
AttmNet: a hybrid Transformer integrating self-attention, Mamba, and multi-layer convolution for enhanced lesion segmentation
Introduction
Breast and skin cancers are among the most significant global health challenges. Breast cancer, in particular, is the most common cancer type worldwide, with notably high incidence rates (1). Ultrasound imaging is a key diagnostic tool for breast cancer due to its non-radiative nature, cost-effectiveness, and rapid imaging. However, challenges such as speckle noise, shadow artifacts, and irregular lesion shapes in ultrasound images make precise lesion segmentation difficult. Similarly, dermoscopy, widely used for skin cancer diagnosis, faces challenges such as hair occlusion and uneven lighting, which add complexity to the segmentation process (2). In addition, the aftermath of the coronavirus disease 2019 (COVID-19) pandemic has introduced new challenges in medical image analysis, particularly for lung infection detection in computed tomography (CT) images (3). Accurate and efficient lesion segmentation is critical for computer-aided diagnosis, significantly improving the accuracy of early diagnosis and treatment planning for both cancer and infectious diseases.
In recent years, convolutional neural networks (CNNs) have achieved notable success in medical image segmentation. A major challenge in this domain is integrating local features with global dependencies. The development of UNet (4) marked a pivotal advancement, with its encoder–decoder architecture effectively capturing contextual and localization information for outstanding segmentation performance. UNet++ (5), inspired by DenseNet (6), introduced dense connectivity, replacing conventional skip connections with dense convolutions to enhance feature depth and diversity. A CNN for Efficient Ultrasound Image Segmentation (EUIS) Net (7) incorporated channel and spatial attention mechanisms in its bottleneck to extract critical contextual features, whereas LeaNet (8) utilized Dilated Efficient Channel Attention and Inverse External Attention modules for efficient global and local feature extraction. For breast ultrasound segmentation, pyramid attention network combining attention mechanism and multi-scale features (AMS-PAN) (9) employed depthwise separable convolutions to construct a multi-scale feature pyramid, while its Global Attention Upsampling module effectively captured edge and texture details in the decoder. Despite these advancements, CNN-based architectures are inherently limited by their constrained receptive fields, which hinder the capture of long-range dependencies and global context. These limitations often result in segmentation errors, such as misclassifying normal tissues resembling lesions, as illustrated in Figure 1.

Although CNNs excel in local feature extraction, Transformers have gained attention for their ability to model global context and long-range dependencies (10). For example, Chen et al. (11) proposed TransUNet, which integrates a Transformer encoder with UNet to enhance global context capture and restore local spatial details. Similarly, Swin-UNet (12), a pure Transformer-based model, leverages the Swin Transformer (13) with shifted windows to achieve enhanced global feature modeling. However, Transformer-based models typically require large-scale datasets to fully realize their potential, which is a challenge in medical imaging due to the high cost of annotation. Additionally, the quadratic complexity of the self-attention mechanism poses significant computational challenges when processing long sequences, limiting their applicability in medical image segmentation.
Mamba (14), based on state-space models (SSM), offers an alternative for efficient long-range dependency modeling. By leveraging a hardware-aware selection mechanism, Mamba improves computational efficiency and scalability. Architectures such as SegMamba (15) use multi-scale tri-directional spatial Mamba modules to efficiently capture global context, outperforming Transformer-based models in some scenarios. Swin-UMamba (16) combines a Swin Transformer with Mamba to enhance segmentation accuracy, whereas U-Mamba (17) merges CNNs and SSM to integrate local feature extraction and long-range dependency modeling. Vision Mamba (18) introduces bidirectional SSM for global visual context modeling, incorporating positional embeddings to boost spatial awareness and achieve more accurate visual understanding. nnMamba (19) further innovates with the Mamba-InConvolution module, incorporating multi-channel spatial coupled learning to advance medical image segmentation performance. Despite these advances, Mamba is less effective than CNNs in capturing fine local details, emphasizing the need for hybrid approaches that combine the strengths of different techniques to overcome their individual limitations.
To tackle these challenges, we propose AttmNet, a hybrid Transformer network specifically designed for medical image segmentation. At its core, AttmNet incorporates the MAM (Multiscale-Convolution, Self-Attention, and Mamba) block in its encoder, which synergistically integrates multi-layer convolution (MLC) for local feature learning, self-attention for global context modeling, and Mamba for efficient long-range dependency capture. This design enables precise pixel-wise interaction and balances local and global feature extraction. The decoder employs bilinear interpolation for upsampling, reducing model complexity without sacrificing accuracy. AttmNet was evaluated against six advanced segmentation methods on four public datasets, demonstrating superior segmentation performance. The main contributions of this work are as follows:
- AttmNet is an innovative medical image segmentation network that integrates CNN, self-attention, and Mamba, consistently outperforming state-of-the-art segmentation models on multiple public datasets.
- A carefully designed MLC module achieves multi-scale receptive field fusion, enhancing the network’s nonlinearity and feature extraction capacity, leading to significant improvements in segmentation accuracy.
- The Transformer-based MAM block combines the MLC with Att-Mamba, effectively capturing long-range pixel interactions for more accurate and comprehensive segmentation outcomes.
The remainder of the manuscript is organized as follows: Methods section outlines the methodology for AttmNet; Results section presents the experimental findings. Finally, the main conclusions, limitations, and future directions of the study are discussed in Discussion and Conclusions section. We present this article in accordance with the TRIPOD+AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-2024-2561/rc).
Methods
The AttmNet architecture, illustrated in Figure 2, consists of three main components: the encoder, decoder, and skip connections. The encoder leverages CNNs, attention mechanisms, and Mamba modules to adaptively highlight critical regions, enhancing feature representation. The decoder uses linear interpolation for up-sampling, offering a lightweight alternative to transposed convolutions. Skip connections preserve fine-grained details from lower layers, enabling multi-level feature fusion and improving the model’s accuracy and robustness.
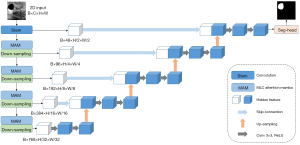
Encoder
As illustrated in Figure 2, the encoder structure of AttmNet comprises a Stem layer and multiple MAM blocks, which collectively serve as the foundation for feature extraction. The Stem layer applies a 7×7 convolution (padding of 3, stride of 2) for initial feature extraction. Given a two-dimensional (2D) input image , where C is the number of input channels, and H and W represent the image height and width, the features extracted by the Stem layer are denoted as . This initial feature map l0 is then sequentially fed into each MAM block, along with its respective down-sampling layer.
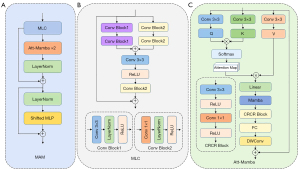
The MAM block, a novel Transformer structure introduced in this work, combines the strengths of the MLC and Att-Mamba modules for robust feature extraction. The MLC module leverages MLC, normalization, and residual connections to capture local features effectively. Meanwhile, the Att-Mamba module integrates a multi-head self-attention mechanism alongside Mamba to capture global information, with depth-wise convolution components further enhancing local detail extraction. Residual connections are incorporated throughout to maintain continuity in the information flow. To ensure optimal efficiency and feature extraction capability, the MAM block incorporates two Att-Mamba modules, which balances computational cost and performance.
Next, we will detail the MAM block’s design. As illustrated in Figure 3A, the computation process for the m-th MAM block is as follows:
where . In the above formulas, MLC, AttM, LN, and SMLP represent the MLC module, Att-Mamba module, layer normalization, and shifted multilayer perceptron, respectively. The specific structure and functions of the MLC, Att-Mamba, and SMLP modules will be explained in detail in the following sections.
MLC module
As shown in Figure 3B, the MLC module consists of two convolutional branches. The first branch uses two 3×3 convolution blocks, each composed of a 3×3 convolution (with padding of 1 and stride of 1), normalization, and a rectified linear unit (ReLU) activation function to capture rich local features. The second branch utilizes two 1×1 convolution blocks to adjust the channel dimensions, enhancing the module’s feature fusion capability. Each of these blocks consists of a 1×1 convolution, normalization, and ReLU activation function. The outputs of both branches are combined through an addition operation, followed by a 3×3 convolution (with padding of 1 and stride of 1) and a ReLU activation function to achieve multi-scale feature fusion. A final 1×1 convolution block adjusts the channel dimensions, compressing information and reducing computational cost. This multi-layer convolutional structure significantly enhances the module’s ability to capture local information. Finally, a skip connection adds the input features back to the output, preserving the original information and improving continuity.
Att-Mamba module
The structure of the Att-Mamba module is shown in Figure 3C. Suppose the input to Att-Mamba is , where B is the batch size, C is the number of channels, and H and C are the height and width of the feature map. First, the input undergoes initial feature processing through a 3×3 convolution (padding of 1, stride of 1). The resulting output is flattened into a 2D patch and normalized with LayerNorm, producing .
Next, is projected into embedding spaces for queries, keys, and values, denoted as . These embeddings are reshaped to to enable parallel computation in the multi-head attention mechanism. In each head, Q is multiplied by the transpose of K, followed by Softmax normalization to yield the context similarity matrix . The matrix P is then used to extract global contextual information from V, defining the attention output as follows:
where, represents the embedding dimension for each attention head, and is the number of attention heads. For optimal performance and computational feasibility, we set , which ensures efficient parallel computation while maintaining high feature representation quality. Following the attention mechanism, the output is processed through a linear layer and then fed into the Mamba module for further enhancement of the attention features. The integration of self-attention and Mamba is designed to leverage the global context captured by self-attention and refine it using Mamba’s structured SSM. Specifically, self-attention captures long-range dependencies, while Mamba introduces temporal dynamics and local interactions, enhancing feature representation robustness.
Mamba, based on a SSM, assumes that the dynamic system’s predictions are driven by a state equation and an output equation, defined as (14):
where A is the state matrix, B is the input matrix, and D is the output matrix. To convert the continuous-time SSM into discrete-time, we apply a zero-order hold for discretization, yielding:
With these discrete parameters, the SSM can be computed either through linear recursion:
or through convolution:
where represents the structured convolution kernel, L is the length of the input sequence u, and * denotes the convolution operation. In this work, we utilize the linear recursion method for computation, which we refer to as .
The Mamba block in this model employs the tri-directional spatial Mamba mechanism from SegMamba (15). Here, 2D input features are flattened into three sequences—processed in forward, reverse, and slice-wise directions—allowing for feature interactions that generate fused 2D features:
where f,r, and s denote forward, reverse, and slice-wise directions, respectively. The Mamba module output is then reshaped to (B,C,H,W) and passed through a Conv-ReLU-Conv-ReLU (CRCR) module, which consists of a 3×3 convolution (padding of 1, stride of 1), ReLU activation, a 1×1 convolution and ReLU activation:
where ϭ denotes the ReLU activation function. The output is further processed by a fully connected (FC) layer and a depth-wise convolution (DWConv) with a 3×3 kernel, stride of 1, and padding of 1 to capture contextual information, enhancing feature representation and spatial information integration:
Shifted multilayer perceptron (MLP) module
The Shifted MLP, initially introduced in UNeXt (20), enhances the model’s focus on specific spatial features by shifting the convolutional feature channels along predefined axes before tokenization. This shift significantly improves the model’s ability to capture fine image structures compared to traditional MLPs. By incorporating these shifts, the Shifted MLP improves local feature awareness and integrates it with a window-based attention mechanism, enabling the effective fusion of local and global information. This design increases flexibility in handling complex image data.
Specifically, the Shifted MLP shifts features along the width in one MLP and along the height in another, dividing them into h partitions and shifting by i positions along the specified axis to create shifted windows. This design improves computational efficiency and speeds up training and inference by processing features independently within each partition. The operation steps are as follows:
where, and denote shifts across the width and height, respectively; and are the features after shifting along width and height, respectively; denotes features that have been tokenized based on features translated horizontally by width and denotes features that have been tokenized based on features translated horizontally by height; stands for multi-layer perceptron, and is an activation function based on the Gaussian error function.
Decoder and skip connections
The AttmNet decoder, based on the classic UNet (4) (illustrated in Figure 2), is designed to progressively upsample image resolution while capturing fine details. Using bilinear interpolation, it doubles the spatial dimensions of the feature map and halves the channel count. This upsampled feature map is then concatenated along the channel dimension with skip connection features from corresponding encoder layers, preserving low-level details and enabling fine-grained information fusion for high-quality reconstruction. Next, the concatenated feature map undergoes refinement through two 3×3 convolutional layers (stride 1, padding 1) with ReLU activation functions. This upsampling process repeats four times, ultimately restoring the feature map to the original input image size and producing the final segmentation prediction.
Results
Datasets
To evaluate the effectiveness and generalizability of our proposed network, we utilized four diverse medical imaging datasets. The first dataset, the breast ultrasound (BUS) dataset (21), was obtained from the UDIAT Diagnostic Center at Parc Taulí Corporation in Sabadell (https://helward.mmu.ac.uk/STAFF/m.yap/dataset.php). It consists of 163 ultrasound images with an average resolution of 760×570 pixels, collected from various patients. This dataset includes 53 malignant tumor cases and 110 benign tumor cases.
The second dataset, the breast ultrasound images (BUSI) dataset (22), was obtained from Baheya Women’s Hospital for Early Detection and Treatment of Cancer (https://scholar.cu.edu.eg/?q=afahmy/pages/dataset). Acquired using LOGIQ E9 ultrasound system and LOGIQ E9 Agile ultrasound systems (GE Healthcare, Chicago, IL, USA), the dataset includes 780 images from 600 female patients aged 25–75 years, with an average resolution of 500×500 pixels. It consists of 437 benign cases, 210 malignant masses, and 133 normal cases. Since this study focuses on lesion segmentation in BUS images, we excluded normal cases from the BUSI dataset to maintain dataset specificity.
The third dataset, the PH2 Dermoscopy Image dataset (23), was collected by the Dermatology Department at Pedro Hispano Hospital using the Tuebinger Mole Analyzer system (http://www2.fc.up.pt/addi). It contains 200 high-resolution color images (768×560 pixels) representing a variety of melanocytic lesions, including 80 common nevi, 80 atypical nevi, and 40 melanomas. Each image is accompanied by pixel-level expert annotations for segmentation tasks and ground-truth diagnoses for classification tasks.
The fourth dataset, the COVID-19 lung CT dataset, was created to address the scarcity of public datasets for infection segmentation in COVID-19 lung CT images (https://drive.google.com/file/d/1FHx0Cqkq9iYjEMN3Ldm9FnZ4Vr1u3p-j/view). To ensure sufficient training samples, the study processed public datasets by slicing and applying uniform sampling, generating 1,277 high-quality CT images (24). These were divided into 894 training images and 383 testing images, all with a resolution of 512×512 pixels.
Experimental details
The experiments in this study were conducted using the PyTorch framework on an NVIDIA GeForce RTX 4090D GPU (NVIDIA, Santa Clara, CA, USA). Model training was performed with the Adam optimizer (25), using a batch size of 8 over 300 epochs. The initial learning rate was set to 0.0001, with a weight decay of 0.0001. Input images were standardized to a size of 128×128 for consistency across training and evaluation. Each dataset was randomly split into 70% for training, 10% for validation, and 20% for testing. However, the COVID-19 Lung dataset followed its original predefined splits for the training and testing sets. To increase the diversity of training data and improve the model’s generalization capability, data augmentation techniques were applied to the training data, including random 90-degree rotations and random flipping.
We employed BceDiceLoss as the loss function, which integrates binary cross-entropy (BCE) loss and Dice loss. This combination aims to balance classification precision and region similarity within segmentation tasks. BCE loss evaluates prediction errors against ground truth labels, whereas Dice loss measures the overlap between predicted and actual segmented regions, enhancing both completeness and consistency in segmentation. The loss function is defined as follows:
where, represents the total number of samples, and denote the true label and predicted value, respectively, and and represent the predicted and ground truth segmentation images. The weight parameters and are both set to 1 by default.
Evaluation metrics
We used six widely adopted metrics to quantitatively evaluate segmentation performance: Dice coefficient, intersection over union (IoU), recall, precision, specificity, and the 95th percentile of the Hausdorff distance (HD95). These metrics are defined as follows:
where TP, FP, FN, and TN represent true positives, false positives, false negatives, and true negatives, respectively, and d(M,N) is the Euclidean distance between points in sets M and N.
The Dice coefficient evaluates the overlap between the predicted and ground truth segmentations, making it ideal for assessing segmentation similarity. IoU measures the degree of overlap and provides a stricter assessment for smaller lesions. Recall indicates the proportion of correctly identified positive pixels, whereas precision assesses the accuracy of positive predictions. Specificity measures the accuracy of identifying negative pixels. HD95 quantifies the error between predicted and true boundaries by capturing the distance within which 95% of boundary errors fall, effectively disregarding the most extreme 5% of outliers. Together, these metrics offer a comprehensive assessment of segmentation accuracy and boundary alignment.
Comparative experiments
We compared AttmNet with several deep learning segmentation models, including UNet (4), TransUNet (11), Attention U-Net (26), nnUNet (27), SegMamba (15), and Swin-UMamba (16). Tables 1-4 summarize the segmentation results for these methods on the BUS, BUSI, PH2, and COVID-19 Lung datasets, respectively. As shown in Table 1, AttmNet outperformed the next best method on the BUS dataset, with a 3.38% improvement in IoU and a 4.54% increase in Dice coefficient. Additionally, its superior HD95 indicates better boundary accuracy. On the BUSI dataset (Table 2), AttmNet’s IoU and Dice coefficients were 1.17% and 3.21% higher than the nearest competitor, respectively. In the PH2 dataset (Table 3), AttmNet outperformed the next best model by 0.25% and 0.25% in IoU and Dice, respectively. Although Swin-UMamba, which combines Swin Transformer’s global context with Mamba’s efficient local feature extraction, performed well on BUS and PH2, AttmNet consistently surpassed all models across datasets, demonstrating its robustness and effectiveness in medical image segmentation. Additionally, on the larger COVID-19 Lung dataset, AttmNet maintained strong performance, achieving IoU and Dice scores slightly higher than those of the next best models, SegMamba and TransUNet (Table 4).
Table 1
| BUS | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| UNet | 0.6702 | 0.7992 | 7.2604 | 0.9884 | 0.8523 | 0.7580 |
| TransUNet | 0.6372 | 0.7362 | 12.6563 | 0.9935 | 0.7410 | 0.7811 |
| Attention U-Net | 0.6641 | 0.7922 | 8.3672 | 0.9835 | 0.9172* | 0.7035 |
| nnUNet | 0.7401 | 0.8233 | 10.8513 | 0.9949 | 0.8428 | 0.8423 |
| SegMamba | 0.7139 | 0.8157 | 10.4181 | 0.9901 | 0.8998 | 0.7740 |
| Swin-UMamba | 0.7524 | 0.8343 | 8.7667 | 0.9945 | 0.8493 | 0.8471 |
| AttmNet | 0.7862* | 0.8797* | 2.6909* | 0.9960* | 0.8620 | 0.9018* |
↑ indicates that higher values are better, while ↓ indicates that lower values are better. The best results are marked by an asterisk (*). BUS, breast ultrasound; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union.
Table 2
| BUSI | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| UNet | 0.6532 | 0.7810 | 5.7400 | 0.9898* | 0.7158 | 0.8844* |
| TransUNet | 0.6009 | 0.7127 | 13.9833 | 0.9797 | 0.7059 | 0.7928 |
| Attention U-Net | 0.6520 | 0.7804 | 4.7203 | 0.9827 | 0.7473 | 0.8348 |
| nnUNet | 0.6755 | 0.7612 | 14.4165 | 0.9790 | 0.7853 | 0.7994 |
| SegMamba | 0.6327 | 0.7660 | 5.0918 | 0.9870 | 0.7073 | 0.8559 |
| Swin-UMamba | 0.6808 | 0.7665 | 14.9387 | 0.9804 | 0.7878* | 0.7885 |
| AttmNet | 0.6925* | 0.8131* | 3.9809* | 0.9840 | 0.7873 | 0.8542 |
↑ indicates that higher values are better, while ↓ indicates that lower values are better. The best results are marked by an asterisk (*). BUSI, breast ultrasound images; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union.
Table 3
| PH2 | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| UNet | 0.8376 | 0.9112 | 1.1657 | 0.9636 | 0.9152 | 0.9121 |
| TransUNet | 0.8648 | 0.9261 | 8.7611 | 0.9372 | 0.9378 | 0.9226 |
| Attention U-Net | 0.8233 | 0.9027 | 1.2000 | 0.9666 | 0.8942 | 0.9153 |
| nnUNet | 0.8920 | 0.9413 | 8.2434 | 0.9727* | 0.9533 | 0.9369 |
| SegMamba | 0.8722 | 0.9315 | 0.8000 | 0.9726 | 0.9328 | 0.9314 |
| Swin-UMamba | 0.8930 | 0.9422 | 7.8832 | 0.9725 | 0.9529 | 0.9380* |
| AttmNet | 0.8955* | 0.9447* | 0.6000* | 0.9717 | 0.9578* | 0.9325 |
↑ indicates that higher values are better, while ↓ indicates that lower values are better. The best results are marked by an asterisk (*). HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union.
Table 4
| COVID-19 lung | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| UNet | 0.6197 | 0.7371 | 4.8813 | 0.9951 | 0.7949 | 0.7239 |
| TransUNet | 0.6723 | 0.7474 | 47.2564 | 0.9965 | 0.7615 | 0.7839 |
| Attention U-Net | 0.6593 | 0.7701 | 3.2289 | 0.9956 | 0.8110* | 0.7716 |
| nnUNet | 0.6119 | 0.6785 | 256.4715 | 0.9936 | 0.7047 | 0.6940 |
| SegMamba | 0.6629 | 0.7716 | 3.0006* | 0.9972* | 0.7641 | 0.8025* |
| Swin-UMamba | 0.6360 | 0.7014 | 251.2436 | 0.9950 | 0.7257 | 0.7161 |
| AttmNet | 0.6724* | 0.7798* | 3.3804 | 0.9957 | 0.7928 | 0.7959 |
↑ indicates that higher values are better, while ↓ indicates that lower values are better. The best results are marked by an asterisk (*). COVID-19, coronavirus disease 2019; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union.
Figure 4 illustrates segmentation outputs across the four datasets. Rows 1 and 2 represent examples from the BUS dataset, rows 3 and 4 from the BUSI dataset, rows 5 and 6 from the PH2 dataset, and rows 7 and 8 from the COVID-19 Lung dataset. In the BUS dataset, traditional CNN-based methods (such as UNet and Attention U-Net) often fail to capture fine details or mistakenly segment non-lesion regions due to their limited ability to model global context. Although Swin-UMamba demonstrated improved performance in capturing both local and global dependencies, its accuracy in delineating complex boundaries was constrained by restricted inter-window information transfer. For the BUSI dataset (rows 3 and 4), all models performed well when lesions exhibited clear boundaries. In the PH2 dataset (rows 5 and 6), the larger and more diffuse lesions required long-range dependency modeling for accurate segmentation, which traditional CNNs struggle to achieve. In the COVID-19 Lung dataset, most methods, except for AttmNet and TransUNet, produced poor segmentation results, with significant under-segmentation observed across the other approaches.
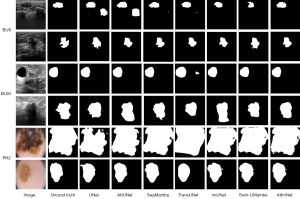
Overall, traditional CNN models, such as UNet and Attention U-Net, often struggle with accurately segmenting lesion boundaries due to their inability to capture long-range dependencies. Although TransUNet introduces Transformers to address this limitation, its segmentation performance remained suboptimal. Swin-UMamba improved multi-scale feature representation but faced difficulties in accurately delineating complex boundaries. AttmNet leverages a synergistic integration of complementary modules to address these challenges effectively, delivering superior segmentation accuracy compared to the other methods (Table 5).
Table 5
| Method | Params (M) | FLOPs (G) | BUS | BUSI | PH2 | COVID-19 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IoU ↑ | Dice ↑ | IoU ↑ | Dice ↑ | IoU ↑ | Dice ↑ | IoU ↑ | Dice ↑ | ||||||
| UNet | 31.04 | 109.47 | 0.6702 | 0.7992 | 0.6532 | 0.7810 | 0.8376 | 0.9112 | 0.6197 | 0.7371 | |||
| TransUNet | 93.23 | 64.43 | 0.6372 | 0.7362 | 0.6009 | 0.7127 | 0.8648 | 0.9261 | 0.6723 | 0.7474 | |||
| Attention U-Net | 34.87 | 133.57 | 0.6641 | 0.7922 | 0.6520 | 0.7804 | 0.8233 | 0.9027 | 0.6593 | 0.7701 | |||
| nnUNet | 143.87 | 209.53 | 0.7401 | 0.8233 | 0.6755 | 0.7612 | 0.8920 | 0.9413 | 0.6119 | 0.6785 | |||
| SegMamba | 23.38 | 78.11 | 0.7139 | 0.8157 | 0.6327 | 0.7660 | 0.8722 | 0.9315 | 0.6629 | 0.7716 | |||
| Swin-UMamba | 55.06 | 351.47 | 0.7524 | 0.8343 | 0.6808 | 0.7665 | 0.8930 | 0.9422 | 0.6360 | 0.7014 | |||
| AttmNet | 28.47 | 87.09 | 0.7862* | 0.8797* | 0.6925* | 0.8131* | 0.8955* | 0.9447* | 0.6724* | 0.7798* | |||
↑ indicates that higher values are better. The best results are marked by an asterisk (*). BUS, breast ultrasound; BUSI, breast ultrasound images; COVID-19, coronavirus disease 2019; FLOP, floating point operations per second; IoU, Intersection over Union; M, millions.
Ablation study
This ablation study, conducted on the BUS dataset, evaluated the effectiveness of key components in AttmNet, including the MLC module, Self-Attention module, Mamba module, and the combined Att-Mamba module. Table 6 compares segmentation performance with various component configurations. The first row shows baseline network results, whereas the second row presents the impact of adding the MLC module. As a foundational feature extraction component, MLC captures local spatial features and refines low-level details through MLCs. With MLC, IoU increased by 12.91%, Dice by 10.80%, and HD95 improved from 4.1331 to 2.7974, underscoring its importance in enhancing segmentation accuracy.
Table 6
| Model configuration | MLC | Self-attention | Mamba | IoU ↑ | Dice ↑ | HD95 ↓ | Params (M) | FLOPs (G) |
|---|---|---|---|---|---|---|---|---|
| 1 | × | × | × | 0.5985 | 0.7310 | 4.1331 | 22.97 | 73.87 |
| 2 | √ | × | × | 0.7276 | 0.8390 | 2.7974 | 26.90 | 82.96 |
| 3 | × | √ | √ | 0.6666 | 0.7931 | 3.2547 | 24.55 | 78.01 |
| 4 | √ | × | √ | 0.7338 | 0.8412 | 4.5456 | 27.29 | 82.96 |
| 5 | √ | √ | × | 0.7516 | 0.8543 | 3.8194 | 28.08 | 87.09 |
| 6 | √ | √ | √ | 0.7863* | 0.8797* | 2.6909* | 28.47 | 87.09 |
↑ indicates that higher values are better, whereas ↓ indicates that lower values are better. The best results are marked by an asterisk (*). BUS, breast ultrasound; FLOP, floating point operations per second; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union; MLC, multi-layer convolution; M, millions.
The third row in Table 6 illustrates the impact of the Att-Mamba module. The self-attention module broadens the model’s receptive field, effectively capturing global context and managing complex backgrounds, whrease the Mamba module enhances multi-scale feature extraction. By integrating self-attention with Mamba, the Att-Mamba module significantly improves segmentation accuracy compared to the baseline model. Additionally, the combination of MLC with the Mamba module (fourth row) and MLC with the self-attention module (fifth row) shows substantial improvements over the baseline in segmentation performance. This improvement can be attributed to MLC’s focus on local feature extraction, which, when combined with the global context modeling of self-attention and Mamba, results in superior segmentation outcomes. In the final row, the integration of MLC, self-attention, and Mamba modules allows the model to effectively handle complex shapes and irregular boundaries, achieving optimal segmentation performance with minimal increases in parameters and computational cost. As a result, AttmNet achieved IoU, Dice, and HD95 scores of 0.7863, 0.8797, and 2.6909, respectively, outperforming all other configurations.
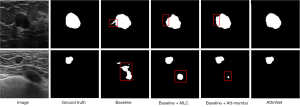
Figure 5 presents a visual comparison of the segmentation results for the baseline, baseline + MLC, baseline + Att-Mamba, and AttmNet on two randomly selected subjects from the BUS dataset. The results show that both Baseline + Att-Mamba and Baseline + MLC effectively improve the performance of the baseline network. However, they still exhibit limitations in boundary precision. In contrast, AttmNet, which combines MLC, self-attention, and Mamba modules, provides the most precise boundary delineation, achieving the highest overall segmentation performance.
Tables 7,8 present the results of the ablation study investigating the impact of the number of heads (num_heads) in Att-Mamba (Table 7) and the number of Att-Mamba layers in the MAM Block (Table 8). Due to GPU memory constraints, the evaluation was limited to configurations where these parameters were set to 1 and 2. The results demonstrate that the model achieved superior performance on the BUS dataset when the num_heads are set to 2 compared to 1. Similarly, the model exhibited better performance when the number of Att-Mamba layers is set to 2 rather than 1.
Table 7
| num_heads | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| 1 | 0.7410 | 0.8397 | 3.9081 | 0.9951 | 0.8186 | 0.8684 |
| 2 | 0.7862* | 0.8797* | 2.6909* | 0.9960* | 0.8620* | 0.9018* |
↑ indicates that higher values are better, whereas ↓ indicates that lower values are better. The best results are marked by an asterisk (*). BUS, breast ultrasound; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union.
Table 8
| Number of Att-Mamba layers | IoU ↑ | Dice ↑ | HD95 ↓ | Specificity ↑ | Recall ↑ | Precision ↑ |
|---|---|---|---|---|---|---|
| 1 | 0.7574 | 0.8563 | 3.5990 | 0.9958 | 0.8258 | 0.8915 |
| 2 | 0.7862* | 0.8797* | 2.6909* | 0.9960* | 0.8620* | 0.9018* |
↑ indicates that higher values are better, whereas ↓ indicates that lower values are better. The best results are marked by an asterisk (*). BUS, breast ultrasound; HD95, the 95th percentile of the Hausdorff distance; IoU, Intersection over Union; MAM, Multiscale-Convolution, Self-Attention and Mamba.
Computational efficiency
As shown in Table 9, the parameter size of our model is 28.47 M, ranking second only to SegMamba’s 23.38 M. Furthermore, the floating point operations per second (FLOPs) of our model are 87.09 G, significantly lower than those of UNet (109.47 G), Attention U-Net (133.57 G), nnUNet (209.53 G), and Swin-UMamba (351.47 G). These results indicate that our model is relatively lightweight, reducing storage and computational resource requirements effectively. Regarding memory usage, our model requires 485.64 MB during training, ranking third, slightly higher than SegMamba’s 404.83 MB and UNet’s 474.18 MB. For inference, the memory usage is 121.78 MB, also placing third, behind SegMamba’s 101.58 MB and UNet’s 118.92 MB. These findings highlight the memory efficiency of our model, particularly in comparison to TransUNet, which requires 1,605.84 MB during training and 402.21 MB during inference.
Table 9
| Networks | Params (M) | FLOPs (G) | TM (MB) | IM (MB) | TT (ms) | IT (ms) |
|---|---|---|---|---|---|---|
| UNet | 31.04 | 109.47 | 474.18 | 118.92 | 21.83 | 5.46 |
| TransUNet | 93.23 | 64.43 | 1,605.84 | 402.21 | 34.63 | 9.11 |
| Attention U-Net | 34.87 | 133.57 | 532.70 | 133.55 | 31.83 | 9.48 |
| nnUNet | 143.87 | 209.53 | 577.50 | 145.87 | 57.94 | 15.09 |
| SegMamba | 23.38 | 78.11 | 404.83 | 101.58 | 37.01 | 9.85 |
| Swin-UMamba | 55.06 | 351.47 | 915.78 | 230.45 | 157.79 | 38.72 |
| AttmNet | 28.47 | 87.09 | 485.64 | 121.78 | 72.08 | 22.37 |
For comparison, training time refers to the duration of one forward and backward pass for a single image, whereas inference time represents the time required for inference on a single image. FLOP, floating point operations per second; IM, inference memory; IT, inference time; M, million; TM, training memory; TT, training time.
However, AttmNet exhibited longer training and inference times, requiring 72.08 ms for training and 22.37 ms for inference, both exceeding those of TransUNet by more than twofold. This can be attributed to the Mamba module’s reliance on structured SSM, which are inherently sequential and restrict parallelism. Additionally, multidirectional scanning, dynamic weighting mechanisms, and the processing of high-resolution intermediate states further contribute to computational overhead and latency.
Discussion
In this study, we proposed AttmNet, a novel network architecture designed to effectively capture multi-scale local and global information through an innovative combination of complementary techniques. The encoder employs the MAM module, which merges MLC and Att-Mamba components to enhance feature extraction and representation. The decoder adopts linear interpolation for upsampling, minimizing model complexity while avoiding additional parameter overhead. Skip connections bridge encoder and decoder features, enabling the effective fusion of high-resolution and low-resolution information to improve segmentation precision. Experimental results confirm that AttmNet achieves state-of-the-art performance in medical image segmentation tasks.
Despite its strong performance, AttmNet has certain limitations. Although the network performs well on the datasets used in this study, its ability to generalize to other lesion types or imaging modalities needs further evaluation. Additionally, the small dataset sizes may limit the model’s robustness when applied to larger and more diverse datasets. Another limitation is the relatively long training and inference times, which could restrict its deployment in real-time applications. Addressing these challenges in future work will involve expanding the network to other medical imaging tasks and modalities with domain-specific priors, refining the architecture for better scalability on large datasets, and developing efficient computational strategies to reduce training and inference times.
Clinical applications may also face obstacles such as annotation variability or insufficient labeled data. For instance, segmentation label accuracy can vary depending on annotators’ expertise, and the limited availability of labeled data may hinder effective training. To address these issues, future research could utilize techniques such as Robust T-Loss (28) to manage label inaccuracies or adopt a Human-in-the-Loop strategy (29) to iteratively improve model performance and adaptability.
Although this study does not directly address three-dimensional (3D) image segmentation tasks, the critical role of 3D segmentation in medical imaging suggests a promising avenue for future research. Compared to 2D segmentation, 3D tasks involve significantly larger data volumes, increasing computational complexity and memory demands. Extending AttmNet to 3D segmentation will likely require advanced computational resources and efficient algorithmic adaptations. Future studies could explore the potential of AttmNet in 3D segmentation tasks, supported by optimized hardware and computational frameworks.
Conclusions
AttmNet demonstrates high segmentation accuracy while offering notable efficiency in model size, computational cost, and memory usage. Its modular and scalable design makes it an effective solution for addressing various medical imaging challenges. During usage, the model requires no user interaction or specific expertise, offering significant potential for real-world applications.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD+AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-2024-2561/rc
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-2024-2561/coif). H.Z. received funding from Humanities and Social Science Fund of Ministry of Education of China (No. 23YJAZH232). K.H. received funding from Zhejiang Provincial Natural Science Foundation of China (No. LZ24F020006). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2024;74:229-63. [Crossref] [PubMed]
- Chowdary GJ, Yathisha GVD, Suganya G, Premalatha M. Automated skin lesion segmentation using multi-scale feature extraction scheme and dual-attention mechanism. 2021 3rd International Conference on Advances in Computing, Communication Control and Networking (ICAC3N). 2021:1763-71.
- Shi F, Wang J, Shi J, Wu Z, Wang Q, Tang Z, He K, Shi Y, Shen D. Review of Artificial Intelligence Techniques in Imaging Data Acquisition, Segmentation, and Diagnosis for COVID-19. IEEE Rev Biomed Eng 2021;14:4-15. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 2015, proceedings, part III 18. 2015:234-41.
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans Med Imaging 2020;39:1856-67. [Crossref] [PubMed]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA; 2017:2261-9.
- Iqbal S, Ahmed H, Sharif M, Hena M, Khan TM, Razzak I. Euis-net: A convolutional neural network for efficient ultrasound image segmentation. arXiv preprint arXiv:12323. 2024.
- Hu B, Zhou P, Yu H, Dai Y, Wang M, Tan S, Sun Y. LeaNet: Lightweight U-shaped architecture for high-performance skin cancer image segmentation. Comput Biol Med 2024;169:107919. [Crossref] [PubMed]
- Lyu Y, Xu Y, Jiang X, Liu J, Zhao X, Zhu X. AMS-PAN: Breast ultrasound image segmentation model combining attention mechanism and multi-scale features. Biomedical Signal Processing Control 2023;81:104425.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Advances in Neural Information Processing Systems. 2017;30:5998-6008.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:04306. 2021.
- Cao H, Wang Y, Chen J, Jiang D, Zhang X, Tian Q, Wang M. Swin-unet: Unet-like pure transformer for medical image segmentation. European conference on computer vision 2022. 2022:205-18.
- Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z, Lin S, Guo B. Swin transformer: Hierarchical vision transformer using shifted windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada; 2021:9992-10002.
- Gu A, Dao T. Mamba: Linear-time sequence modeling with selective state spaces. 2023. arXiv preprint arXiv: 2312.00752.
- Xing Z, Ye T, Yang Y, Liu G, Zhu L. Segmamba: Long-range sequential modeling mamba for 3d medical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2024:578-88.
- Liu J, Yang H, Zhou HY, Xi Y, Yu L, Li C, Liang Y, Shi G, Yu Y, Zhang S, Zheng H, Wang S. Swin-umamba: Mamba-based unet with imagenet-based pretraining. International Conference on Medical Image Computing and Computer-Assisted Intervention 2024. 2024:615-25.
- Ma J, Li F, Wang B. U-mamba: Enhancing long-range dependency for biomedical image segmentation. 2024. arXiv preprint arXiv:2401.04722.
- Zhu L, Liao B, Zhang Q, Wang X, Liu W, Wang X. Vision mamba: Efficient visual representation learning with bidirectional state space model. 2024. arXiv preprint arXiv: 2401.09417.
- Gong H, Kang L, Wang Y, Wan X, Li H. nnMamba: 3D Biomedical Image Segmentation, Classification and Landmark Detection with State Space Model. 2024. arXiv preprint arXiv: 2402.03526.
- Valanarasu JMJ, Patel VM. Unext: Mlp-based rapid medical image segmentation network. MICCAI 2022;2022:23-33.
- Yap MH, Pons G, Marti J, Ganau S, Sentis M, Zwiggelaar R, Davison AK, Marti R. Moi Hoon Yap, Pons G, Marti J, Ganau S, Sentis M, Zwiggelaar R, Davison AK, Marti R. Automated Breast Ultrasound Lesions Detection Using Convolutional Neural Networks. IEEE J Biomed Health Inform 2018;22:1218-26. [Crossref] [PubMed]
- Al-Dhabyani W, Gomaa M, Khaled H, Fahmy A. Dataset of breast ultrasound images. Data in brief 2020;28:104863. [Crossref] [PubMed]
- Mendonça T, Ferreira PM, Marques JS, Marcal AR, Rozeira J. PH 2-A dermoscopic image database for research and benchmarking. 2013 35th annual international conference of the IEEE engineering in medicine and biology society (EMBC). 2013:5437-40.
- Zhao X, Jia H, Pang Y, Lv L, Tian F, Zhang L, Sun W, Lu H. M2SNet: Multi-scale in Multi-scale Subtraction Network for Medical Image Segmentation. 2023. arXiv preprint arXiv: 2303.10894.
- Kingma DP, Ba J. Adam: A method for stochastic optimization. 2014. arXiv preprint arXiv:1412.6980.
- Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D. Attention U-net: Learning where to look for the pancreas. 2018. arXiv preprint arXiv:1804.03999.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Gonzalez-Jimenez A, Lionetti S, Gottfrois P, Gröger F, Pouly M, Navarini AA. Robust T-loss for medical image segmentation. MICCAI 2023;2023:714-24.
- Kerdvibulvech C, Li Q. Empowering Zero-Shot Object Detection: A Human-in-the-Loop Strategy for Unveiling Unseen Realms in Visual Data. International Conference on Human-Computer Interaction 2024. 2024:235-44.


