
FCFDiff-Net: full-conditional feature diffusion embedded network for 3D brain tumor segmentation
Introduction
Brain tumors are diseases caused by the abnormal growth of cells in or near the brain. Since it usually occurs in or near the brain tissue, it can easily affect the brain, which poses a major threat to the life and health of patients. According to statistics, the number of brain tumor cases in China in 2022 alone will be as many as 87,500, and the number of deaths will be as many as 56,600 (1). Consequently, the early identification and management of brain tumors are major clinical priorities. However, the complexity of their location and structure within brain tissue, along with the heterogeneity and variability across different tumors, significantly complicates their diagnosis and treatment (2). Among various imaging techniques, magnetic resonance imaging (MRI) is the main diagnostic method for brain tumors by integrating fluid-attenuated inversion recovery (FLAIR), T1-weighted (T1), post-contrast T1-weighted (T1ce), and T2-weighted (T2) sequences to observe and analyze the progression of the disease. Specifically, brain tumor segmentation (BraTS) requires the identification of key regions such as enhancing tumor (ET), necrosis and non-enhancing tumor (NCR/NET), and peritumoral edema (ED)/infiltration. Figure 1 illustrates a three-dimensional (3D) MRI with multiple modalities for BraTS. Therefore, how to accurately segment the brain tumor region from the MRI data, to allow doctors to accurately analyze the patient’s condition and make the correct judgment, is the key point of BraTS. Conventional BraTS not only demands extensive expertise and experience but also is time-consuming and highly susceptible to subjective factors, which can lead to inconsistencies and reduced accuracy (3,4). So, researchers have tried to solve this problem by using computer-aided diagnosis.

With the rapid development of computer technology, researchers are increasingly leveraging deep learning techniques to address these challenges, with promising results (5,6). Since the proposal of fully convolutional network (FCN) (7) and U-Net (8), a series of variants of U-Net structure, such as Attention U-Net (9), 3D-UNet (10), V-Net (11), U-Net++ (12), dilated multi-scale residual attention U-net (DMRAU-Net) (13), and DAU-Net (14), have achieved remarkable results in BraTS. SegResNet (15) significantly improves the feature extraction capability of the model by adding reconstruction branches using a variational auto-encoder (VAE). Isensee et al. (16) applied no new net (nnUNet) to BraTS with several targeted processing, which effectively improved the accuracy of BraTS. The success of these models demonstrates convolutional neural networks’ (CNNs) power in learning semantic information. However, the limitations of convolutional operations limit the usefulness of spatial dependencies and do not allow for good learning of the connection between global and local information, which is crucial in BraTS (17,18). The Transformer structure has been widely used in image segmentation because of its unique self-attention mechanism and ability to efficiently capture global contextual information (19). TransBTS (20) uses 3D CNN to extract local contextual information and then uses Transformer for global feature modeling. TransUNet (21) combines the advantages of CNN and Transformer to alleviate the limitations of CNN in long-distance dependency modeling. UNEt TRansformers (UNETR) (22) employs a vision transformer (ViT) as the encoder to capture global features, followed by a CNN-based decoder to produce segmentation results. Swin UNETR (23) utilizes a Swin Transformer as the encoder to extract multi-scale features, and similarly, a CNN-based decoder is used to generate the final output. CKD-TransBTS (24) designs two modules to extract multimodal information and fuse the features of CNN and Transformer at the decoder. Although the aforementioned methods have yielded promising results, automatic segmentation remains challenging due to the computational complexity of Transformers and their variants, as well as the inherent heterogeneity of brain tumors in terms of appearance, shape, and histology (25).
Recently, the swift advancement of deep learning techniques has driven significant success in denoising diffusion models, particularly in their application to medical image segmentation and other image processing tasks. The diffusion model added Gaussian noise to the segmentation label and embedded the original image into the network as a condition to guide the segmentation. Through multiple iterations of denoising, the final predicted segmentation label was mapped to the corresponding image space (26,27). For example, Wolleb et al. (28) used a diffusion model to solve the problem of two-dimensional (2D) medical image segmentation by embedding the original image into a diffusion model to guide the network in generating segmentation labels. In contrast, the sampling process uses summation to merge the results of multiple predictions, thus improving the robustness of the segmentation results. Wu et al. (29) proposed MedSegDiff to design dynamic conditional coding to enhance the feature extraction of the denoising process and used the Fourier transform to suppress the influence of high-frequency noise. In the subsequent MedSegDiff-V2 (30), Wu et al. combined the Transformer with a diffusion model and designed several modules to simulate the interactions between noise and semantic features, improving the segmentation performance of the model. Bozorgpour et al. (31) designed DermoSegDiff to improve segmentation performance by introducing a new loss function that allows the model to prioritize learning the boundary information of the region to be segmented and integrating noise and semantic information within the network through a new U-Net network architecture. Chen et al. (32) introduced BerDiff, which is the first method to utilize Bernoulli noise as the diffusion kernel to boost the segmentation capabilities of the model, generating varied segmentation labels through random sampling to enhance performance. Xing et al. (33) proposed Diff-UNet to integrate the diffusion model into the U-Net architecture to efficiently extract semantic information from multimodal images, while to enhance the robustness of the prediction results of the diffusion model, an uncertainty-based fusion module is used in the sampling stage to merge to obtain the final segmentation results.
Despite the promising performance of diffusion-based approaches in segmentation tasks, their application to medical imaging—particularly BraTS—remains challenging due to fundamental limitations in conditional embedding. As a generative framework, diffusion models incorporate the original image as a conditional prior within the denoising process to guide segmentation label generation. However, medical imaging presents unique challenges, including extreme tumor heterogeneity (e.g., varying sizes, irregular shapes, and indistinct boundaries) and domain-specific noise (e.g., MRI artifacts, intensity inhomogeneity). Traditional conditional embedding techniques, such as direct feature concatenation or global pooling, struggle to retain fine-grained anatomical structures and to effectively separate tumor-relevant features from noise throughout the iterative denoising process. These shortcomings manifest in two major failure modes: (I) false negatives, where small or low-contrast tumors are omitted due to insufficient feature retention; and (II) false positives, where noisy artifacts are misclassified as pathological tissue due to unresolved feature ambiguity. Notably, although diffusion models theoretically learn a robust data distribution, their reliance on suboptimal conditional priors can hinder accurate feature refinement in the presence of medical image variability. This underscores the necessity for advanced conditional embedding mechanisms specifically designed for medical diffusion models—mechanisms that ensure precise alignment between noisy latent features and diagnostically critical anatomical structures, ultimately enhancing segmentation accuracy and robustness.
To solve the above problems, we propose a novel full-conditional feature diffusion (FCFD) embedded network for 3D BraTS, named FCFDiff-Net. To guide the diffusion model in predicting the true and complete segmentation labels more accurately, we designed the full-conditional feature embedding (FCFE) module. This module uses the original image as a condition to guide the diffusion model in predicting the segmentation labels. In addition, we also perform feature mapping between the decoder side of the FCFE to the encoder side of the denoising module to enhance the network’s ability to model the segmentation target and the segmentation boundary. Meanwhile, to further optimize the model performance and reduce the feature differences between the conditional features and denoising module, we designed a multi-head attention residual fusion (MHARF) module. This module integrates the feature information of the FCFE with the information with noise in the denoising network, extracts the context information, and realizes the feature alignment of semantic information and noise information. This design allows the model to generate more robust and accurate segmentation results. Extensive experiments were conducted using the BraTS 2020 and BraTS 2021 multimodal BraTS datasets. Comparisons with several state-of-the-art BraTS models demonstrate that our method outperforms existing state-of-the-art approaches in accurately segmenting brain tumor images.
Our contributions are summarized as follows:
- To solve the problem of medical image segmentation in which the network extracts insufficient features due to problems such as image blurring and noise interference, which makes the segmentation label appear empty and incorrectly segmented, we designed the FCFE method. This method improves the network’s feature extraction capabilities by conducting multi-level feature extraction on the original image and thoroughly incorporating conditional feature information, leading to more precise segmentation.
- To reduce the effect of noise in conditional embedding and denoising networks, we designed the MHARF module. This module integrates the conditional features from full-conditional embedding with noise features in the denoising network, using a multi-head attention mechanism to capture their correlations. This approach enhances the model’s expressive capability and further improves the network’s feature extraction performance.
- Comparative experiments were conducted on the BraTS 2020 and BraTS 2021 brain tumor datasets, as well as with multiple state-of-the-art segmentation models, to thoroughly evaluate the performance of our FCFDiff-Net. The results show that our method achieves the highest segmentation accuracy.
We present this article in accordance with the CLEAR reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2300/rc).
Methods
Overall architecture
The overall workflow of our proposed FCFDiff-Net is shown in Figure 2. The network is different from traditional medical image segmentation. It takes the original image I, and the noise segmentation mask as the input of the network, learns the denoising process by training the network, and finally generates a clear segmentation mask to achieve the goal of image segmentation.

For ease of understanding, we describe the overall flow of the network at t steps. At step t, the segmentation label can be obtained as a noisy segmentation mask through a forward process, and for the diffusion model to generate a meaningful segmentation mask , we need to embed the original image as a condition to guide the generation in the network. Therefore, we concatenate and the original image I at the channel level to obtain , and then use as the input of the denoising network . On the one hand, we input the original image I into the network according to FCFE, which can obtain several segmentation features at the decoder side. The sum operation of and the denoising network of is carried out, that is, . Through the above operations, more complete conditional features can be embedded into the denoising network, to better introduce the original image data, enhance the perception ability of the diffusion model to the segmentation target, and reduce the diffusion variance. On the other hand, when the conditional features are embedded into the denoising network, the presence of Gaussian noise will make a certain semantic gap between them, and this difference will affect the performance of the model. Therefore, we designed the MHARF module, through which the network ignores the semantic gap between them, to reduce the difference between the conditional feature and the noise in the denoising network. By the above method, the final segmentation label can be obtained.
The general diffusion model is trained by predicting the noise added before, whereas for diffusion models to complete the image segmentation task better, our FCFDiff-Net directly predicts the final segmentation label (33). Therefore, the loss functions used during training are binary cross entropy (BCE) loss, mean squared error (MSE) loss, and DICE loss. The total loss for model training is defined as:
FCFE module
Diffusion models, as generative models applied in image segmentation, must be guided appropriately to achieve the purpose of image segmentation. Without effective generation guidance during the training and sampling phases, the generated results may be meaningless (28). To address this issue, researchers have explored various methods to incorporate the original image I into the model as conditional features, guiding the generation of a meaningful segmentation mask .
Wolleb et al. (28) proposed a method to guide the denoising network to generate the segmentation mask by treating the noise segmentation mask as an additional channel of the original image I. As shown in Figure 3A, this approach introduces original feature information at the channel level but may result in the generation of meaningless segmentation masks since the denoising network does not extract enough feature information. Xing et al. (33) designed a feature encoder on their Diff-UNet model to add features from the original image I to the denoising network. They sum up the features extracted by the feature encoder with the features of corresponding size in the denoising network, and then add the fused features as inputs to the decoder of the denoising network, thus realizing the effective guidance of the generation process. As shown in Figure 3B, this approach embeds the condition directly into the noisy information of the denoising network, which may cause confusion in the bootstrapping process and lead to the generation of incorrect segmentation masks. However, MedSegDiff-V2 (30), proposed by Wu et al., adopts a novel conditional embedding method, as shown in Figure 3C. They used a standard U-Net as the conditional model to extract image features, which were then input into a designed Transformer module alongside features of matching size from the denoising network. This approach helps to mitigate the disparity between noise and conditional embeddings. This approach effectively guided the generative process of the diffusion model. However, incorporating the Transformer module also increases the model’s computational complexity and may be constrained by available computational resources.

However, when faced with smaller or fuzzy segmentation labels, these conditional fusion methods often fail to extract useful information due to limited feature extraction, resulting in empty or wrong segmentation. To solve the above problems, we propose FCFE, which comprehensively improves the embedding of original image features to effectively guide the denoising network in generating segmentation labels . The structure is shown in Figure 4.
We combine the above conditional fusion methods by letting the original image I and the noisy segmentation mask be channel spliced as the input to the denoising network , which gives us . Meanwhile, the original image I is fed into a standard U-Net structure, and then the obtained features at the decoder side and at the encoder side of the denoising network are fused, and then the remaining convolutional operation is employed, which is expressed by the formula:
where , , denotes a series of convolution operations and denotes a pixel-by-pixel addition operation.
We use this approach to guide the denoising network to correctly generate the segmentation label , and the boundary information of the segmentation label obtained in this way is more complete.
MHARF module
During diffusion models training, we utilize the denoising network to predict the added noise and integrate the image information into the model through FCFE to guide it in generating accurate segmentation labels. However, a mismatch between the semantic information embedded in the features and the noise in the denoising network can disrupt the model’s training process, potentially impacting the network’s segmentation performance (30). Therefore, our main goal was to explore how to effectively minimize the negative impact of this discrepancy on network performance.
To address this issue, our main goal was to explore strategies for minimizing the negative impact of such discrepancies on network performance. During the training of the denoising network, it is crucial not only to predict the noise distribution but also to accurately capture the feature distribution of the segmentation target. Given the multi-head attention mechanism’s effectiveness in processing multi-feature information in transformer architectures, we developed a multi-head attention residual fusion (MHARF) module. This module fuses the feature of the original image with the denoising feature of the denoising network as input. The overall structure is shown in Figure 5.
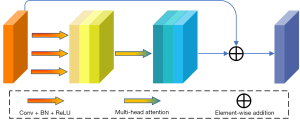
We planned to utilize a multi-head attention mechanism to fit the conditional features and the noise so that the network sees both as a unified overall feature. Specifically, the fused feature is first passed through three convolutional layers of different scales to extract local information gradually, and then batch normalization and rectified linear unit (ReLU) activation functions are used to enhance the expressive power of the feature, which can be expressed as follows: . Then, the multi-head attention mechanism is used to integrate these processed features. This helps to reduce the feature difference between and , and extract higher-level semantic information, which can be expressed as: . Finally, these features are summed with the original input features to prevent the gradient vanishing problem. In summary, it is expressed in Eq. [3] as:
Each of these operations contains batch normalization and ReLU activation functions. The MHARF module integrates the features of the original image with the noise in the denoising network, allowing the network to disregard the differences between them and thus enhancing its performance in segmentation tasks. In addition, the final output features also contain more advanced semantic information, which can better guide the denoising network to generate more accurate segmentation labels.
Experiments
Datasets
In this experiment, we used two datasets. The first dataset used for the experiment was provided by the BraTS 2020 Challenge (34-36). Since the validation and test data of this dataset are not visible, we split its training set (369 3D MRIs) into a train set, validation set, and test set with a ratio of 7:1:2. Each MRI sample consists of four modes, namely T1, T2, T1ce, and FLAIR. The label consisted of Gd-ET (label 4), peritumoral edema (label 2), necrotic tumor core (TC, label 1), and background (label 0). These four labels can be clustered into three easily segmentable regions for performance evaluation, namely TC (labels 1 and 4), ET (label 4), and whole tumor (WT, labels 1, 2, and 4). Another dataset is from the BraTS 2021 Challenge (34,35,37). Its train set consists of 1,251 3D MRIs. The two datasets (BraTS 2020 and BraTS 2021) are identical except for the number of samples.
Evaluation metrics
We used the Dice similarity coefficient (DSC), Hausdorff distance at the 95th percentile (HD95), specificity, and false positive rate (FPR) to comprehensively evaluate the segmentation performance of the network. Eq. [4] represents the spatial overlap between the true segmentation label and the predicted segmentation label , defined as the DSC value:
where denotes the number of overlapping pixels between the prediction and ground truth, and , are the total pixels in the predicted and true labels, respectively.
The Hausdorff distance (HD) is represented by Eq. [5], which measures the shape similarity between the true segmentation label and the predicted segmentation label by computing the boundary distance.
where a and b denote boundary pixels of and , respectively. To mitigate sensitivity to outliers. we report the HD95.
Additionally, specificity and FPR were computed to assess the model’s robustness in avoiding false positives. Specificity measures the proportion of true negative pixels correctly identified:
where represents the complement of the true label, and denotes the true negative pixels. Conversely, the FPR reflects the rate of background pixels misclassified as positive:
These metrics provide complementary insights: DSC and HD95 emphasize spatial overlap and boundary accuracy, whereas specificity and FPR evaluate the model’s precision in preserving true negatives and suppressing false activations.
Implementation details
Our FCFDiff-Net is implemented using PyTorch and MONAI on 2× 40 G Nvidia A100 GPUs (Nvidia, Santa Clara, CA, USA), each with a memory capacity of 40 G. During training, we adopt BCE Loss, MSE Loss, and DICE Loss as the total loss function. The optimization is performed using the AdamW optimizer with a weight decay of 10−5, whereas the learning rate follows a cosine annealing schedule. The batch size is set to 4, and the model is trained for 300 epochs.
The original resolution of BraTS scans is 240×240×155. Instead of resizing, we adopt a patch-based training strategy, where n patches of 96×96×96 are randomly sampled from the full volume. This approach ensures that the original aspect ratio is maintained while allowing the model to capture diverse brain regions across different training iterations. To enhance data diversity and improve generalization, we apply random flips (horizontal, vertical, and depth-wise), random rotations (within ±15°), intensity scaling (0.9–1.1), and intensity shifts (−0.1 to 0.1) for data augmentation.
During inference, we set the number of denoising diffusion implicit models (DDIMs) sampling steps to 10, and each generated sample has a size of 96×96×96. A sliding window strategy with an overlap rate of 0.5 is employed to ensure full-volume prediction, preventing the loss of critical brain structures.
The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Results
To validate the effectiveness of FCFDiff-Net in 3D brain tumor image segmentation, we compared it with the current state-of-the-art models on the BraTS 2020 and BraTS 2021 datasets. These segmentation models include CNN-based methods such as Attention-UNet (9), DAUnet (14), SegResNet (15) and nnUNet (16), Transformer-based methods such as TransBTS (20), TransUNet (21), UNETR (22), SwinUNETR (23), SwinBTS (38), and CKD-TransBTS (24), and the diffusion model-based method FDiff-Fusion (39). To ensure the fairness of the comparison, all models involved, including our FCFDiff-Net, were trained and tested under the same computer hardware and dataset conditions. The quantitative results of the experiments are presented in Tables 1-4.
Table 1
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Attention-UNet (9) | 0.845 | 15.174 | 0.782 | 16.380 | 0.716 | 9.095 | 0.781 | 13.549 | |||
| DAUnet (14) | 0.898 | 5.400 | 0.830 | 9.800 | 0.786 | 27.600 | 0.838 | 14.267 | |||
| SegResNet (15) | 0.915 | 3.275 | 0.836 | 3.769 | 0.730 | 3.486 | 0.827 | 3.494 | |||
| nnUNet (16) | 0.912 | 3.781 | 0.842 | 7.771 | 0.765 | 18.230 | 0.840 | 9.927 | |||
| TransBTS (20) | 0.911 | 3.360 | 0.836 | 2.986 | 0.740 | 3.403 | 0.829 | 3.249 | |||
| TransUNet (21) | 0.892 | 3.146 | 0.825 | 2.891 | 0.758 | 3.621 | 0.825 | 3.219 | |||
| UNETR (22) | 0.902 | 4.305 | 0.813 | 5.740 | 0.732 | 4.643 | 0.816 | 4.896 | |||
| SwinUNETR (23) | 0.917* | 2.856 | 0.826 | 4.314 | 0.749 | 4.503 | 0.830 | 3.891 | |||
| SwinBTS (38) | 0.891 | 8.560 | 0.804 | 15.780 | 0.774 | 26.840 | 0.823 | 17.060 | |||
| CKD-TransBTS (24) | 0.898 | 2.419 | 0.841 | 3.447 | 0.770 | 3.018 | 0.836 | 2.961 | |||
| FDiff-Fusion (39) | 0.905 | 2.207 | 0.844 | 3.311 | 0.776 | 2.714 | 0.842 | 2.473 | |||
| FCFDiff-Net | 0.916 | 1.917* | 0.860* | 2.571* | 0.786* | 2.581* | 0.854* | 2.356* | |||
Higher DSC scores (↑) indicate better segmentation, whereas lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; HD95, Hausdorff distance at the 95th percentile; TC, tumor core; WT, whole tumor.
Table 2
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | ||||
| Attention-UNet (9) | 0.985 | 0.015 | 0.980 | 0.020 | 0.970 | 0.030 | 0.978 | 0.022 | |||
| DAUnet (14) | 0.987 | 0.013 | 0.980 | 0.020 | 0.975 | 0.025 | 0.981 | 0.020 | |||
| SegResNet (15) | 0.990 | 0.010 | 0.985 | 0.015 | 0.980 | 0.020 | 0.985 | 0.015 | |||
| nnUNet (16) | 0.988 | 0.012 | 0.982 | 0.018 | 0.976 | 0.024 | 0.982 | 0.018 | |||
| TransBTS (20) | 0.991 | 0.009 | 0.985 | 0.015 | 0.978 | 0.022 | 0.985 | 0.015 | |||
| TransUNet (21) | 0.990 | 0.010 | 0.985 | 0.015 | 0.977 | 0.023 | 0.984 | 0.016 | |||
| UNETR (22) | 0.985 | 0.015 | 0.980 | 0.020 | 0.973 | 0.027 | 0.979 | 0.021 | |||
| SwinUNETR (23) | 0.992 | 0.008 | 0.990 | 0.010 | 0.985 | 0.015 | 0.989 | 0.011 | |||
| SwinBTS (38) | 0.987 | 0.013 | 0.980 | 0.020 | 0.974 | 0.026 | 0.980 | 0.020 | |||
| CKD-TransBTS (24) | 0.990 | 0.010 | 0.985 | 0.015 | 0.978 | 0.022 | 0.984 | 0.016 | |||
| FDiff-Fusion (39) | 0.992 | 0.008 | 0.990 | 0.010 | 0.985 | 0.015 | 0.989 | 0.011 | |||
| FCFDiff-Net | 0.998* | 0.002* | 0.999* | 0.001* | 0.999* | 0.001* | 0.999* | 0.001* | |||
Higher specificity scores (↑) and lower FPR (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; ET, enhancing tumor; FPR, false positive rate; TC, tumor core; WT, whole tumor.
Table 3
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Attention-UNet (9) | 0.910 | 8.990 | 0.869 | 6.572 | 0.841 | 5.302 | 0.873 | 6.952 | |||
| DAUnet (14) | 0.899 | 6.700 | 0.844 | 6.600 | 0.776 | 14.400 | 0.839 | 9.233 | |||
| SegResNet (15) | 0.917 | 6.113 | 0.896 | 5.790 | 0.861 | 5.423 | 0.891 | 5.781 | |||
| nnUNet (16) | 0.926 | 3.550 | 0.874 | 10.560 | 0.837 | 22.440 | 0.879 | 12.183 | |||
| TransBTS (20) | 0.920 | 4.980 | 0.882 | 4.860 | 0.795 | 16.320 | 0.866 | 8.720 | |||
| TransUNet (21) | 0.919 | 6.162 | 0.877 | 7.340 | 0.818 | 13.090 | 0.872 | 8.860 | |||
| UNETR (22) | 0.890 | 14.423 | 0.847 | 10.221 | 0.825 | 8.785 | 0.854 | 11.142 | |||
| SwinUNETR (23) | 0.926 | 5.831 | 0.885 | 3.770 | 0.858 | 6.016 | 0.890 | 5.206 | |||
| SwinBTS (38) | 0.918 | 3.650 | 0.848 | 14.510 | 0.832 | 16.030 | 0.866 | 11.397 | |||
| CKD-TransBTS (24) | 0.923 | 4.230 | 0.881 | 4.390 | 0.848 | 3.160 | 0.884 | 3.927 | |||
| FCFDiff-Net | 0.926* | 2.156* | 0.903* | 1.834* | 0.869* | 1.583* | 0.899* | 1.858* | |||
Higher DSC scores (↑) indicate better segmentation, while lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; HD95, Hausdorff distance at the 95th percentile; TC, tumor core; WT, whole tumor.
Table 4
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | Specificity↑ | FPR↓ | ||||
| Attention-UNet (9) | 0.980 | 0.020 | 0.975 | 0.025 | 0.970 | 0.030 | 0.975 | 0.025 | |||
| DAUnet (14) | 0.985 | 0.015 | 0.980 | 0.020 | 0.975 | 0.025 | 0.980 | 0.020 | |||
| SegResNet (15) | 0.990 | 0.010 | 0.985 | 0.015 | 0.980 | 0.020 | 0.985 | 0.015 | |||
| nnUNet (16) | 0.989 | 0.011 | 0.986 | 0.014 | 0.980 | 0.020 | 0.985 | 0.015 | |||
| TransBTS (20) | 0.991 | 0.009 | 0.986 | 0.014 | 0.982 | 0.018 | 0.986 | 0.014 | |||
| TransUNet (21) | 0.990 | 0.010 | 0.987 | 0.013 | 0.981 | 0.019 | 0.986 | 0.014 | |||
| UNETR (22) | 0.988 | 0.012 | 0.983 | 0.017 | 0.975 | 0.025 | 0.982 | 0.018 | |||
| SwinUNETR (23) | 0.992 | 0.008 | 0.990 | 0.010 | 0.985 | 0.015 | 0.989 | 0.011 | |||
| SwinBTS (38) | 0.989 | 0.011 | 0.982 | 0.018 | 0.978 | 0.022 | 0.983 | 0.017 | |||
| CKD-TransBTS (24) | 0.993 | 0.007 | 0.988 | 0.012 | 0.986 | 0.014 | 0.989 | 0.011 | |||
| FCFDiff-Net | 0.999* | 0.001* | 0.999* | 0.001* | 0.999* | 0.001* | 0.999* | 0.001* | |||
Higher specificity scores (↑) and lower false positive rates (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; ET, enhancing tumor; FPR, false positive rates; FPR, false positive rate; TC, tumor core; WT, whole tumor.
On the BraTS 2020 dataset, as shown in Table 1, FCFDiff-Net achieves state-of-the-art performance, with an average DSC score of 0.854 and an HD95 of 2.356 mm, outperforming all competing methods across all tumor sub-regions. In the particularly challenging ET region, our model surpassed the second-best FDiff-Fusion by improving the DSC score from 0.776 to 0.786 while also reducing HD95 from 2.714 to 2.581 mm. Additionally, the exceptional specificity scores presented in Table 2 (with an average of 0.999) and the minimal FPRs (with an average FPR of 0.001) confirm FCFDiff-Net’s reliability in clinical applications by effectively avoiding erroneous lesion detection.
The advantage of FCFDiff-Net becomes even more evident on the BraTS 2021 benchmark (Tables 3,4), where it establishes new state-of-the-art results with an average DSC score of 0.899 and an HD95 of 1.858 mm. Notably, our model maintains strong consistency across all tumor sub-regions, achieving the best HD95 values in every category (WT: 2.156 mm, TC: 1.834 mm, ET: 1.583 mm), demonstrating a significant reduction in segmentation error compared to the second-best CKD-TransBTS, particularly in the ET region. Furthermore, its near-perfect specificity (0.999) and negligible FPR (0.001) across all tumor regions (Table 4) further validate its precision in tumor boundary delineation for clinical applications.
Compared to CNN-based approaches, FCFDiff-Net achieves a noticeably higher average DSC score than SegResNet on BraTS 2020 while also significantly reducing HD95. When compared to Transformer architectures, our model demonstrates superior HD95 performance over SwinUNETR while maintaining comparable DSC scores. Moreover, FCFDiff-Net outperforms the existing diffusion-based FDiff-Fusion, achieving both higher DSC scores and lower HD95 values on BraTS 2020. These results collectively highlight the effectiveness of our method in delivering superior segmentation accuracy and precise tumor boundary delineation.
Figure 6 provides us with an intuitive view of the segmentation effect of FCFDiff-Net on the BraTS dataset in comparison to several other segmentation models. We chose the flair image as the background of the segmentation to observe the segmentation effect more clearly. In the first and second rows, the segmentation results of FCFDiff-Net significantly reduce the erroneous segmentation regions compared to other models.

Specifically, the segmentation boundaries of FCFDiff-Net are closer to the boundaries of Ground Truth, reducing the occurrence of false positives. In the third row, all models showed some degree of false positive results, namely, incorrectly identifying non-tumor regions as tumor regions. However, FCFDiff-Net has significantly fewer false-positive results, indicating that it has higher accuracy in distinguishing tumor from non-tumor regions. In the fourth row, the segmentation results generated by FCFDiff-Net are closer to the Ground Truth through its unique FCFE method and MHARF module, whereas other methods have a certain degree of false negative, that is, the tumor region is wrongly identified as a non-tumor region. Moreover, the noise is also significantly reduced, reducing the possible errors in the segmentation process.
Figure 7 compares the 3D stereo segmentation results of FCFDiff-Net and the Ground Truth. It can be seen from the figure that the segmentation results of FCFDiff-Net are very close to the Ground Truth, which further verifies its ability to segment brain tumors in 3D space.

Experiments show that FCFDiff-Net obtains more accurate segmentation results regarding segmentation boundaries and details. Compared with other segmentation models, FCFDiff-Net can identify and segment the tumor region more effectively, significantly reducing the appearance of false positives and false negatives and providing clearer and more accurate segmentation results.
Discussion
Ablation experiments on major modules
To gain a deeper understanding of the role of the FCFE and the MHARF module in FCFDiff-Net, we performed ablation experiments. The quantitative results of these experiments are displayed in Tables 5,6, respectively. In particular, we note that the conditional embedding adopted by the “basic” model is in Figure 3B.
Table 5
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Basic | 0.915 | 1.866 | 0.847 | 3.018 | 0.783 | 2.800 | 0.848 | 2.561 | |||
| Basic + FCFE | 0.914 | 1.910 | 0.854 | 2.727 | 0.782 | 2.912 | 0.850 | 2.516 | |||
| Basic + MHARF | 0.915 | 1.883 | 0.848 | 2.961 | 0.784 | 2.756 | 0.849 | 2.533 | |||
| FCFDiff-Net | 0.916* | 1.917 | 0.860* | 2.571* | 0.786* | 2.581* | 0.854* | 2.356* | |||
Higher DSC scores (↑) indicate better segmentation, whereas lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; FCFE, full-conditional feature embedding; HD95, Hausdorff distance at the 95th percentile; MHARF, multi-head attention residual fusion; TC, tumor core; WT, whole tumor.
Table 6
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Basic | 0.923 | 2.740 | 0.892 | 1.890 | 0.861 | 1.645 | 0.892 | 2.092 | |||
| Basic + FCFE | 0.925 | 2.591 | 0.900 | 1.779 | 0.867 | 1.589 | 0.897 | 1.986 | |||
| Basic + MHARF | 0.924 | 2.451 | 0.898 | 1.772 | 0.860 | 1.628 | 0.894 | 1.950 | |||
| FCFDiff-Net | 0.926* | 2.156* | 0.903* | 1.835 | 0.868* | 1.583* | 0.899* | 1.858* | |||
Higher DSC scores (↑) indicate better segmentation, while lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; FCFE, full-conditional feature embedding; HD95, Hausdorff distance at the 95th percentile; MHARF, multi-head attention residual fusion; TC, tumor core; WT, whole tumor.
In these quantitative experiments, we can see that “basic + FCFE” significantly outperforms “basic” in terms of average DSC and average HD95 values on all three regions, WT, TC, and ET. It shows that our FCFE method, as a feature embedding approach, can introduce more image information into the diffusion model, thus improving the segmentation performance of the model.
Meanwhile, the experimental results of “basic + MHARF” were also better than “basic”, which indicates that the MHARF module can not only effectively make up for the difference in information between feature embedding and denoising network, but also capture more information about the image and further improve the segmentation performance. This shows that the MHARF module can not only effectively compensate for the difference between feature embedding and denoising network but also capture more image information and further improve the segmentation performance. Meanwhile, the experimental results of “basic + MHARF” were also better than “basic”, which indicates that the MHARF module can not only effectively compensate for the noise difference between feature embedding and denoising network, but also capture more image information and further improve the segmentation performance.
This shows that the MHARF module not only can effectively compensate for the noise difference between feature embedding and denoising network but also can capture more image information and further improve the segmentation performance. When both modules are applied together, this is our FCFDiff-Net. Its average DSC and average HD95 values were significantly better than all other combination methods. This shows that by combining these two modules, our method can embed more image information into the denoising network and thus segment the target region more accurately, significantly improving the segmentation accuracy of the segmentation network.
Ablation experiments with conditional embedding
To enable diffusion models to perform image segmentation tasks, researchers have explored various approaches. Among them, the three most widely used conditional embedding methods are (I) conditional fusion; (II) feature encoder; and (III) conditional embedding, as illustrated in Figure 3. To compare the performance difference between these three methods and our proposed FCFE, we conducted experiments on the BraTS 2020 dataset. The quantitative results of these experiments are presented in Table 7.
Table 7
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Conditional fusion | 0.912 | 3.190 | 0.840 | 5.611 | 0.780 | 4.014 | 0.844 | 4.272 | |||
| Feature encoder | 0.915* | 1.866* | 0.847 | 3.018 | 0.783* | 2.800* | 0.848 | 2.561 | |||
| Conditional embedding | 0.910 | 2.040 | 0.850 | 2.816 | 0.780 | 2.772 | 0.847 | 2.543 | |||
| FCFDiff-Net | 0.914 | 1.910 | 0.854* | 2.727* | 0.782 | 2.912 | 0.850* | 2.516* | |||
Higher DSC scores (↑) indicate better segmentation, while lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; FCFDiff-Net, full-conditional feature diffusion embedded network; HD95, Hausdorff distance at the 95th percentile; TC, tumor core; WT, whole tumor.
The data in Table 7 show that the average DSC and HD95 values of methods feature encoder, conditional embedding, and our proposed FCFE are significantly better than those of method conditional fusion across the three tumor regions of WT, TC, and ET. This shows that the conditional embedding method can capture more image information than simple feature fusion, thereby improving segmentation accuracy. Furthermore, our proposed FCFE outperformed methods feature encoder and conditional embedding in all evaluation metrics, demonstrating that our full-conditional feature embedding approach captures richer feature information for better segmentation performance.
Ablation experiments of the MHARF MODULEs in different conditional embedding
To investigate the ability of the MHARF module to deal with the difference between condition information and noise under different conditional embedding ways, we conducted experiments on the BraTS 2020 dataset. Specifically, we integrated the MHARH module into Figure 3B method feature encoder and method conditional embedding, respectively, and conducted experimental comparisons on these improved models. Table 8 displays the quantitative results of the experiments.
Table 8
| Methods | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| Feature encoder + MHARF | 0.915 | 1.883* | 0.848 | 2.961 | 0.784 | 2.756 | 0.849 | 2.533 | |||
| Conditional embedding + MHARF | 0.911 | 2.130 | 0.854 | 2.730 | 0.783 | 2.719 | 0.849 | 2.526 | |||
| FCFDiff-Net | 0.916* | 1.917 | 0.860* | 2.571* | 0.786* | 2.581* | 0.854* | 2.356* | |||
Higher DSC scores (↑) indicate better segmentation, while lower HD95 values (↓) indicate better performance. The top result is marked with asterisk (*). BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; FCFDiff-Net, full-conditional feature diffusion embedded network; HD95, Hausdorff distance at the 95th percentile; MHARF, multi-head attention residual fusion; TC, tumor core; WT, whole tumor.
By analyzing these data, we can see that the average DSC, as well as the average HD95 values on the three regions of WT, TC, and ET, were improved after the addition of the MHARF module for both methods feature encoder and conditional embedding compared to the pre-addition period. This suggests that the MHARF module can effectively compensate for the difference between the noise in the conditional embedding and denoising network, thus capturing more semantic information and improving the model’s segmentation performance.
Ablation experiments for robustness to noise and outliers
In this experiment, we evaluated the performance of the model under different types of noise to assess its robustness. The experiments were conducted using the BraTS 2020 dataset, which was divided into training, validation, and testing sets in a 7:1:2 ratio. The training and validation sets were used to train the model on clean data, whereas the model’s performance was evaluated on a test set under different noise conditions.
The testing set was subjected to four different noise conditions: (I) clean images; (II) images with 0.01 salt-and-pepper noise; (III) images with 0.01 Gaussian noise; and (IV) images with 0.05 Gaussian noise. The purpose of this experiment was to evaluate how well the model could generalize to noisy data, especially under the presence of Gaussian and salt-and-pepper noise.
The results presented in Table 9 show the model’s performance (in terms of accuracy, DSC coefficient, and other relevant metrics) under each noise condition. It is shown that although noise and outliers adversely affect model performance, the model’s robustness improves with increasing noise levels, particularly when exposed to both salt-and-pepper and Gaussian noise.
Table 9
| Dataset | WT | TC | ET | Average | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | DSC↑ | HD95↓ (mm) | ||||
| (I) | 0.916 | 1.917 | 0.860 | 2.571 | 0.786 | 2.581 | 0.854 | 2.356 | |||
| (II) | 0.899 | 4.233 | 0.827 | 4.979 | 0.770 | 3.960 | 0.832 | 4.391 | |||
| (III) | 0.905 | 2.291 | 0.838 | 3.729 | 0.759 | 4.389 | 0.834 | 3.470 | |||
| (IV) | 0.878 | 5.419 | 0.812 | 6.740 | 0.733 | 5.903 | 0.808 | 6.021 | |||
Higher DSC scores (↑) indicate better segmentation, while lower HD95 values (↓) indicate better performance. (I) Clean images; (II) images with 0.01 saltand-pepper noise; (III) images with 0.01 Gaussian noise; and (IV) images with 0.05 Gaussian noise. BraTS, brain tumor segmentation; DSC, Dice similarity coefficient; ET, enhancing tumor; HD95, Hausdorff distance at the 95th percentile; TC, tumor core; WT, whole tumor.
As shown in Table 9, the model’s performance decreased when subjected to noisy test sets. However, the decrease in performance was relatively small in the case of Gaussian noise with a standard deviation of 0.01, indicating that the model maintained its ability to generalize to moderate noise. The performance drop became more noticeable when the Gaussian noise level increased to 0.05, demonstrating that higher levels of noise negatively impacted the model’s robustness. Overall, the experiments suggest that the model is relatively robust to noise and outliers, especially under mild noise conditions.
Conclusions
This paper proposes a full-conditional feature diffusion embedded network for 3D BraTS called FCFDiff-Net. This network aims to solve the problems of brain tumor image segmentation in which false positives (misdetection of non-tumor regions) and false negatives (omission of tumor regions) occur due to the uncertainty of segmentation boundaries and image blurring. We introduce the FCFE technique, which extracts features from the original image using a U-Net network and thoroughly integrates these features into the model. This approach greatly enhances the model’s capability to handle uncertain boundaries and fuzzy regions, effectively reducing both false positives and false negatives. We also designed the MHARF module to reduce the difference between the conditional feature information and the noise by fusing the feature information processed by multiple convolutions and multi-head attention mechanisms. This not only increases the nonlinear capability of the model but also improves the robustness of the model to noise and outliers. The experimental results on two publicly available BraTS datasets, BraTS 2020 and BraTS 2021, showed that FCFDiff-Net outperformed existing state-of-the-art methods in segmentation performance. This demonstrates that our method provides a more effective solution for 3D brain tumor image segmentation.
Although FCFDiff-Net has achieved remarkable success in the task of 3D brain tumor image segmentation, we are aware of some limitations. On the one hand, our method necessitates a substantial amount of high-quality labeled data for the training process. The performance of the model may be affected when data is scarce and data quality is low. On the other hand, if we use a complex diffusion model, FCFDiff-Net requires high computational resources in the training and sampling phases. This may limit the performance of the model when computational resources are limited. To overcome these challenges, we plan to increase the generalizability of the model in future research, enabling it to be applied to a wider range of medical image segmentation tasks. Additionally, we will investigate lightweight modifications to the model to ensure it performs well in environments with limited computational resources without compromising accuracy.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the CLEAR reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-2300/rc
Funding: This work was supported in part by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-2300/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted using the publicly available BraTS 2020 and BraTS 2021 datasets, which contain de-identified MRI scans of glioma patients. The datasets were provided as part of the BraTS challenges, which adhere to ethical guidelines for data sharing and anonymization. No additional patient data collection nor human subject involvement was required for this study. The study was conducted in accordance with the Declaration of Helsinki and its subsequent amendments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Han B, Zheng R, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J. Cancer incidence and mortality in China, 2022. J Natl Cancer Cent 2024;4:47-53. [Crossref] [PubMed]
- Wu X, Tan G, Pu B, Duan M, Cai W DH-GAC. Deep hierarchical context fusion network with modified geodesic active contour for multiple neurofibromatosis segmentation. Neural Comput Appl 2022;37:7511-26. [Crossref]
- Liu J, Li M, Wang J, Wu F, Liu T, Pan Y. A survey of MRI-based brain tumor segmentation methods. Tsinghua Sci Technol 2014;19:578-95. [Crossref]
- Sun K, Ding J, Li Q, Chen W, Zhang H, Sun J, Jiao Z, Ni X. CMAF-Net: a cross-modal attention fusion-based deep neural network for incomplete multi-modal brain tumor segmentation. Quant Imaging Med Surg 2024;14:4579-604. [Crossref] [PubMed]
- Krishnapriya S, Karuna Y. A survey of deep learning for MRI brain tumor segmentation methods: Trends, challenges, and future directions. Health and Technology 2023;13:181-201. [Crossref]
- Pu B, Lu Y, Chen J, Li S, Zhu N, Wei W, Li K. MobileUNet-FPN: A Semantic Segmentation Model for Fetal Ultrasound Four-Chamber Segmentation in Edge Computing Environments. IEEE J Biomed Health Inform 2022;26:5540-50. [Crossref] [PubMed]
- Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640-51. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional Networks for Biomedical Image Segmentation. In: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer, Cham, 2015:234-241.
- Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B. Attention U-Net: Learning Where to Look for the Pancreas. Medical Imaging with Deep Learning. Proceedings of Machine Learning Research, 2018.
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In: Ourselin S, Joskowicz L, Sabuncu M, Unal G, Wells W. editors. Medical Image Computing and Computer-Assisted Intervention - MICCAI 2016. MICCAI 2016. Lecture Notes in Computer Science(), vol 9901. Springer, Cham.
- Milletari F, Navab N, Ahmadi SA. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. 2016 Fourth International Conference on 3D Vision (3DV); 2016 25-28 Oct. doi:
10.1109/3DV.2016.79 .10.1109/3DV.2016.79 - Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: A Nested U-Net Architecture for Medical Image Segmentation. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2018) 2018;11045:3-11. [Crossref] [PubMed]
- Zhang L, Li Y, Liang Y, Xu C, Liu T, Sun J. Dilated multi-scale residual attention (DMRA) U-Net: three-dimensional (3D) dilated multi-scale residual attention U-Net for brain tumor segmentation. Quant Imaging Med Surg 2024;14:7249-64. [Crossref] [PubMed]
- Feng Y, Cao Y, An D, Liu P, Liao X, Yu B. DAUnet: A U-shaped network combining deep supervision and attention for brain tumor segmentation. Knowledge-Based Syst 2024;285:285111348. [Crossref]
- Myronenko A, editor. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; 2019; Cham: Springer International Publishing.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Magadza T, Viriri S. Brain Tumor Segmentation Using Partial Depthwise Separable Convolutions. IEEE Access 2022;10:124206-16.
- Liang S, Hua Z, Li J. Transformer-based multi-scale feature fusion network for remote sensing change detection. J Appl Remote Sens 2022;16:046509. [Crossref]
- Shamshad F, Khan S, Zamir SW, Khan MH, Hayat M, Khan FS, Fu H. Transformers in medical imaging: A survey. Med Image Anal 2023;88:102802. [Crossref] [PubMed]
- Wang W, Chen C, Ding M, Yu H, Zha S, Li J. TransBTS: Multimodal Brain Tumor Segmentation Using Transformer. Medical Image Computing And Computer Assisted Intervention - MICCAI 2021, PT I 2021;12901109-119.
- Chen J, Mei J, Li X, Lu Y, Yu Q, Wei Q, Luo X, Xie Y, Adeli E, Wang Y, Lungren MP, Zhang S, Xing L, Lu L, Yuille A, Zhou Y. TransUNet: Rethinking the U-Net architecture design for medical image segmentation through the lens of transformers. Med Image Anal 2024;97:103280. [Crossref] [PubMed]
- Hatamizadeh A, Tang Y, Nath V, Yang D, Myronenko A, Landman B, Roth HR, Xu D. UNETR: Transformers for 3D Medical Image Segmentation. 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 2022;1748-58. doi:
10.1109/WACV51458.2022.00181 .10.1109/WACV51458.2022.00181 - Hatamizadeh A, Nath V, Tang Y, Yang D, Roth HR, Xu D. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. In: Crimi A, Bakas S. editors. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2021. Lecture Notes in Computer Science, vol 12962. Springer, Cham.
- Lin J, Lin J, Lu C, Chen H, Lin H, Zhao B, Shi Z, Qiu B, Pan X, Xu Z, Huang B, Liang C, Han G, Liu Z, Han C. CKD-TransBTS: Clinical Knowledge-Driven Hybrid Transformer With Modality-Correlated Cross-Attention for Brain Tumor Segmentation. IEEE Trans Med Imaging 2023;42:2451-61. [Crossref] [PubMed]
- Jyothi P, Singh AR. Deep learning models and traditional automated techniques for brain tumor segmentation in MRI: a review. Artif Intell Rev 2023;56:2923-69. [Crossref]
- Croitoru FA, Hondru V, Ionescu RT, Shah M. Diffusion Models in Vision: A Survey. IEEE Trans Pattern Anal Mach Intell 2023;45:10850-69. [Crossref] [PubMed]
- Yang L, Zhang Z, Song Y, Hong S, Xu R, Zhao Y, Zhang W, Cui B, Yang MH. Diffusion Models: A Comprehensive Survey of Methods and Applications. ACM Comput Surv 2024;56:1-39. [Crossref]
- Wolleb J, Sandkühler R, Bieder F, Valmaggia P, Cattin PC. Diffusion Models for Implicit Image Segmentation Ensembles. In: Wolleb J, Sandkühler R, Bieder F et al., editors. Proceedings of The 5th International Conference on Medical Imaging with Deep Learning; Proceedings of Machine Learning Research: PMLR; 2022. p. 1336-1348.
- Wu J, Fu R, Fang H, Zhang Y, Yang Y, Xiong H, Liu H, Xu Y. MedSegDiff: Medical Image Segmentation with Diffusion Probabilistic Model. Medical Imaging with Deep Learning. Proceedings of Machine Learning Research 2024;227:1623-39.
- Wu J, Ji W, Fu H, Xu M, Jin Y, Xu Y. MedSegDiff-V2: Diffusion-Based Medical Image Segmentation with Transformer. Proceedings of the AAAI Conference on Artificial Intelligence 2024;38:6030-8. [Crossref]
- Bozorgpour A, Sadegheih Y, Kazerouni A, Azad R, Merhof D. DermoSegDiff: A Boundary-Aware Segmentation Diffusion Model for Skin Lesion Delineation. Predictive Intelligence in Medicine; 2023. Cham: Springer Nature Switzerland, 2023.
- Chen T, Wang C, Shan H. BerDiff: Conditional Bernoulli Diffusion Model for Medical Image Segmentation. In: Greenspan H. Medical Image Computing and Computer Assisted Intervention - MICCAI 2023. Cham: Springer Nature Switzerland, 2023.
- Xing Z, Wan L, Fu H, Yang G, Zhu L. Diff-UNet: A Diffusion Embedded Network for Volumetric Segmentation. [Preprint]. 2023 [cited 2025 April 17]. Available online: https://arxiv.org/abs/2303.10326
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans Med Imaging 2015;34:1993-2024. [Crossref] [PubMed]
- Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 2017;4:170117. [Crossref] [PubMed]
- Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the BRATS Challenge. [Preprint]. 2018 [cited 2025 April 17]. Available online: https://arxiv.org/abs/1811.02629
- Baid U, Ghodasara S, Mohan S, Bilello M, Calabrese E, Colak E, et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. [Preprint]. 2021 [cited 2025 April 17]. Available online: https://arxiv.org/abs/2107.02314
- Jiang Y, Zhang Y, Lin X, Dong J, Cheng T, Liang J. SwinBTS: A Method for 3D Multimodal Brain Tumor Segmentation Using Swin Transformer. Brain Sci 2022;12:797. [Crossref] [PubMed]
- Ding W, Geng S, Wang H, Huang J, Zhou T. FDiff-Fusion: Denoising diffusion fusion network based on fuzzy learning for 3D medical image segmentation. Inf Fusion 2024;112:102540. [Crossref]



