) | |
| Microlobulated | 0 (0) | 4 (2.4) | 8 (2.0) | 31 (10.5) | 127 (45.2) | |
| Orientation | <0.001 | |||||
| Parallel | 38 (90.5) | 165 (98.8) | 364 (91.2) | 241 (81.4) | 191 (68.0) | |
| Not parallel | 4 (9.5) | 2 (1.2) | 35 (8.8) | 55 (18.6) | 90 (32.0) | |
| Posterior feature | <0.001 | |||||
| No posterior feature | 25 (59.5) | 38 (22.8) | 33 (8.3) | 19 (6.4) | 14 (5.0) | |
| Enhancement sound | 8 (19.0) | 31 (18.6) | 41 (10.3) | 20 (6.8) | 8 (2.8) | |
| Shadowing | 6 (14.3) | 14 (8.4) | 102 (25.6) | 71 (24.0) | 54 (19.2) | |
| Combined pattern | 3 (7.1) | 84 (50.3) | 223 (55.9) | 186 (62.8) | 205 (73.0) | |
| Calcification | <0.001 | |||||
| In a mass | 2 (4.8) | 4 (2.4) | 22 (5.5) | 61 (20.6) | 112 (39.9) | |
| Outside of a mass | 0 (0) | 1 (0.6) | 0 (0) | 0 (0) | 2 (0.7) | |
| Intraductal calcification | 0 (0) | 0 (0) | 0 (0) | 2 (0.7) | 0 (0) | |
| None | 40 (95.2) | 162 (97.0) | 377 (94.5) | 233 (78.7) | 167 (59.4) | |
| Vascularity distribution | <0.001 | |||||
| Absent | 22 (52.4) | 78 (46.7) | 156 (39.1) | 74 (25.1) | 20 (7.1) | |
| Vessels in rim | 4 (9.5) | 33 (19.8) | 64 (16.0) | 48 (16.2) | 35 (12.5) | |
| Internal | 16 (38.1) | 56 (33.5) | 179 (44.9) | 174 (58.7) | 226 (80.4) | |
| Vascularity grade | <0.001 | |||||
| Grade I | 22 (52.4) | 78 (46.7) | 155 (38.8) | 74 (25.0) | 21 (7.5) | |
| Grade II | 10 (23.8) | 58 (34.7) | 149 (37.3) | 109 (36.8) | 88 (31.3) | |
| Grade III | 3 (7.1) | 26 (15.6) | 61 (15.3) | 77 (26.0) | 116 (41.3) | |
| Grade IV | 7 (16.7) | 5 (3.0) | 34 (8.5) | 36 (12.2) | 56 (19.9) | |
| Tumor size (mm) | 14.08±7.16 | 14.59±5.87 | 17.29±8.71 | 20.91±10.84 | 25.90±12.0 | <0.001 |
| TBP (mm) | 8.10±2.97 | 7.71±2.73 | 8.05±3.32 | 8.48±3.74 | 9.20±3.83 | <0.001 |
| RPT | 0.52±0.12 | 0.51±0.12 | 0.52±0.14 | 0.51±0.15 | 0.49±0.23 | 0.453 |
| RPF | 1.73±1.01 | 1.79±1.55 | 1.93±1.70 | 1.84±1.86 | 1.60±1.82 | 0.207 |
| CBC | ||||||
| B1 | 2 (4.8) | 15 (9.0) | 15 (3.8) | 11 (3.7) | 1 (0.4) | <0.001 |
| B2 | 36 (85.7) | 135 (80.8) | 335 (84.0) | 166 (56.1) | 42 (14.9) | |
| B3 | 3 (7.1) | 10 (6.0) | 12 (3.0) | 23 (7.8) | 9 (3.2) | |
| B5 | 1 (2.4) | 7 (4.2) | 37 (9.3) | 96 (32.4) | 229 (81.5) | |
Data are presented as mean ± standard deviation or number (%). BI-RADS, Breast Imaging Reporting and Data System; BMI, body mass index; CBC, core needle biopsy category; RPF, thickness ratio of breast parenchyma to mammary fat; RPT, thickness ratio of breast parenchyma to tissue before pectoralis fascia; TBP, anteroposterior thickness of breast parenchyma.
Selection of features, construction, and performance of models
Following the best subset analysis, 10 features were selected for model construction, including age, BMI, shape, weight, orientation, margin, tumor size, BI-RADS category, vascularity distribution, and vascularity grade. No statistically significant differences were observed in the clinical characteristics or ultrasound features between the training and validation cohorts (Table S1).
These features were used to build models through five machine learning algorithms. The AUC values for each model are presented in Table 2. According to the AUC results, RF was the optimal algorithm, achieving the highest AUC of 0.943 [95% confidence interval (CI): 0.930–0.956]. The AUC values for the remaining algorithms were as follows: MLP, 0.916 (95% CI: 0.898–0.934); LR, 0.472 (95% CI: 0.435–0.509); KNN, 0.828 (95% CI: 0.802–0.854); and SVM, 0.909 (95% CI: 0.891–0.928).
Table 2
| Algorithms | AUC (95% CI) |
|---|---|
| MLP | 0.916 (0.898–0.934) |
| RF | 0.943 (0.930–0.956) |
| LR | 0.472 (0.435–0.509) |
| KNN | 0.828 (0.802–0.854) |
| SVM | 0.909 (0.891–0.928) |
AUC, area under the curve; CI, confidence interval; KNN, k-nearest neighbor; LR, logistic regression; MLP, multilayer perceptron; RF, random forest; SVM, support vector machine.
The feature importance ranking derived from RF model is reported in Figure 3. Based on the length of the bars in the bar chart in Figure 3, the most important feature was age, followed in descending order by tumor size, BMI, weight, BI-RADS, margin, vascularity distribution, vascularity grade, shape, and orientation.

Probability of disease in BI-RADS category 3 and 4A lesions
The CBC prediction was calculated for each lesion, and these predictions were used to adjust (upgrade or downgrade) the BI-RADS categories. The number and rate of accurate and missed upgrades or downgrades are summarized in Table 3.
Table 3
| Upgrade or downgrade | BI-RADS category 3 (n=42) |
BI-RADS category 4A (n=167) |
|---|---|---|
| Upgrade one category | 4 | 18 |
| Downgrade one category | 38 | 149 |
| Missed upgrade | 1 (2.4) | 5 (3.0) |
| Accurate upgrade | 3 (7.1) | 13 (7.8) |
| Missed downgrade | 1 (2.4) | 4 (2.4) |
| Accurate downgrade | 37 (88.1) | 145 (86.8) |
The data are presented as number or number (%). BI-RADS, Breast Imaging Reporting and Data System.
Among the 42 BI-RADS category 3 lesions, 4 (9.5%) were upgraded, with 3 being accurate, while 38 cases (90.5%) were downgraded, with 37 being accurate. The accurate upgrade rate (7.1%) was higher than the missed upgrade rate (2.4%), and the accurate downgrade rate (88.1%) was significantly higher than the missed downgrade rate (2.4%) (Figure 4).

For the 167 BI-RADS category 4A lesions, 149 (89.2%) cases were downgraded, with 145 being accurate, and 18 (10.8%) cases were upgraded, with 13 being accurate. The accurate upgrade rate (7.8%) was higher than the missed upgrade rate (3.0%), and the accurate downgrade rate (86.8%) was significantly higher than the missed downgrade rate (2.4%) (Figure 4).
Discussion
Our study identified RF as the optimal machine learning algorithm for predicting CBC. We used the RF model to enhance the diagnostic performance of BI-RADS categories 3 and 4A, thereby reducing unnecessary biopsies for patients with category 4A lesions, as demonstrated in Table 3. First, for BI-RADS categories 3 and 4A, the misgrading rates were low (4.8% and 5.4%, respectively), with most of the cases being accurately graded (95.2% and 94.6%, respectively). The accurate upgrade and downgrade rates were significantly higher than were the missed rates. Meucci et al. analyzed the distribution of MRI features in CBC lesions but only included 61 cases (23). Meanwhile, Giuliani et al. analyzed multiple clinical and sonographic characteristics in 102 B3 lesions; however, their study did not comprehensively evaluate the association between B3 lesions and individual BI-RADS lexicon features (24). Our previous study also identified RF as the optimal machine learning algorithm for predicting CBC; however, we did not integrate RF into the practical diagnostic workflow to refine BI-RADS categorization (25). In the present study, we confirmed RF as the optimal algorithm using a larger sample size (1,185 nodules), rendering our findings more robust and practical as compared to those reported previously studies (23-25). Furthermore, among the 167 BI-RADS category 4A cases, 149 (89.2%) were downgraded, with 145 being correctly downgraded. These results suggest that our approach can avoid a significant number of unnecessary biopsies.
Although Wang et al. and Wei et al. used computer-aided diagnosis (CAD) systems to improve the performance of BI-RADS (26,27), their studies had several limitations. Their AUC values were lower than ours (0.91 and 0.906 vs. 0.945, respectively), and they only classified breast masses into malignant and benign categories, failing to address the issue of unnecessary biopsies for some nonmalignant masses (e.g., atypical lesions). Additionally, Wang et al.’s study had a small sample size, comprising only 54 malignant and 162 benign lesions (26). Meanwhile, Wei et al. used BI-RADS categories 4A and 4B as the cutoff for their CAD software (27); however, BI-RADS categorization is highly subjective and should not be used as the sole basis for setting diagnostic thresholds. In contrast, our study included a larger sample size (1,185 nodules) and refined the BI-RADS classification based on the objective standard of CBC, which are grounded in breast histological types. Consequently, our study can be considered more objective and to have greater clinical applicability.
Both the benign-or-malignant classification system and the CBC system are based on histological classification. However, the CBC system provides more precise guidance for the clinical management of breast lesions following biopsy, making it superior to the benign-or-malignant system. In practice, the clinical goal of both CBC and BI-RADS is to serve as the foundation for breast mass management. However, BI-RADS classification is subject to variability due to its reliance on human experience. Therefore, we believe that CBC is better suited for refining and improving the BI-RADS system.
Machine learning is highly advantageous for constructing predictive models and plays a pivotal role in radiological research (13,28). Among the various types of machine learning algorithms, the five employed in our study are widely used. Although MLP, KNN, and SVM had an inferior performance to that of RF, they were satisfactory, with AUC values exceeding 0.8. In contrast, LR had poor performance (AUC =0.472). LR, as a linear classifier, has several limitations: (I) it is unsuitable to solving nonlinear problems; (II) it is sensitive to multicollinearity in data; (III) it struggles to handle imbalanced datasets; and (IV) its accuracy is limited due to its simplistic structure, making it difficult to capture the true distribution of the data. Consequently, LR failed to achieve the classification objectives in our study. Both our previous and current studies identified RF as the optimal method for predicting CBC (25). In this study, we successfully integrated RF into the practical diagnostic workflow, achieving excellent performance. RF operates through ensemble learning, aggregating predictions from multiple decision trees and determining the final output category via majority voting among individual tree outputs. This ensemble approach grants RF a significant advantage in classification tasks.
Our study demonstrated strong reproducibility for several reasons. First, the sample size was substantially large, enhancing the reliability of our findings. Second, the results were obtained using SPSS Modeler software, which ensures robustness due to its fixed seed number for randomization.
Three cases (7.1%) of BI-RADS category 3 lesions were accurately upgraded, representing a prevention of misdiagnoses that would require biopsy. However, 4 cases (2.4%) of BI-RADS category 4A lesions were incorrectly downgraded. This discrepancy may be attributed to the highly ambiguous features in these cases, such as younger age, regular shape, absence of calcification, and other confounding factors. Consequently, the RF model requires further refinement so that its performance in reclassifying BI-RADS category 4A lesions can be improved.
Our study involved several limitations that should be addressed. First, it was conducted at a single center, and the sample size for the B3 category was relatively small (n=42). The generalizability of our RF model requires validation in larger, multicenter cohorts. Second, because of the inherent limitations of retrospective data, other clinical risk factors (e.g., serological markers, family history, and menopausal status) were not included. Future prospective studies with comprehensive datasets are recommended to address this deficiency. Third, the assessment of histological features relied on subjective analysis, potentially introducing bias. To mitigate this, the incorporation of objective parameters, such as ultrasound radiomics, should be explored to enhance the model’s accuracy. Finally, although RF demonstrated the potential to improve upon the BI-RADS classification, further research is needed to integrate it into electronic ultrasound systems for practical clinical application.
Conclusions
Based on a comprehensive array of clinical and ultrasonographic characteristics, machine learning algorithms were employed to assess CBC in solid breast lesions. Among the evaluated models, RF demonstrated superior performance in predicting CBC, achieving the highest AUC. Subsequently, this predictive model was effectively used to refine the BI-RADS category classification for 3 and 4A lesions. Our findings indicate that the model-assisted approach can significantly enhance grading accuracy, reduce the number of unnecessary biopsy procedures, and minimize the misdiagnosis of malignant tumors.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD+AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-2070/rc
Funding: This work was supported by the National Center for Inheritance and Innovation of Traditional Chinese Medicine Research Special Project (No. 2022QN18).
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2070/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments. Ethical approval for this study was granted by the institutional review board of Guangdong Provincial People’s Hospital (No. KY2023-1069-01), who waived the requirement for informed consent due to the retrospective nature of the analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Neal L, Sandhu NP, Hieken TJ, Glazebrook KN, Mac Bride MB, Dilaveri CA, Wahner-Roedler DL, Ghosh K, Visscher DW. Diagnosis and management of benign, atypical, and indeterminate breast lesions detected on core needle biopsy. Mayo Clin Proc 2014;89:536-47. [Crossref] [PubMed]
- Rungruang B, Kelley JL 3rd. Benign breast diseases: epidemiology, evaluation, and management. Clin Obstet Gynecol 2011;54:110-24. [Crossref] [PubMed]
- Sardanelli F, Boetes C, Borisch B, Decker T, Federico M, Gilbert FJ, et al. Magnetic resonance imaging of the breast: recommendations from the EUSOMA working group. Eur J Cancer 2010;46:1296-316. [Crossref] [PubMed]
- Yap YS, Lu YS, Tamura K, Lee JE, Ko EY, Park YH, Cao AY, Lin CH, Toi M, Wu J, Lee SC. Insights Into Breast Cancer in the East vs the West: A Review. JAMA Oncol 2019;5:1489-96. [Crossref] [PubMed]
- Shen S, Zhou Y, Xu Y, Zhang B, Duan X, Huang R, Li B, Shi Y, Shao Z, Liao H, Jiang J, Shen N, Zhang J, Yu C, Jiang H, Li S, Han S, Ma J, Sun Q. A multi-centre randomised trial comparing ultrasound vs mammography for screening breast cancer in high-risk Chinese women. Br J Cancer 2015;112:998-1004. [Crossref] [PubMed]
- Chae EY, Cha JH, Shin HJ, Choi WJ, Kim HH. Reassessment and Follow-Up Results of BI-RADS Category 3 Lesions Detected on Screening Breast Ultrasound. AJR Am J Roentgenol 2016;206:666-72. [Crossref] [PubMed]
- Barr RG, Zhang Z, Cormack JB, Mendelson EB, Berg WA. Probably benign lesions at screening breast US in a population with elevated risk: prevalence and rate of malignancy in the ACRIN 6666 trial. Radiology 2013;269:701-12. [Crossref] [PubMed]
- Weng L, Yu M. Diagnosis of Benign and Malignant BI-RADS 4 Breast Masses by Contrastenhanced Ultrasound Combined with Shear Wave Elastography. Curr Med Imaging 2023; Epub ahead of print. [Crossref]
- Zhao XB, Yao JY, Zhou XC, Hao SY, Mu WJ, Li LJ, Zhong WJ, Hui Z. Strain Elastography: A Valuable Additional Method to BI-RADS? Ultraschall Med 2018;39:526-34. [Crossref] [PubMed]
- Lee A, Anderson N, Carder P, Cooke J, Deb R, Ellis IO, Howe M, Jenkins JA, Knox F, Stephenson T. Guidelines for non-operative diagnostic procedures and reporting in breast cancer screening. London, UK: The Royal College of Pathologists; 2016.
- Achilonu OJ, Fabian J, Bebington B, Singh E, Eijkemans MJC, Musenge E. Predicting Colorectal Cancer Recurrence and Patient Survival Using Supervised Machine Learning Approach: A South African Population-Based Study. Front Public Health 2021;9:694306. [Crossref] [PubMed]
- Yu KH, Lee TM, Yen MH, Kou SC, Rosen B, Chiang JH, Kohane IS. Reproducible Machine Learning Methods for Lung Cancer Detection Using Computed Tomography Images: Algorithm Development and Validation. J Med Internet Res 2020;22:e16709. [Crossref] [PubMed]
- Zhang B, Tian J, Pei S, Chen Y, He X, Dong Y, Zhang L, Mo X, Huang W, Cong S, Zhang S. Machine Learning-Assisted System for Thyroid Nodule Diagnosis. Thyroid 2019;29:858-67. [Crossref] [PubMed]
- Bitencourt AGV, Gibbs P, Rossi Saccarelli C, Daimiel I, Lo Gullo R, Fox MJ, Thakur S, Pinker K, Morris EA, Morrow M, Jochelson MS. MRI-based machine learning radiomics can predict HER2 expression level and pathologic response after neoadjuvant therapy in HER2 overexpressing breast cancer. EBioMedicine 2020;61:103042. [Crossref] [PubMed]
- Narula S, Shameer K, Salem Omar AM, Dudley JT, Sengupta PP. Machine-Learning Algorithms to Automate Morphological and Functional Assessments in 2D Echocardiography. J Am Coll Cardiol 2016;68:2287-95. [Crossref] [PubMed]
- Nanayakkara S, Fogarty S, Tremeer M, Ross K, Richards B, Bergmeir C, Xu S, Stub D, Smith K, Tacey M, Liew D, Pilcher D, Kaye DM. Characterising risk of in-hospital mortality following cardiac arrest using machine learning: A retrospective international registry study. PLoS Med 2018;15:e1002709. [Crossref] [PubMed]
- Sutton EJ, Onishi N, Fehr DA, Dashevsky BZ, Sadinski M, Pinker K, Martinez DF, Brogi E, Braunstein L, Razavi P, El-Tamer M, Sacchini V, Deasy JO, Morris EA, Veeraraghavan H. A machine learning model that classifies breast cancer pathologic complete response on MRI post-neoadjuvant chemotherapy. Breast Cancer Res 2020;22:57. [Crossref] [PubMed]
- Wu T, Sultan LR, Tian J, Cary TW, Sehgal CM. Machine learning for diagnostic ultrasound of triple-negative breast cancer. Breast Cancer Res Treat 2019;173:365-73. [Crossref] [PubMed]
- Panourgias E, Karampotsis E, Douma N, Bourgioti C, Koutoulidis V, Rigas G, Moulopoulos L, Dounias G. Accuracy of distinguishing benign, high-risk lesions and malignancies with inductive machine learning models in BIRADS 4 and BIRADS 5 lesions on breast MR examinations. Eur J Radiol 2024;181:111801. [Crossref] [PubMed]
- Bahl M, Barzilay R, Yedidia AB, Locascio NJ, Yu L, Lehman CD. High-Risk Breast Lesions: A Machine Learning Model to Predict Pathologic Upgrade and Reduce Unnecessary Surgical Excision. Radiology 2018;286:810-8. [Crossref] [PubMed]
- Lin X, Zhuang S, Yang S, Lai D, Chen M, Zhang J. Development and internal validation of a conventional ultrasound-based nomogram for predicting malignant nonmasslike breast lesions. Quant Imaging Med Surg 2022;12:5452-61. [Crossref] [PubMed]
- Adler DD, Carson PL, Rubin JM, Quinn-Reid D. Doppler ultrasound color flow imaging in the study of breast cancer: preliminary findings. Ultrasound Med Biol 1990;16:553-9. [Crossref] [PubMed]
- Meucci R, Pistolese Chiara A, Perretta T, Vanni G, Portarena I, Manenti G, Ryan Colleen P, Castrignanò A, Di Stefano C, Ferrari D, Lamacchia F, Pellicciaro M, Materazzo M, Buonomo Oreste C. MR imaging-guided vacuum assisted breast biopsy: Radiological-pathological correlation and underestimation rate in pre-surgical assessment. Eur J Radiol Open 2020;7:100244. [Crossref] [PubMed]
- Giuliani M, Rinaldi P, Rella R, D’Angelo A, Carlino G, Infante A, Romani M, Bufi E, Belli P, Manfredi R. A new risk stratification score for the management of ultrasound-detected B3 breast lesions. Breast J 2018;24:965-70. [Crossref] [PubMed]
- Liang T, Shen J, Wang J, Liao W, Zhang Z, Liu J, Feng Z, Pei S, Liu K. Ultrasound-based prediction of preoperative core biopsy categories in solid breast tumor using machine learning. Quant Imaging Med Surg 2023;13:2634-46. [Crossref] [PubMed]
- Wang Y, Tang L, Chen P, Chen M. The Role of a Deep Learning-Based Computer-Aided Diagnosis System and Elastography in Reducing Unnecessary Breast Lesion Biopsies. Clin Breast Cancer 2023;23:e112-21. [Crossref] [PubMed]
- Wei Q, Yan YJ, Wu GG, Ye XR, Jiang F, Liu J, Wang G, Wang Y, Song J, Pan ZP, Hu JH, Jin CY, Wang X, Dietrich CF, Cui XW. The diagnostic performance of ultrasound computer-aided diagnosis system for distinguishing breast masses: a prospective multicenter study. Eur Radiol 2022;32:4046-55. [Crossref] [PubMed]
- Lu CF, Hsu FT, Hsieh KL, Kao YJ, Cheng SJ, Hsu JB, Tsai PH, Chen RJ, Huang CC, Yen Y, Chen CY. Machine Learning-Based Radiomics for Molecular Subtyping of Gliomas. Clin Cancer Res 2018;24:4429-36. [Crossref] [PubMed]


