
A comparative analysis of sagittal, coronal, and axial magnetic resonance imaging planes in diagnosing anterior cruciate ligament and meniscal tears via a deep learning model: emphasizing the unexpected importance of the axial plane
Introduction
Anterior cruciate ligament (ACL) and meniscal injuries represent the most prevalent knee disorders in the general population, particularly those engaged in physical activities (1,2). These injuries manifest through characteristic symptoms such as pain, joint instability, and functional impairment, directly reflecting structural damage to ligamentous and meniscal tissues (3). Accurate diagnosis remains critical for both athletes and the general population to prevent secondary complications (4).
Since its clinical adoption in the 1980s, magnetic resonance imaging (MRI) has served as the cornerstone modality for evaluating ACL and meniscal injuries, with extensive studies validating and critically examining its diagnostic accuracy (ACC), sensitivity (SEN), and specificity (SPE) (5-7). The sagittal and coronal planes constitute the primary MRI planes for ACL and meniscal injury assessment, whereas the axial plane provides supplementary diagnostic value, particularly for meniscal pathology (8,9). Multi-plane integration enhances diagnostic accuracy through comprehensive structural visualization (9-11).
Artificial intelligence (AI) has driven transformative advancements in medical image analysis over decades (12,13), with MRI-based computer-aided diagnosis (CAD) systems demonstrating clinical utility across diverse anatomical contexts (14-17). Accumulated evidence confirms that well-optimized CAD systems can significantly enhance radiological workflow efficiency by automating preliminary MRI screening and classification tasks (18,19).
Regarding auxiliary diagnoses of ACL and meniscal injuries conducted via magnetic resonance (MR) images, numerous studies have focused on deep learning (DL)-based image preprocessing, segmentation, classification, and data augmentation methods (20-26). These studies have primarily aimed to increase the quality of images and improve the diagnostic or grading ACC of their models (27-32). Conversely, most AI datasets for ACL/meniscal injuries predominantly rely on single-plane MRI, with no existing studies have systematically investigated the differential contributions of sagittal, coronal, and axial planes in AI-based diagnostics (20,31). Despite the technical superiority of three-dimensional (3D) MRI, two-dimensional (2D) multi-planar sequences (sagittal/coronal/axial) remain the predominant clinical protocol for knee injury diagnosis due to cost-effectiveness and workflow efficiency (33,34).
To systematically evaluate the contributions of individual MRI planes, we developed a DL framework capable of processing flexible combinations of sagittal, coronal, and axial inputs. This design enables direct comparison between AI-driven multi-plane integration strategies and established radiological diagnostic logic. We present this article in accordance with the TRIPOD+AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1808/rc).
Methods
Dataset
The MRNet dataset is currently the only publicly available dataset that is suitable for research on ACL and meniscus tears, providing corresponding MR images in the sagittal, coronal, and axial planes for each case. The MRNet dataset contains 1,370 knee MR images from Stanford University Medical Center collected between January 2001 and December 2012. Each case was labeled to reflect whether the knee had an ACL tear, a meniscal tear, or other abnormalities, such as osteoarthritis or ligament injuries. In the MRNet dataset, the MR examination of each patient includes three distinct planes: sagittal T2-weighted, sagittal proton density-weighted, coronal T1-weighted, and axial proton density-weighted images. These images were uniformly formatted to a resolution of 256×256 pixels, with the number of slices per scan ranging from 17 to 61 (29).
The dataset includes 1,104 abnormal cases, 319 ACL tear cases, and 508 meniscal tear cases. Standard preprocessing techniques such as intensity standardization and normalization have been applied to enhance the image quality of this dataset. We utilized the publicly accessible portion of the dataset, comprising 1,130 training cases and 120 validation cases, to develop and validate our proposed model (29). Stratified random sampling was used to construct the training and validation sets, ensuring that each set included at least 50 positive examples for each label (ACL tear and meniscal tear).
The distribution of the data in the training and validation sets was as follows: in the training set, 208 cases (18.4%) are ACL tear cases, 397 cases (35.1%) are meniscal tear cases, and 125 cases (11.1%) have both ACL and meniscal tears. In the validation set, 54 cases (45.0%) are ACL tear cases, 52 cases (43.3%) are meniscal tear cases, and 31 cases (25.8%) have both ACL and meniscal tears. These distributions are detailed in Table 1, which also shows the demographic characteristics of the patients, including the number of female patients, age distribution, and the prevalence of different injury types. All patients underwent the same imaging procedures.
Table 1
| Statistic | Training | Validation |
|---|---|---|
| Number of exams | 1,130 | 120 |
| Number of patients | 1,088 | 111 |
| Number of female patients (%) | 480 (42.5) | 50 (41.7) |
| Age (years), mean (SD) | 38.3 (16.9) | 36.3 (16.9) |
| Number with ACL tear (%) | 208 (18.4) | 54 (45.0) |
| Number with meniscal tear (%) | 397 (35.1) | 52 (43.3) |
| Number with ACL and meniscal tear (%) | 125 (11.1) | 31 (25.8) |
The training set was used to optimize model parameters and the validation set was to select the best model. ACL, anterior cruciate ligament; SD, standard deviation.
MRNet model, a DL classification system based on convolutional neural networks (CNNs), was introduced alongside the MRNet dataset. The MRNet model is composed of three distinct single-plane models, each of which is based on a modified version of the AlexNet architecture (29). The modified AlexNet architecture consists of five convolutional layers followed by three fully connected layers. Each model in the MRNet framework processes single-plane MRI images, with each MRI sequence comprising multiple 256×256 pixel slices. The input data undergoes histogram normalization and is expanded to three channels to meet AlexNet’s input requirements. Feature vectors generated after activation and pooling are passed through fully connected layers and a sigmoid function, producing probabilities between 0 and 1. The prediction probabilities from the three models are combined using a simple logistic regression model, yielding a final classification probability as a weighted sum of the probabilities from each plane. A 0.5 threshold is then applied to convert this probability into a classification outcome. The model is pre-trained on ImageNet, enabling optimized weight initialization for the specific dataset, followed by fine-tuning on the MRNet dataset for classification tasks.
The original MRNet model was designed as a high-performance classification diagnostic model, emphasizing both accuracy and efficiency. In contrast, our study aimed to compare the roles and impacts of sagittal, coronal, and axial MRI planes in DL-based diagnostic models. To better align with our research objectives, we have made several modifications to the original MRNet model.
Our retrospective study was performed using publicly available data for which no institutional review board approval was required. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Model
In our study, we designed a DL model, TripleMRNet, which allows for the selection of 1–3 MRI planes from the sagittal, coronal, and axial planes. This led to three categories and seven data combinations: single-plane images (sagittal, coronal, or axial), two-plane images (sagittal + coronal, sagittal + axial, or coronal + axial), and three-plane images. For the submodels using two or three planes, feature-level fusion is applied, where features extracted from multiple planes are combined to make the final decision. We named our model TripleMRNet and adapted it to include seven submodels, each corresponding to one of the seven plane combinations (Figure 1).

The MRNet was pre-trained on ImageNet, with AlexNet’s weights initialized using those trained on the ImageNet dataset. The model was then fine-tuned on the MRNet dataset for binary classification tasks, including the detection of ACL tears and meniscal tears. Unlike natural images in ImageNet, knee MRI images exhibit unique intensity patterns and anatomical structures. To avoid potential confounding effects on the analysis across three imaging planes, our TripleMRNet was trained from scratch without using transfer learning. This approach ensures that the features learned by the model are derived entirely from knee MRI images.
The MRNet network employed a parameter-sharing strategy across the models for the sagittal, coronal, and axial planes, where the convolutional and fully connected layers shared the same weights. This approach reduced the total number of parameters, thereby lowering computational demands and accelerating the training process (29). However, it might have limited each model’s ability to capture plane-specific information. In our study, to preserve the independence of each plane model during training, we removed parameter sharing. This adjustment allowed the model to better extract and utilize unique features from each plane. We hypothesized that this would improve performance on tasks requiring detailed plane-specific analysis (35,36).
When merging model outputs, our approach differs from the original MRNet, which employs a simple logistic regression for probability-level fusion, weighting the probabilities generated by each plane-specific model. In contrast, our method employs feature-level fusion by concatenating feature vectors from different plane-specific models. While training TripleMRNet submodels with two- or three-plane combinations, the feature vectors from each plane-specific model are concatenated into a new feature vector that integrates information from multiple planes. The final prediction is made based on this unified feature vector, closely resembling the clinical scenario where doctors synthesize MRI images. A 0.5 threshold is then applied to convert this probability into a classification outcome.
There is a class imbalance in the sample distribution between categories in the training and validation datasets. Our model, in similarity with the MRNet, was optimized using binary cross-entropy loss. Each example’s loss was scaled inversely to its class prevalence, with the class prevalence calculated as the proportion of samples in the dataset. The reciprocal of this proportion was used as the class weight to address the issue of class imbalance.
During training, the loss gradient for each training example was calculated using backpropagation, and the parameters were updated in the direction opposite to the gradient. The model parameters were saved after each complete pass through the training data, and the model with the lowest average loss on the tuning set was selected for evaluation on the validation set.
We argue that a DL-based CAD system for knee MRI screening should prioritize minimizing false negatives to avoid missing injuries, while also reducing false positives to conserve medical resources. Therefore, we recorded the performance of each of the seven submodels at their highest F1 score on the validation set during training as the optimal performance. The F1 score, balancing precision (minimizing false positives) and recall (reducing false negatives), serves as a comprehensive performance metric.
In addition to the F1 score, we also evaluated the submodels using ACC, SEN, and SPE. ACC measures the proportion of correct predictions (both true positives and true negatives) against the total number of cases. SEN, or recall, measures the model’s ability to correctly identify positive cases (ACL or meniscal tears), whereas SPE evaluates the model’s ability to correctly identify negative cases (non-tears). We also documented the false-positive and false-negative predictions, along with their validation set indices, to further investigate the roles of the three planes in the classification model.
At the outset, we trained the three-plane model and evaluated its performance based on the highest F1 score, comparing ACC, SPE, and SEN with the original MRNet model, and hyperparameters for the model were selected through incremental adjustments informed by prior studies and iterative experimentation. This comparison aimed to show that these modifications did not degrade overall performance. To ensure fair comparisons of diagnostic performance for ACL and meniscal tears across the seven data combinations, we used the consistent hyperparameters (including the learning rate, batch size, L2 regularization parameter, and dropout rate) for each model (37). This approach enabled a more objective analysis of the impact of different plane combinations on model performance, ensuring the reliability of the results (38).
To evaluate the variation exhibited by our estimates, we calculated 95% Wilson score confidence intervals (95% CI) for metrics such as ACC, SEN, F1 score, and SPE. Since we performed multiple comparisons in this study to evaluate the diagnostic performance of different plane combinations in our model, we controlled the overall false discovery rate at 0.05. We report both unadjusted P values and adjusted q values, with q values <0.05 providing a more reliable assessment of statistical significance. A two-sided Fisher’s exact test was conducted, followed by independent corrections within the ACL and meniscal tear detection tasks, to determine whether significant differences in ACC, SEN, and SPE existed among the models.
Each model was trained for 50 iterations on the training set with a 24-GB NVIDIA GeForce RTX 3090 GPU (NVIDIA, Santa Clara, CA, USA). TripleMRNet was implemented with Python 3.10.13 (The Python Software Foundation, Wilmington, DE, USA) and PyTorch 2.0.1 (Meta AI, Menlo Park, CA, USA). All histograms were generated via GraphPad Prism 9.5.1 (GraphPad Software, San Diego, CA, USA).
Interpretation methods
There is a need for an interpretable AI model to facilitate efficient human–machine collaboration, as gaining insights into the model’s decision-making across different MRI planes is crucial for continuously refining its support in clinical diagnostics (39). In this study, our goal was to understand the AI’s decision-making logic when analyzing images from three different planes. The visualization of the model training process can, to some extent, serve as a reference for the system’s diagnostic logic, providing valuable insights for future improvements and more in-depth research on the model (40).
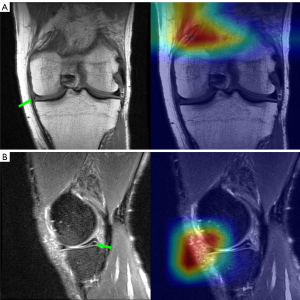
In medical image processing tasks, gradient-weighted class activation mapping (Grad-CAM) is widely used for interpreting and visualizing DL models. During training, the CNN progressively extracts features from the image through multiple convolutional layers. These features are passed layer by layer to the fully connected layers, where the final classification is made. Grad-CAM can be applied to various CNN models by utilizing gradient information to generate finer-grained activation maps. These maps are weighted based on the gradients, producing a weighted feature map. This map is then passed through a rectified linear unit (ReLU) activation function to obtain the final Grad-CAM heatmap, which is overlaid on the original image to highlight the regions the model focuses on during decision-making (41).
Grad-CAM also has its limitations, and the explanations it provides are somewhat limited. It is typically applied to the last convolutional layer of CNNs. Although convolutional layers capture local features, higher-level abstract features may be better represented in deeper layers. Grad-CAM helps visualize which areas are most important for the model’s prediction, but it does not directly explain why these areas have such a significant impact on the final prediction. However, in research, generating Grad-CAM heatmaps for DL models can offer valuable insights into the regions that most influence the model’s output. This information can be useful for model optimization or guiding subsequent research. Grad-CAM has been applied in studies involving pelvic X-rays, brain MR images, and chest computed tomography (CT) scans (42,43). In research involving the use of DL models to process knee MR images, multiple studies have utilized the Grad-CAM method for interpreting and visualizing their models (2,44).
In our study, we generated Grad-CAM heatmaps for each slice of three plane images from the best-performing models in the validation set for both ACL tear detection and meniscal tear detection tasks. These heatmaps highlighted the regions that significantly contribute to the model’s decision for diagnosing both ACL and meniscal tears. We then overlaid the corresponding heatmaps onto the original images, providing a more intuitive visualization of the model’s decision-making process (Figure 2). Furthermore, after training the model on the seven different plane data groupings and recording the false-positive and false-negative predictions obtained for each group in the validation set, we examined the heatmaps of these incorrect predictions and compared them with heatmaps of correctly predicted images.
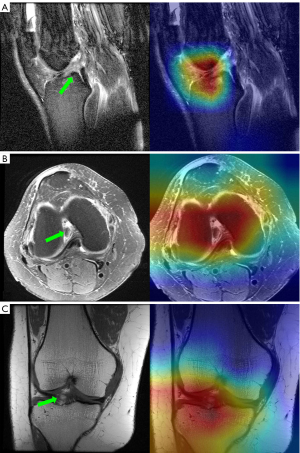
Results
We initially trained the 3-plane model of TripleMRNet, which achieved the highest F1 scores for ACL tear diagnosis (0.919) and meniscal tear diagnosis (0.725). Since the original MRNet model did not report F1 scores, we compared the ACC, SEN, and SPE of the TripleMRNet model with those of the MRNet model across both diagnostic tasks.
Our three-plane model outperformed the MRNet model in all metrics except for SPE in ACL tear detection. Notably, our model exhibited a significant improvement in SEN for ACL tear detection (0.944 vs. 0.759) (Tables 2,3). This indicates that the three-plane model of TripleMRNet is indeed more effective at extracting features from the images of each plane compared to the MRNet.
Table 2
| Model | Plane | Accuracy | Sensitivity | Specificity | F1 score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 95% CI | P value | q value | (95% CI) | P value | q value | (95% CI) | P value | q value | |||||
| MRNet | 3 planes | 0.867 (0.794–0.916) |
– | – | 0.759 (0.635–0.850) |
– | – | 0.968 (0.870–0.991) |
– | – | – | ||
| Our approach | 3 planes | 0.925 (0.864–0.960) |
– | – | 0.944 (0.849–0.981) |
– | – | 0.909 (0.816–0.958) |
– | – | 0.919 (0.856–0.956) |
||
| Sagittal + coronal | 0.892 (0.823–0.936) |
0.372 | 0.669 | 0.870 (0.756–0.936) |
0.184 | 0.552 | 0.909 (0.816–0.958) |
1.000 | 1.000 | 0.879 (0.808–0.926) |
|||
| Sagittal + axial | 0.917 (0.853–0.954) |
0.642 | 0.770 | 0.926 (0.825–0.971) |
0.695 | 0.834 | 0.909 (0.816–0.958) |
1.000 | 1.000 | 0.909 (0.844–0.949) |
|||
| Coronal + axial | 0.858 (0.785–0.910) |
0.007 | 0.042 | 0.833 (0.713–0.910) |
0.004 | 0.036 | 0.879 (0.779–0.937) |
0.318 | 0.572 | 0.841 (0.765–0.896) |
|||
| Sagittal | 0.892 (0.823–0.936) |
0.372 | 0.669 | 0.870 (0.756–0.936) |
0.184 | 0.552 | 0.909 (0.816–0.958) |
1.000 | 1.000 | 0.879 (0.808–0.926) |
|||
| Coronal | 0.842 (0.766–0.896) |
0.001 | 0.018 | 0.859 (0.734–0.923) |
0.053 | 0.212 | 0.833 (0.726–0.904) |
0.007 | 0.042 | 0.829 (0.752–0.886) |
|||
| Axial | 0.867 (0.794–0.916) |
0.021 | 0.113 | 0.815 (0.692–0.896) |
0.002 | 0.027 | 0.909 (0.816–0.958) |
1.000 | 1.000 | 0.846 (0.771–0.900) |
|||
The accuracy, sensitivity, specificity, and F1 score of the MRNet and seven distinct TripleMRNet models in detecting ACL tears, along with their corresponding 95% confidence intervals. For accuracy, specificity, and sensitivity, we conducted a two-sided Fisher’s exact test to evaluate potential differences between the six (single- and dual-plane) models and the three-plane model of our approach. For these metrics, we report both unadjusted P values and adjusted q values. A q value <0.05 indicates statistical significance. ACL, anterior cruciate ligament; CI, confidence interval.
Table 3
| Model | Plane | Accuracy | Sensitivity | Specificity | F1 score | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (95% CI) | P value | q value | (95% CI) | P value | q value | (95% CI) | P value | q value | |||||
| MRNet | 3 planes | 0.725 (0.639–0.797) |
– | – | 0.710 (0.587–0.808) |
– | – | 0.741 (0.616–0.837) |
– | – | – | ||
| Our approach | 3 planes | 0.783 (0.702–0.848) |
– | – | 0.731 (0.598–0.832) |
– | – | 0.824 (0.716–0.896) |
– | – | 0.745 (0.660–0.815) |
||
| Sagittal + coronal | 0.675 (0.587–0.752) |
0.021 | 0.126 | 0.692 (0.557–0.801) |
0.612 | 0.734 | 0.662 (0.543–0.763) |
0.006 | 0.054 | 0.649 (0.560–0.728) |
|||
| Sagittal + axial | 0.750 (0.666–0.819) |
0.318 | 0.477 | 0.712 (0.577–0.817) |
0.754 | 0.820 | 0.779 (0.667–0.862) |
0.318 | 0.477 | 0.712 (0.625–0.785) |
|||
| Coronal + axial | 0.750 (0.666–0.819) |
0.318 | 0.477 | 0.712 (0.577–0.817) |
0.754 | 0.820 | 0.779 (0.667–0.862) |
0.318 | 0.477 | 0.712 (0.625–0.785) |
|||
| Sagittal | 0.633 (0.544–0.714) |
<0.001 | 0.018 | 0.750 (0.618–0.848) |
0.820 | 0.820 | 0.545 (0.426–0.657) |
<0.001 | 0.018 | 0.639 (0.550–0.720) |
|||
| Coronal | 0.750 (0.666–0.819) |
0.318 | 0.477 | 0.731 (0.598–0.832) |
0.984 | 0.984 | 0.765 (0.651–0.850) |
0.612 | 0.734 | 0.717 (0.631–0.790) |
|||
| Axial | 0.775 (0.692–0.841) |
0.820 | 0.820 | 0.731 (0.598–0.832) |
0.984 | 0.984 | 0.809 (0.670–0.885) |
0.820 | 0.820 | 0.738 (0.653–0.808) |
|||
The accuracy, sensitivity, specificity, and F1 score of the MRNet and seven distinct TripleMRNet models in detecting meniscus tears, along with their corresponding 95% confidence intervals. For accuracy, specificity, and sensitivity, we conducted a two-sided Fisher’s exact test to evaluate potential differences between the six (single- and dual-plane) models and the three-plane model of our approach. For these metrics, we report both unadjusted P values and adjusted q values. A q value <0.05 indicates statistical significance. CI, confidence interval.
Using these same hyperparameters, we trained and validated the submodels for the other six plane combinations, calculating the performance metrics (ACC, SEN, SPE, and F1 score) for each combination in both ACL and meniscal tear detection.
ACL tear classification
For ACL tear detection, we trained and validated seven submodels of TripleMRNet. The three-plane model achieved the highest performance in terms of ACC (0.925), SEN (0.944), SPE (0.909), and F1 score (0.919). The sagittal + axial model showed the second-best performance across all four metrics, closely matching the three-plane model. Compared to the three-plane model, the coronal + axial model exhibited significantly lower ACC (0.858, q value =0.042) and SEN (0.833, q value =0.036). The coronal model showed a significant decline in ACC (0.842, q value =0.018) and SPE (0.833, q value =0.042), whereas the axial model demonstrated a notable reduction in SEN (0.815, q value =0.027). Among the three single-plane models, the sagittal model outperformed the others (Table 2).
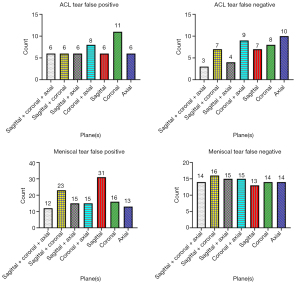
In terms of false positives, the coronal model produced the highest number (11), whereas the axial model produced the most false negatives (10) (Figure 3). Overall, the sagittal plane was the most effective for classifying ACL tears, and images from the axial plane complemented the sagittal plane. The coronal model, however, had the weakest performance and resulted in more false positives (Figure 4).
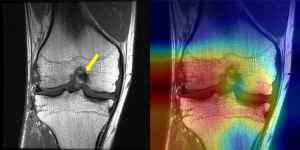
Meniscal tear classification
For meniscal tear detection, we trained and validated seven submodels of TripleMRNet and documented the optimal performance for each configuration. The sensitivity of these submodels showed little variation. The three-plane model achieved the best ACC (0.783), SPE (0.824), and F1 score (0.745). The axial model closely followed, surpassing the three two-plane models with an ACC of 0.775, SPE of 0.809, and F1 score of 0.738. In contrast, the sagittal model performed the worst, with a significant decline in both ACC (0.663, q value =0.018) and SPE (0.545, q value =0.018) compared to the three-plane model (Table 3).
In meniscal tear classification, the sagittal model and sagittal + coronal model had significantly higher false-positive counts (31 and 23, respectively) compared to the other models, which ranged from 12 to 16. The false negatives across all models were relatively consistent, ranging from 13 to 16 (Figure 3). Overall, sagittal images performed the worst, leading to a substantial number of incorrect results (Figure 5).
Discussion
The majority of existing DL studies on ACL and meniscal tear detection primarily focus on single-plane MRI analysis. We designed TripleMRNet, a DL model that selects 1–3 MRI planes (sagittal, coronal, axial) from seven possible combinations to diagnose the two injuries. Our TripleMRNet significantly modifies MRNet to meet our research objectives. The key improvements include the following: (I) avoiding transfer learning by training the network from scratch to prevent biases from non-knee MRI data; (II) not sharing parameters across planes, preserving independent feature extraction for each plane; and (III) adopting feature-level fusion instead of probability-level fusion.
In clinical diagnostics, selecting and examining MR images is time-consuming and labor-intensive for physicians (45,46). With AI’s rapid development, its integration with knee joint MRI has become a hot topic, focusing on classification, grading, and segmentation of knee injuries (16,18,22,29,36,47). Our study provides insights into the ability of the three MRI planes to assess these two types of knee injuries from the perspective of DL models.
Sagittal images are the most commonly used for diagnosing meniscal tears in clinical practice, followed by coronal images (9). However, in our model, axial images provided the best results, whereas sagittal images performed the worst. Existing studies often show lower classification performance for meniscal tears compared to ACL tears (20,48), which we hypothesize may be due to the lack of axial images in the datasets. Further research with larger datasets and standardized imaging protocols is needed to validate this hypothesis.
We generated Grad-CAM heatmaps for the model’s operation on the three-plane image slices, offering insights into its decision-making logic during classification. However, Grad-CAM reflects the model’s focus on local features in the final convolutional layer and does not explain deeper, abstract features. Although it highlights the regions the model prioritizes, it does not clarify their significance. Variations in MRI slice quantity and quality, along with the lack of lesion segmentation for simulating a clinical scenario, may limit Grad-CAM’s interpretability in this study.
TripleMRNet performed better in detecting ACL tears than it did for meniscal tears, with heatmaps for meniscal classification being more scattered and inconsistent. This difference may be partly due to dataset imbalances, despite our efforts to address it by adjusting the loss function with class weights based on the dataset’s class distribution.
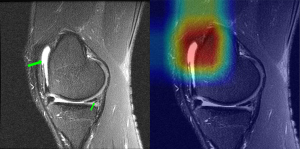
Our study has several limitations. First, several studies based on the MRNet dataset have employed more complex and advanced models, although their primary aim has generally been to improve classification performance (22,29,30,32,49). In future studies, we plan to use more advanced models to conduct similar research. Second, we observed that certain features in knee MR images, such as effusion, swelling, and bone contusions, could influence the focus areas of the model, leading to misdiagnoses of ACL and meniscal tears (Figure 6). Third, the MRNet dataset has certain limitations, primarily its small size and the fact that images from the three planes come from different sequences, which may introduce bias and limit the generalizability of the findings. For instance, the coronal images are T1-weighted, not T2-weighted, which may explain their poorer performance in our study. Despite these limitations, the MRNet dataset is the only publicly available resource for our research, providing 2D MR images across all three planes. In future research, collecting and compiling a more comprehensive and clinically relevant dataset will be a crucial task.
Conclusions
For ACL tears, the sagittal plane exhibited the highest diagnostic performance, whereas the coronal plane demonstrated the lowest. In contrast, for meniscal tears, the axial model achieved performance comparable to the three-plane model, whereas the sagittal model yielded the poorest results, leading to a notably higher false positive rate.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD+AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-1808/rc
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1808/coif). Q.L. reports that this work was supported by the Tianjin Natural Science Foundation (grant No. 22JCZDJC00220) and the Foundation of State Key Laboratory of Ultrasound in Medicine and Engineering (grant No. 2022KFKT004). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Culvenor AG, Girdwood MA, Juhl CB, Patterson BE, Haberfield MJ, Holm PM, Bricca A, Whittaker JL, Roos EM, Crossley KM. Rehabilitation after anterior cruciate ligament and meniscal injuries: a best-evidence synthesis of systematic reviews for the OPTIKNEE consensus. Br J Sports Med 2022;56:1445-53. [Crossref] [PubMed]
- Luo A, Gou S, Tong N, Liu B, Jiao L, Xu H, Wang Y, Ding T. Visual interpretable MRI fine grading of meniscus injury for intelligent assisted diagnosis and treatment. NPJ Digit Med 2024;7:97. [Crossref] [PubMed]
- Thrush C, Porter TJ, Devitt BM. No evidence for the most appropriate postoperative rehabilitation protocol following anterior cruciate ligament reconstruction with concomitant articular cartilage lesions: a systematic review. Knee Surg Sports Traumatol Arthrosc 2018;26:1065-73. [Crossref] [PubMed]
- Poulsen E, Goncalves GH, Bricca A, Roos EM, Thorlund JB, Juhl CB. Knee osteoarthritis risk is increased 4-6 fold after knee injury - a systematic review and meta-analysis. Br J Sports Med 2019;53:1454-63. [Crossref] [PubMed]
- Koch JEJ, Ben-Elyahu R, Khateeb B, Ringart M, Nyska M, Ohana N, Mann G, Hetsroni I. Accuracy measures of 1.5-tesla MRI for the diagnosis of ACL, meniscus and articular knee cartilage damage and characteristics of false negative lesions: a level III prognostic study. BMC Musculoskelet Disord 2021;22:124. [Crossref] [PubMed]
- De Smet AA, Mukherjee R. Clinical, MRI, and arthroscopic findings associated with failure to diagnose a lateral meniscal tear on knee MRI. AJR Am J Roentgenol 2008;190:22-6. [Crossref] [PubMed]
- Krakowski P, Nogalski A, Jurkiewicz A, Karpiński R, Maciejewski R, Jonak J. Comparison of Diagnostic Accuracy of Physical Examination and MRI in the Most Common Knee Injuries. Applied Sciences 2019;9:4102.
- Tarhan NC, Chung CB, Mohana-Borges AV, Hughes T, Resnick D. Meniscal tears: role of axial MRI alone and in combination with other imaging planes. AJR Am J Roentgenol 2004;183:9-15. [Crossref] [PubMed]
- Mancino F, Kayani B, Gabr A, Fontalis A, Plastow R, Haddad FS. Anterior cruciate ligament injuries in female athletes: risk factors and strategies for prevention. Bone Jt Open 2024;5:94-100. [Crossref] [PubMed]
- Choi SH, Bae S, Ji SK, Chang MJ. The MRI findings of meniscal root tear of the medial meniscus: emphasis on coronal, sagittal and axial images. Knee Surg Sports Traumatol Arthrosc 2012;20:2098-103. [Crossref] [PubMed]
- Bajaj S, Chhabra A, Taneja AK. 3D isotropic MRI of ankle: review of literature with comparison to 2D MRI. Skeletal Radiol 2024;53:825-46. [Crossref] [PubMed]
- Cheng J, Gao M, Liu J, Yue H, Kuang H, Liu J, Wang J. Multimodal Disentangled Variational Autoencoder With Game Theoretic Interpretability for Glioma Grading. IEEE J Biomed Health Inform 2022;26:673-84. [Crossref] [PubMed]
- Cho JH, Kim M, Nam HS, Park SY, Lee YS. Age and medial compartmental OA were important predictors of the lateral compartmental OA in the discoid lateral meniscus: Analysis using machine learning approach. Knee Surg Sports Traumatol Arthrosc 2024;32:1660-71. [Crossref] [PubMed]
- Verma PR, Bhandari AK. Role of Deep Learning in Classification of Brain MRI Images for Prediction of Disorders: A Survey of Emerging Trends. Archives of Computational Methods in Engineering 2023;30:4931-57.
- Wang J, Luo J, Liang J, Cao Y, Feng J, Tan L, Wang Z, Li J, Hounye AH, Hou M, He J. Lightweight Attentive Graph Neural Network with Conditional Random Field for Diagnosis of Anterior Cruciate Ligament Tear. J Imaging Inform Med 2024;37:688-705. [Crossref] [PubMed]
- Choi ES, Sim JA, Na YG, Seon JK, Shin HD. Machine-learning algorithm that can improve the diagnostic accuracy of septic arthritis of the knee. Knee Surg Sports Traumatol Arthrosc 2021;29:3142-8. [Crossref] [PubMed]
- Richardson ML, Ojeda PI A. "Bumper-Car" Curriculum for Teaching Deep Learning to Radiology Residents. Acad Radiol 2022;29:763-70. [Crossref] [PubMed]
- Eresen A. Diagnosis of meniscal tears through automated interpretation of medical reports via machine learning. Acad Radiol 2022;29:488-9. [Crossref] [PubMed]
- Guermazi A, Omoumi P, Tordjman M, Fritz J, Kijowski R, Regnard NE, Carrino J, Kahn CE Jr, Knoll F, Rueckert D, Roemer FW, Hayashi D, How AI. May Transform Musculoskeletal Imaging. Radiology 2024;310:e230764. [Crossref] [PubMed]
- Shetty ND, Dhande R, Unadkat BS, Parihar P. A Comprehensive Review on the Diagnosis of Knee Injury by Deep Learning-Based Magnetic Resonance Imaging. Cureus 2023;15:e45730. [Crossref] [PubMed]
- Andriollo L, Picchi A, Sangaletti R, Perticarini L, Rossi SMP, Logroscino G, Benazzo F. The Role of Artificial Intelligence in Anterior Cruciate Ligament Injuries: Current Concepts and Future Perspectives. Healthcare (Basel) 2024.
- Tran A, Lassalle L, Zille P, Guillin R, Pluot E, Adam C, Charachon M, Brat H, Wallaert M, d'Assignies G, Rizk B. Deep learning to detect anterior cruciate ligament tear on knee MRI: multi-continental external validation. Eur Radiol 2022;32:8394-403. [Crossref] [PubMed]
- Cheng Q, Lin H, Zhao J, Lu X, Wang Q. Application of machine learning-based multi-sequence MRI radiomics in diagnosing anterior cruciate ligament tears. J Orthop Surg Res 2024;19:99. [Crossref] [PubMed]
- Li MD, Deng F, Chang K, Kalpathy-Cramer J, Huang AJ. Automated Radiology-Arthroscopy Correlation of Knee Meniscal Tears Using Natural Language Processing Algorithms. Acad Radiol 2022;29:479-87. [Crossref] [PubMed]
- Li W, Xiao Z, Liu J, Feng J, Zhu D, Liao J, Yu W, Qian B, Chen X, Fang Y, Li S. Deep learning-assisted knee osteoarthritis automatic grading on plain radiographs: the value of multiview X-ray images and prior knowledge. Quant Imaging Med Surg 2023;13:3587-601. [Crossref] [PubMed]
- Hu J, Zheng C, Yu Q, Zhong L, Yu K, Chen Y, Wang Z, Zhang B, Dou Q, Zhang X. DeepKOA: a deep-learning model for predicting progression in knee osteoarthritis using multimodal magnetic resonance images from the osteoarthritis initiative. Quant Imaging Med Surg 2023;13:4852-66. [Crossref] [PubMed]
- Li J, Qian K, Liu J, Huang Z, Zhang Y, Zhao G, Wang H, Li M, Liang X, Zhou F, Yu X, Li L, Wang X, Yang X, Jiang Q. Identification and diagnosis of meniscus tear by magnetic resonance imaging using a deep learning model. J Orthop Translat 2022;34:91-101. [Crossref] [PubMed]
- Namiri NK, Flament I, Astuto B, Shah R, Tibrewala R, Caliva F, Link TM, Pedoia V, Majumdar S. Deep Learning for Hierarchical Severity Staging of Anterior Cruciate Ligament Injuries from MRI. Radiol Artif Intell 2020;2:e190207. [Crossref] [PubMed]
- Bien N, Rajpurkar P, Ball RL, Irvin J, Park A, Jones E, et al. Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 2018;15:e1002699. [Crossref] [PubMed]
- Yu C, Wang M, Chen S, Qiu C, Zhang Z, Zhang X. Improving anterior cruciate ligament tear detection and grading through efficient use of inter-slice information and simplified transformer module. Biomedical Signal Processing and Control 2023;86:105356.
- Kunze KN, Rossi DM, White GM, Karhade AV, Deng J, Williams BT, Chahla J. Diagnostic Performance of Artificial Intelligence for Detection of Anterior Cruciate Ligament and Meniscus Tears: A Systematic Review. Arthroscopy 2021;37:771-81. [Crossref] [PubMed]
- Smolle MA, Goetz C, Maurer D, Vielgut I, Novak M, Zier G, Leithner A, Nehrer S, Paixao T, Ljuhar R, Sadoghi P. Artificial intelligence-based computer-aided system for knee osteoarthritis assessment increases experienced orthopaedic surgeons’ agreement rate and accuracy. Knee Surg Sports Traumatol Arthrosc 2023;31:1053-62. [Crossref] [PubMed]
- Thakur U, Gulati V, Shah J, Tietze D, Chhabra A. Anterior cruciate ligament reconstruction related complications: 2D and 3D high-resolution magnetic resonance imaging evaluation. Skeletal Radiol 2022;51:1347-64. [Crossref] [PubMed]
- Cheraya G, Chhabra A. Cruciate and Collateral Ligaments: 2-Dimensional and 3-Dimensional MR Imaging-Aid to Knee Preservation Surgery. Semin Ultrasound CT MR 2023;44:271-91. [Crossref] [PubMed]
- Mangone M, Diko A, Giuliani L, Agostini F, Paoloni M, Bernetti A, Santilli G, Conti M, Savina A, Iudicelli G, Ottonello C, Santilli V. A Machine Learning Approach for Knee Injury Detection from Magnetic Resonance Imaging. Int J Environ Res Public Health 2023;20:6059. [Crossref] [PubMed]
- Kara A, Hardalaç F. Detection and Classification of Knee Injuries from MR Images Using the MRNet Dataset with Progressively Operating Deep Learning Methods. Machine Learning and Knowledge Extraction 2021;3:1009-29.
- M MM. T R M, V VK, Guluwadi S. Enhancing brain tumor detection in MRI images through explainable AI using Grad-CAM with Resnet 50. BMC Med Imaging 2024;24:107. [Crossref] [PubMed]
- Rimal Y, Sharma N, Alsadoon A. The accuracy of machine learning models relies on hyperparameter tuning: student result classification using random forest, randomized search, grid search, bayesian, genetic, and optuna algorithms. Multimed Tools Appl 2024;83:74349-64.
- Makino T, Jastrzębski S, Oleszkiewicz W, Chacko C, Ehrenpreis R, Samreen N, et al. Differences between human and machine perception in medical diagnosis. Sci Rep 2022;12:6877. [Crossref] [PubMed]
- Jeon Y, Yoshino K, Hagiwara S, Watanabe A, Quek ST, Yoshioka H, Feng M. Interpretable and Lightweight 3-D Deep Learning Model for Automated ACL Diagnosis. IEEE J Biomed Health Inform 2021;25:2388-97. [Crossref] [PubMed]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. International Journal of Computer Vision 2020;128:336-59.
- Cheng CT, Ho TY, Lee TY, Chang CC, Chou CC, Chen CC, Chung IF, Liao CH. Application of a deep learning algorithm for detection and visualization of hip fractures on plain pelvic radiographs. Eur Radiol 2019;29:5469-77. [Crossref] [PubMed]
- Alshazly H, Linse C, Barth E, Martinetz T. Explainable COVID-19 Detection Using Chest CT Scans and Deep Learning. Sensors (Basel) 2021.
- Schiratti JB, Dubois R, Herent P, Cahané D, Dachary J, Clozel T, Wainrib G, Keime-Guibert F, Lalande A, Pueyo M, Guillier R, Gabarroca C, Moingeon P. A deep learning method for predicting knee osteoarthritis radiographic progression from MRI. Arthritis Res Ther 2021;23:262. [Crossref] [PubMed]
- Lecouvet F, Van Haver T, Acid S, Perlepe V, Kirchgesner T, Vande Berg B, Triqueneaux P, Denis ML, Thienpont E, Malghem J. Magnetic resonance imaging (MRI) of the knee: Identification of difficult-to-diagnose meniscal lesions. Diagn Interv Imaging 2018;99:55-64. [Crossref] [PubMed]
- Ahn JH, Jeong SH, Kang HW. Risk Factors of False-Negative Magnetic Resonance Imaging Diagnosis for Meniscal Tear Associated With Anterior Cruciate Ligament Tear. Arthroscopy 2016;32:1147-54. [Crossref] [PubMed]
- Wang Q, Yao M, Song X, Liu Y, Xing X, Chen Y, Zhao F, Liu K, Cheng X, Jiang S, Lang N. Automated Segmentation and Classification of Knee Synovitis Based on MRI Using Deep Learning. Acad Radiol 2024;31:1518-27. [Crossref] [PubMed]
- Santomartino SM, Kung J, Yi PH. Systematic review of artificial intelligence development and evaluation for MRI diagnosis of knee ligament or meniscus tears. Skeletal Radiol 2024;53:445-54. [Crossref] [PubMed]
- Xue Y, Yang S, Sun W, Tan H, Lin K, Peng L, Wang Z, Zhang J. Approaching expert-level accuracy for differentiating ACL tear types on MRI with deep learning. Sci Rep 2024;14:938. [Crossref] [PubMed]


