
Multitask Swin Transformer for classification and characterization of pulmonary nodules in CT images
Introduction
Pulmonary cancer is one of the most prevalent and deadliest types of cancer, presenting significant challenges for prevention and treatment (1). Early detection and diagnosis of benign and malignant pulmonary nodules can significantly improve patient survival rates (2). Computed tomography (CT) is the primary tool to detect pulmonary nodules, leveraging the varying transmission of X-rays in multiple directions to generate cross-sectional or 3D images through computer processing (3). Radiologists use the size, calcification, interstitial structure, and texture of nodules on CT images to differentiate between benign and malignant cases (4). However, this diagnostic process is complex, requiring a comprehensive assessment of multiple features such as nodule size and morphology. Additionally, cognitive variability and physician fatigue can limit their ability to consistently interpret CT images, increasing the risk of misdiagnosis (5).
Computer-aided diagnosis (CAD) systems utilize computer technology to address medical diagnostic challenges. For pulmonary nodule diagnosis, CAD systems employ digital image processing to analyze medical images and evaluate nodule malignancy (6). Moreover, CAD systems can process large volumes of images rapidly, enhancing diagnostic efficiency and providing decision support (5). Machine learning algorithms are essential to the performance of CAD systems (7). Conventional machine learning methods have been applied to CAD systems; for example, de Carvalho Filho et al. (8) used a distance-based phylogenetic diversity index as a texture descriptor for pulmonary nodule classification with support vector machine (SVM) and genetic algorithms for benign-malignant classification. Wang et al. (9) developed a feature model for CT nodule regions and applied a semi-supervised extreme learning machine (SS-ELM) for classification. However, these early CAD approaches faced limitations such as small datasets and the need for manual feature extraction.
Deep-learning algorithms offer several advantages over traditional machine learning, such as automatic feature extraction, handling large-scale data, robustness, and managing nonlinear relationships (7). Hua et al. (10) were the first to apply deep learning for the benign-malignant classification of pulmonary nodules, achieving sensitivities of 73.4% and 73.3% and specificities of 82.2% and 78.7% using deep belief networks (DBN) and convolutional neural networks (CNN), respectively. Lin et al. (11) employed a hierarchical semantic CNN for multilevel benign-malignant classification, achieving 74% accuracy. Sahu et al. (12) used a lightweight multisection CNN for binary classification and reported 93.18% accuracy. Masood et al. (13) created a deep, fully convolutional network (DFCNet)-based CAD system that achieved 84.2% accuracy. In summary, deep learning has shown broad applicability and improved diagnostic support in CAD systems. While CNNs are the primary deep learning method used, some studies have also explored feedforward neural networks (FNNs) and generative adversarial networks (GANs) (14,15).
Owing to the ongoing advancements in deep learning algorithms, Transformer models have garnered increasing attention in recent years. In 2017, Vaswani et al. (16) proposed the Transformer model, an end-to-end sequential processing model based on a self-attention mechanism. Transformer models are widely used in machine translation, text generation, and speech recognition applications (17). In 2017, Google introduced the Google Neural Machine Translation (GNMT) system based on the Transformer model that significantly improved translation accuracy (18). OpenAI’s GPT-4 model is an excellent text-generating model (19). A speech recognition system based on Google’s Transformer model, Starting Transducer, was released in 2019. Compared to conventional speech recognition models, transducers have better accuracy and efficiency (20).
The Swin Transformer, proposed by Liu et al. (21), is an enhanced version of the Transformer that excels in image recognition tasks. Wang et al. (22) introduced a deep learning model that integrated a Swin Transformer and a graph convolutional network (GCN) to extract image features and learn label dependencies for multilabel image recognition, resulting in better convergence efficiency and classification performance compared to CNN models. Zhao et al. (23) developed a self-supervised Swin Transformer model for music classification that learned meaningful representations from large amounts of unlabeled music data, outperforming existing models in both music-type classification and tagging tasks. Ayas et al. (24) applied the Swin Transformer model to hyperspectral image classification and demonstrated improved performance over existing models in both quantitative and visual evaluations. Swin Transformers have gradually found applications in CAD (17). Iqbal et al. (25) developed a CAD system for breast tumor segmentation and classification using a Swin Transformer network, achieving an area under the curve (AUC) of 0.944 for breast tumor classification. Huang et al. (17) combined a Swin Transformer with 2D convolutional layers to reconstruct magnetic resonance imaging (MRI) images, resulting in higher-quality reconstruction compared to CNNs. Given its strong performance in image classification, the application of the Swin Transformer has been extended across various fields, including some CAD areas. However, its use in pulmonary nodule classification CAD is still limited.
In this study, we propose a multitask Swin Transformer (MTST) for classifying benign and malignant pulmonary nodules, which outputs nodule features as classification criteria. The pulmonary nodule images extracted from the CT images were input into a U-Net GAN for data augmentation. The resulting images and the images obtained from conventional data augmentation were used to train the model. After the nodule-centered images were input to the MTST, the feature maps were simultaneously output for benign and malignant nodule classification and nodule feature prediction through the shared layer and different task layers of the model. The entire model was trained by minimizing the weighted sum of each task loss function.
The contributions of this paper are threefold:
- Considering most medical datasets suffer from imbalances in benign and malignant sample proportions, significant data structure variability, and long data acquisition periods (26), a U-Net GAN was used in this study to generate images of pulmonary nodule sections to effectively expand the training dataset and improve the model’s generalization ability. This approach is beneficial for training complex deep-learning models on limited datasets for benign and malignant nodule diagnosis.
- Considering the excellent performance of the Swin Transformer model in image classification, this study applies the Swin Transformer model to the classification of pulmonary nodules. When classifying pulmonary nodules, owing to the different sizes and shapes of pulmonary nodules, the conventional CNN model may suffer from problems, such as information loss and ignoring the correlation between the local and the whole. In contrast, the Swin Transformer, built on the shifted window multi-head self-attention mechanism, effectively captures nodule features, improving classification accuracy.
- An interpretable network structure was designed using a multitask learning approach, along with a loss function for joint task layer training. This framework outputs benign and malignant classification results and nodule characteristics essential for clinical diagnosis. This dual-output system aids clinicians by providing classification results and additional diagnostic context through detailed nodule characteristics.
We present this article in accordance with the TRIPOD + AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1619/rc).
Methods
Study framework
In this study, we propose an image generation and multitask learning-based method for assessing benign and malignant pulmonary nodules. This approach aims to enhance the accuracy of pulmonary nodule classification through data augmentation and multitasking learning. Figure 1 presents the overall research framework.

As shown in Figure 1, the research framework comprises of data augmentation part and MTST model part. Following data augmentation using both image generation methods and conventional data augmentation techniques, pulmonary nodule images are fed into the shared layer of the MTST model. The multitask evaluation module outputs multiple nodule features, including classifications of benign and malignant.
Data augmentation based on image generation method
In this study, a U-Net GAN was used to enhance the dataset. This network has demonstrated strong performance in medical image reconstruction and generation; its structural diagram is depicted in Figure 2 (27).

Figure 2 illustrates the U-Net GAN structure, which comprises a generator and a discriminator. The generator synthesizes images from random noise, while the discriminator aims to differentiate between generated and real images. By continuously optimizing the adversarial interaction between the generator and the discriminator, the GAN can produce high-quality images. However, because training the U-Net GAN directly on 64×64 nodule images yielded suboptimal performance, the nodule images were upscaled to 512×512 pixels before input into the U-Net GAN for training.
In the U-Net GAN used in this study, the generator is composed of a U-Net network, as shown in Figure 3.
As depicted in Figure 3, the generator uses a U-Net architecture and employs skip connections to link the convolutional feature maps in the downsampling layers to those in the upsampling layers. These skip connections preserve low-level features such as edges and spots from the initial feature maps. This design choice enables the model to effectively capture both local and global information in the input images, resulting in accurate and robust outcomes.
The structure of the discriminator is shown in Figure 4.
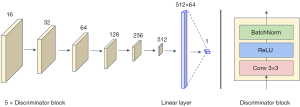
Figure 4 demonstrates that the discriminator consists of five discriminator blocks, each of which contains a 3×3 convolution layer, a ReLU activation function, and BatchNorm layer. The number of channels in the discriminator doubles with each successive block. Finally, the discriminant results were obtained using the linear layer.
Swin Transformer layer
As shown in Figure 1, the backbone model used in this study mainly consisted of multiple Swin Transformer layers. Each Swin Transformer layer includes Patch Merging and a Swin Transformer block.
Patch merging
The patch-merging operation is used for downsampling. After each patch-merging operation, the width and height of the feature map are halved, while the number of channels is doubled. Specifically, downsampling is accomplished by selecting every alternate element in both the row and column directions to create four patches. These patches are then reassembled into single tensors. The resulting tensor has a channel dimension (C) four times larger than that of the original feature map, with the height (H) and width (W) each reduced by half. Finally, a fully connected layer is applied to adjust the channel dimension C to double its original size.
Swin Transformer block
The Swin Transformer block comprises a LayerNorm (LN) layer, a (Shifted) window multi-head self-attention (W-MSA or SW-MSA) layer, and a 2-layer multi-layer perceptron (MLP) with GELU non-linearity in between. Each Swin Transformer block includes two residual connections. Additionally, W-MSA and SW-MSA are alternately used in pairs of two Swin Transformer blocks. The specific structure is shown in Figure 5.

Each pair of Swin Transformer blocks is defined as follows:
In Eq. [1], the Swin Transformer blocks with W-MSA output features as , whereas those with SW-MSA output features as .
(S)W-MSA
The standard Transformer module uses MSA to compute global self-attention across all tokens, operating over the entire image space. The computational complexity of the MSA is as follows:
(S)W-MSA computes the self-attention for each window. First, a feature map of size is divided into non-overlapping windows of size , and self-attention is then computed for each window. The computational complexity of the (S)W-MSA is as follows:
In Eq. [3], (S)W-MSA only computes self-attention for a portion within each window. Compared to Eq. [2], where the computational complexity scales quadratically with the product of the width and height, the complexity of (S)W-MSA scales linearly, indicating lower computational requirements. However, if window partitioning remains static and the feature map windows do not change, information between different windows can become disconnected. Therefore, shifting the windows is necessary to facilitate inter-window communication.
In (S)W-MSA, for each non-overlapping window , self-attention is computed with , where are the shared projection matrices across all windows. Query Q, key K, value V, and learnable relative position encoding B (learnable relative position encoding B, feature map size is ) were used to compute the local self-attention within each window, as follows:
Multitask learning model
The MTST model employs a multitasking learning method for hard parameter sharing, and the overall architecture is shown in Figure 6.
As shown in Figure 6, the shared model layer consists of a convolutional layer, Patch Embedding, and multiple serial Patch Merging and Swin Transformer Blocks. This constitutes the shared layer of the multi-task model, where the input image is represented as , with H, W, and C denoting the height, width, and number of channels of the image, respectively. The image passes through the shared layer of the model, resulting in the output feature map . This can be expressed mathematically as:
After passing through the shared layer, the feature maps are directed to the specific task layers, each composed of multiple Swin Transformer blocks for the nine nodule features. Ultimately, the model outputs evaluation values for these nine nodule features, which can be represented as:
where each represents a specific task’s Swin Transformer Block branch, and denotes the prediction results for the t-th task. These tasks include classifications for features such as Malignancy, Subtlety, Internal Structure, Calcification, Sphericity, Margin, Spiculation, Texture, and Lobulation.
The entire model can be represented by the following formula:
The loss function adopted for training the nine specific task layers of the MTST is cross-entropy loss (). To each specific task layer loss function multiplied by the weighting sum, to get the total loss function (L), the formula is as follows:
In Eq. [8], represents the weight of each feature (including benign and malignant cases), and is the cross-entropy loss for a specific task layer corresponding to each feature. The weight values used in this study were adjusted based on experience and guided by the settings outlined in Marques et al.’s study (28). The weights for each feature were set as follows: 1.4 for malignancy, 1.8 for sphericity, 1.2 for calcification, 0.9 for margin, 0.7 for lobulation, spiculation, and subtlety, and 1 for all other features.
Experiment
Dataset
The Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI) dataset was used in this study (29). This publicly available dataset contains 1,018 low-dose lung CT cases and can be accessed online. The CT images are provided in DCM format, with each case accompanied by an annotation file in XML format. The LIDC-IDRI dataset was annotated through a combination of manual blind and non-blind readings. During the blind reading phase, each radiologist independently reviewed the CT scans. In the subsequent unblinded reading phase, each radiologist independently reviewed their own markers alongside the anonymous markers from three other radiologists to form a final opinion.
The annotations in this dataset include information such as the coordinates and quantitative scores for characteristics of pulmonary nodules larger than 3 mm in diameter, including their degree of malignancy. The dataset provides detailed characteristics and ratings, such as malignancy, subtlety, internal structure, calcification, sphericity, margin, spiculation, texture, and lobulation. Apart from calcification (graded 1–6) and Internal Structure (graded 1–4), each characteristic is rated on a scale from 1 to 5. In this dataset, nodule malignancy is classified as Highly Unlikely [1], Moderately Unlikely [2], Indeterminate [3], Moderately Suspicious [4], or Highly Suspicious [5]. To ensure consistency in evaluation and model comparison, similar to previous studies, we excluded nodules with an indeterminate malignancy score [3] since these nodules do not provide definitive benign or malignant information (12,30).
Data preprocessing
Nodule screening
Benign and malignant nodules were extracted based on annotations provided by the LIDC-IDRI dataset (Figure 7).
As shown in Figure 7, the nodule extraction involved four steps.
- Step 1: after converting the CT image format from DCM to JPG, the pulmonary parenchyma was separated from the image. In this study, images of pulmonary nodules were used as training data. Segmenting the pulmonary parenchyma ensured that the nodule images did not contain other pulmonary tissues.
- Step 2: using the nodule coordinates provided by the XML file annotations in the dataset, nodules with a diameter between 3 mm and 30 mm were extracted. The reason for selecting this range is that the database includes nodules sized from 3 mm to 30 mm (29).
- Step 3: annotations for all features of the extracted pulmonary nodules, including benign and malignant classifications, were obtained.
- Step 4: each nodule and its corresponding features were sorted and converted into labels that could be input into the model.
Data augmentation and data splitting
From the extracted nodules, we selected 3,157 nodules with an average malignancy of 1, 2, 4, and 5 as the training set and then conducted data augmentation on these data. First, we used conventional data augmentation methods, including flipping, rotating, and adjusting the brightness, contrast, and image scaling, and added enhanced data to the training set. After preprocessing, the augmented data were input into the U-Net GAN to generate additional training data. The training set was subsequently divided into five equal parts, with four parts used as training sets and one part as a validation set. To compare the effects of data augmentation, we retained data without data augmentation. Among the nodule images outside the training set, 1,600 pulmonary nodule images were selected as the test set. The specific divisions of the experimental data are listed in Table 1.
Table 1
| Malignancy level | Training (80%) and validation (20%) set | Testing set | |||
|---|---|---|---|---|---|
| Initial numbers | Conventional augmentation | Image generation | Total | ||
| 1 | 1,002 | 1,013 | 985 | 3,000 | 400 |
| 2 | 756 | 823 | 1,421 | 3,000 | 400 |
| 3 | – | – | – | – | – |
| 4 | 634 | 712 | 1,654 | 3,000 | 400 |
| 5 | 764 | 823 | 1,413 | 3,000 | 400 |
| Total | 3,156 | 3,371 | 5,473 | 12,000 | 1,600 |
As shown in Table 1, nodules with a malignancy level of 3 were excluded from this study. After data augmentation in the training and test sets, the number of nodules at each malignancy level reached 3,000. In addition, image generation using the U-Net GAN is based on only four types of benign and malignant nodules and does not include other image features. Therefore, the image generation results are mainly applied to the classification of benign and malignant nodules and cannot be used for the output of low-level features of pulmonary nodules. When the generated images were used to train the classification of nodule malignancy, the weights of the low-level feature classification module were frozen.
Experimental setting
For the GAN model based on U-Net, the Adam optimizer was used to update all parameters in the network, and the learning rate was set to 5×10−4. The training was repeated four times, each time focusing solely on generating pulmonary nodule images with varying degrees of malignancy.
Transfer learning techniques were employed for the MTST model. We initialized the parameters of the feature extraction module in our network using parameters from the shared layer pretrained on the ImageNet dataset. The Adam optimizer was used to update all parameters within the network. The learning rate of the model was initially set at 3×10−6. The model was trained for 50 epochs with a batch size of eight. Five experiments were conducted, and the final results were calculated as the average of these five trials.
The model training process was conducted using PyTorch on a computer equipped with an R7 4800H CPU, 16 GB of RAM, and an RTX 2060 GPU with 6 GB of memory.
Results
Image generation
The U-Net GAN model was trained for image generation, and the generated images are shown in Figure 8.
As shown in Figure 8, the pulmonary nodule image generated by the U-Net GAN is highly similar to the original nodule image. The generated images and those obtained using other data augmentation methods were sorted into the training data.
Feature evaluation results
There are notable differences in the classification of each feature. Calcification was categorized into six classes, while malignancy and subtlety were divided into five classes, internal structure was divided into four classes, sphericity and texture were divided into three classes, and the remaining characteristics were divided into two classes. To compare our model’s performance with other studies, we employed the absolute distance error metric to evaluate prediction results, defined as follows (31):
In Eq. [9], n is the total number of samples, represents the predicted value of a certain feature for nodule i, and is the ground-truth value of that feature for nodule i. We compared our prediction results with those from previous feature evaluation models, as shown in Table 2.
Table 2
| Models | Sub | Int | Cal | Sph | Mar | Spi | Tex | Lob |
|---|---|---|---|---|---|---|---|---|
| MTST (ours) | 0.67 | 0.03 | 0.52 | 0.41 | 0.36 | 0.45 | 0.26 | 0.37 |
| MTR (32) | 0.75 | 0.04 | 0.48 | 0.81 | 0.86 | 0.8 | 0.58 | 0.87 |
| LASSO (32) | 1.25 | 0.02 | 2.18 | 1.25 | 1.13 | 0.89 | 1.04 | 0.95 |
| EN (32) | 1.2 | 0.14 | 1.44 | 1.09 | 0.98 | 0.86 | 1.24 | 0.96 |
| MTMR-Net (31) | 0.54 | 0.03 | 0.56 | 0.59 | 0.54 | 0.49 | 0.44 | 0.54 |
MTST, multitask Swin Transformer; MTR, Multi-Task Regression model; LASSO, least absolute shrinkage and selection operator; EN, Elastic Net; MTMR-Net, Multi-Task deep model with Margin Ranking loss for Lung Nodule Analysis; Sub, subtlety; Int, internalStructure; Cal, calcification; Sph, sphericity; Mar, margin; Spi, spiculation; Tex, texture; Lob, lobulation.
As presented in Table 2, we compared the IB, MTR, lasso regression, MTMR-Net, and EN networks. These results indicate that for most features, the absolute distance error results of the proposed MTST model are superior to those of previous studies on pulmonary nodule feature evaluation. Among these feature evaluation tasks, the “Internal Structure” prediction was particularly accurate. This can be attributed to the significant imbalance in the dataset for the “Internal Structure” feature, where most nodules were rated with a score of 1, implying that this feature had a minimal impact on the malignancy of the nodules.
Benign and malignant binary classification
The binary classification of benign and malignant nodules is a widely explored topic in current research on pulmonary nodule classification (14,15,26). For this binary classification task, nodules classified as Highly Unlikely [1] and Moderately Unlikely [2] in terms of malignancy were considered benign, whereas nodules rated as Moderately Suspicious [4] and Highly Suspicious [5] were considered malignant.
While accuracy is a primary evaluation metric for classification models, relying solely on it may not provide a complete evaluation of model performance. Additional metrics such as accuracy (ACC), sensitivity (SEN), specificity (SPE), and the area under the receiver operating characteristic curve (AUC) were used to offer a comprehensive assessment. The ACC, SEN, and SPE metrics are defined as follows:
AUC values range from 0.5 to 1.0, where values closer to 1.0 indicate higher authenticity of the classification method. An AUC value of 0.5 represents the lowest authenticity and holds no practical value.
The MTST model was also tested using different training data based on the standard evaluation metrics for the binary classification models mentioned above. The results of the binary classification experiment are listed in Table 3.
Table 3
| Data augmentation | ACC (%) | SEN (%) | AUC | SPE (%) |
|---|---|---|---|---|
| Conventional + U-Net GAN generated | 93.74 | 91.55 | 0.985 | 96.09 |
| Conventional | 93.21 | 89.67 | 0.981 | 95.36 |
| None | 89.58 | 86.2 | 0.969 | 92.13 |
U-Net GAN, U-shaped network generative adversarial network; ACC, accuracy; SEN, sensitivity; AUC, area under the curve; SPE, specificity.
As shown in Table 3, models trained with both conventional data augmentation and image generation outperformed those trained solely with conventional data augmentation or without any augmentation, demonstrating the significant impact of image generation on enhancing medical image datasets.
In the training process of the MTST, Figure 9 shows the training loss and accuracy as well as the change function of the epoch.
As shown in Figure 9, the training and validation losses were almost identical and approached zero, and the difference in accuracy between training and validation was minimal. From Figure 9A,9B, it can be concluded that the proposed model did not overfit the training dataset.
CT images of pulmonary nodules as good/poor examples of the model’s prediction As shown in Figure 10.

To further assess the performance of the MTST model in benign and malignant binary classification tasks, we compared it to other state-of-the-art deep learning-based binary classification systems. The comparison results, based on the same dataset (LIDC-IDRI) and similar test data sizes (more than 1,000 nodule images in the test set), are displayed in Table 4.
Table 4
| CADx system | Training/validation/test set | ACC (%) | SEN (%) | AUC | SPE (%) |
|---|---|---|---|---|---|
| MT-Swin Transformer (our) | 9,600/2,400/1,600 nodules | 93.74 | 91.55 | 0.985 | 96.09 |
| Sahu et al. (12) | 1,174/NA/131 nodules | 93.18 | 89.4 | 0.98 | 95.61 |
| Jiang et al. (33) | 1,004 nodules are used | 90.24 | 92.04 | 0.933 | – |
| Masood et al. (13) | 465,504/NA/1,700 nodules | 86.02 | 89.01 | – | 83.2 |
| Shen et al. (34) | 88,948 nodules are used | – | – | 0.899±0.018 | – |
| Sun et al. (35) | 2,126/1,063/1,063 nodules | 84.2 | 70.5 | 0.856 | 88.9 |
MT-Swin Transformer, multitask Swin Transformer; CADx, computer-aided diagnosis; U-Net GAN, U-shaped network generative adversarial network; ACC, accuracy; SEN, sensitivity; AUC, area under the curve; SPE, specificity.
As shown in Table 4, under the same dataset and similar test data size, the accuracy, sensitivity, AUC, and specificity of our model are superior to those of previous studies on the benign and malignant binary classification of pulmonary nodules.
Benign and malignant multilevel classification
In the clinical diagnosis of pulmonary nodules, it is essential not only to classify nodules as benign or malignant but also to further distinguish the degree of malignancy. This section investigates the multilevel classification of benign and malignant pulmonary nodules, with classification labels defined as follows: Highly Unlikely [1], Moderately Unlikely [2], Moderately Suspicious [4], and Highly Suspicious [5].
To evaluate multilevel classification results, previous studies have employed the off-by-one accuracy index, as represented by the following Eq. [11]:
In Eq. [11], n represents the total number of samples, represents the predicted malignancy level for nodule i, and represents the ground-truth malignancy value for nodule i.
Following the approach used for binary classification, we tested the MTST model with different parameters for multilevel classification. The results of this experiment are presented in Table 5.
Table 5
| CADx system | Training/validation/test set | Off-by-one accuracy (%) |
|---|---|---|
| MT-Swin Transformer (ours) | 48,000/12,000/1,600 images | 95.73 |
| Sahu et al. (12) | 1,174/NA/131 nodules | 93.79 |
| Hussein et al. (36) (multitask learning) | 1,029/NA/115 nodules | 91.26 |
| Hussein et al. (37) | 1,030/NA/115 nodules | 82.47 |
| Buty et al. (38) | 5,155 images are used | 82.4 |
| Hussein et al. (36) | 1,029/NA/115 nodules | 80.08 |
CADx, computer-aided diagnosis; MT-Swin Transformer, multitask Swin Transformer.
As shown in Table 5, our model achieved a higher off-by-one accuracy in the multi-classification task, outperforming previous related studies.
Discussion
This study generates pulmonary nodule images based on deep learning and combines traditional data augmentation methods to significantly improve the quality of the training set. The proposed MTST model outperforms traditional machine learning models in tasks such as nodule feature evaluation, binary classification of nodule malignancy, and multilevel classification.
In the aspect of pulmonary nodule image generation, deep learning models can generate new data by learning from existing datasets. In the context of data augmentation, these networks can be used to generate new images similar to existing images but exhibiting unique variations, thus increasing the diversity of the dataset (39-41). In this study, the U-Net GAN was utilized for image generation to expand the training dataset and improve the model’s generalization ability. The U-Net is renowned for its strong performance in medical image segmentation and is widely used in the field (42). From the perspective of model complexity, U-Net uses a small convolution kernel and few parameters to train the model faster, and its model complexity is low. Additionally, U-Net uses multilevel feature extraction to retain more details of the original image, and a GAN based on U-Net can generate more realistic pulmonary nodules images. Although data augmentation addresses the issue of limited annotated data in medical training sets, which can hinder the development of complex deep learning models, it is crucial to carefully review newly generated images. The use of an inappropriate image generation model may result in unrealistic or meaningless images.
In the aspect of nodule feature evaluation, although current computer-aided diagnostic methods achieve excellent results in classifying benign and malignant nodules, it remains challenging to interpret the rationale behind these classifications (26). This limitation often necessitates doctors to rely on their expertise to assess the characteristics of pulmonary nodules on CT images. Therefore, a computer-aided diagnostic system capable of accurately evaluating benign and malignant nodules while simultaneously providing information on other nodule characteristics (such as morphology, texture, and density) holds significant potential. Such a system can supply detailed nodule characteristics to assist doctors, enhance diagnostic efficiency, and reduce the risk of misdiagnosis.
In the aspect of binary classification of nodule malignancy, in previous studies, CNNs were predominantly used for the binary classification of benign and malignant pulmonary nodules. Sahu et al. (12) proposed a lightweight multisection CNN architecture that, after processing the nodules, obtained multiple views of the nodules from different perspectives. This model aggregated the information through a view-pooling layer to encode the nodule’s volume information into a compact representation. Jiang et al. (33) developed an attentive ensemble 3D dual-path network for pulmonary nodule classification. This network structure incorporated a context attention mechanism to simulate the correlation between adjacent positions, enhancing the representativeness of deep features. Additionally, the network employed a spatial attention mechanism to automatically identify regions crucial for nodule classification. These studies achieved accuracy rates exceeding 90% for the binary classification of benign and malignant pulmonary nodules.
In this study, we utilized a Swin Transformer to construct a feature-extraction module. This model can better focus on important regions and features of the nodule image and has high accuracy and robustness. Our model outperforms previous CNN studies in terms of both benign and malignant binary classification accuracy (93.24%) and feature evaluation. Possible reasons for the superior image classification performance of our Swin Transformer-based model compared to CNN models include the following: Due to the limited size of convolutional kernels in CNNs, fine details and edge information in the image can be missed. Additionally, pooling layers in CNNs can result in information loss and overlook the correlation between local and global features (17,25). Conversely, the Swin Transformer model incorporates innovative techniques such as W-MSA and cross-stage feature integration, enabling it to better capture both global and local information within the image and process a large number of features more effectively (17,25). However, Swin Transformers may still face challenges with position information encoding and fixed window sizes (17,25).
In the aspect of multilevel classification, Sahu et al. (12) proposed a lightweight multisection CNN architecture that explored pulmonary nodule multilevel alongside classification. Hussein et al. (36) developed a CADx system using a 3D CNN that employed volumetric CT data to generate 3D models of nodules, preserving more information. This system employs volumetric data from CT scans to generate 3D models of nodules, thereby preserving more nodule information. This system extracted relevant feature representations for six different nodule features and combined them through multitask learning. Transfer learning was utilized to obtain highly discriminative features to mitigate the substantial data requirements of CNNs. Hussein et al. (37) also introduced an end-to-end trainable multi-view deep CNN that used median intensity projection to create 2D patches for each dimension, forming a three-channel input tensor. Finally, the study used a trained network to extract features from the input image and Gaussian process regression to obtain the malignancy score. Buty et al. (38) proposed a CADx system based on a CNN that can quantitatively evaluate differences in appearance and 3D surface changes. The system first modeled and parameterized nodule shapes using spherical harmonics, extracted appearance features using a deep CNN, and finally estimated the malignancy of the nodules using a random forest classifier.
As indicated by these research outcomes, previous studies on the multilevel classification of pulmonary nodules have predominantly focused on CNNs. The optimization of CNN models has progressively improved accuracy from 82.40% to 93.79%. In this study, the Swin Transformer was employed for the multilevel classification of pulmonary nodules, achieving an accuracy of 95.73%, surpassing that of previous CNN-based models. In an overview of the current research on pulmonary nodule classification, the research on pulmonary nodule classification pays relatively more attention to the binary classification of benign and malignant nodules, and relatively few studies focus on multiple classifications. However, from the perspective of the clinical application of CAD, the multilevel classification of benign and malignant pulmonary nodules is in line with the requirements of clinical practice. Therefore, future research on the auxiliary diagnosis of pulmonary nodules should place greater emphasis on multilevel classification.
Despite achieving excellent results in pulmonary nodule image generation, nodule feature evaluation, binary and multilevel classification of nodule malignancy, this study still has certain limitations. The limitations of this study are as follows. First, due to limited computational resources during the experiment, a relatively simple structure was selected for the image generation model. In future work, it will be essential to develop more complex image generation models to enhance performance. Second, while the detection, segmentation, and classification of pulmonary nodules form a comprehensive process to assist doctors in diagnosis, this study focused solely on classification. Future research should aim to integrate detection and segmentation procedures to create a more complete diagnostic tool. Thirdly, this study used data from the LIDC and the IDRI. However, the nodules in these databases are assessed based on the “malignancy probability” evaluated by experienced radiologists, rather than confirmed benign or malignant results from pathology or follow-up. Consequently, this may affect the primary findings and conclusions of our study.
Conclusions
In this study, we proposed a Swin Transformer model integrated into a multitask learning framework for classifying benign and malignant pulmonary nodules alongside the output of nodule feature evaluation results. To enhance generalization, pulmonary nodule images generated by the U-Net GAN model were used for model training. The Swin Transformer block demonstrated exceptional feature extraction capabilities for pulmonary nodule classification. The MTST model, trained by minimizing the weighted sum of each task loss function, effectively outputs both nodule malignancy and feature evaluations simultaneously. The experimental results validate the proposed MTST model’s effectiveness in binary classification of benign and malignant nodules, multiclass classification, and feature evaluation.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Footnote
Reporting Checklist: The authors have completed the TRIPOD + AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-1619/rc
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1619/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work to ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, Miettinen OS. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med 2006;355:1763-71. [Crossref] [PubMed]
- Goldman LW. Principles of CT and CT technology. J Nucl Med Technol 2007;35:115-28; quiz 129-30. [Crossref] [PubMed]
- Gould MK, Donington J, Lynch WR, Mazzone PJ, Midthun DE, Naidich DP, Wiener RS. Evaluation of individuals with pulmonary nodules: when is it lung cancer? Diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013;143:e93S-e120S.
- Agnes SA, Anitha J. Appraisal of Deep-Learning Techniques on Computer-Aided Lung Cancer Diagnosis with Computed Tomography Screening. J Med Phys 2020;45:98-106. [Crossref] [PubMed]
- Doi K. Diagnostic imaging over the last 50 years: research and development in medical imaging science and technology. Phys Med Biol 2006;51:R5-27. [Crossref] [PubMed]
- Yanase J, Triantaphyllou E. A systematic survey of computer-aided diagnosis in medicine: Past and present developments. Expert Syst Appl 2019;138:112821.
- de Carvalho Filho AO, Silva AC, Cardoso de Paiva A, Nunes RA, Gattass M. Computer-Aided Diagnosis of Lung Nodules in Computed Tomography by Using Phylogenetic Diversity, Genetic Algorithm, and SVM. J Digit Imaging 2017;30:812-22. [Crossref] [PubMed]
- Wang Z, Xin J, Sun P, Lin Z, Yao Y, Gao X. Improved lung nodule diagnosis accuracy using lung CT images with uncertain class. Comput Methods Programs Biomed 2018;162:197-209. [Crossref] [PubMed]
- Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [Crossref] [PubMed]
- Lin Y, Wei L, Han SX, Aberle DR, Hsu W. EDICNet: An end-to-end detection and interpretable malignancy classification network for pulmonary nodules in computed tomography. Proc SPIE Int Soc Opt Eng 2020;11314:113141H.
- Sahu P, Yu D, Dasari M, Hou F, Qin H. A Lightweight Multi-Section CNN for Lung Nodule Classification and Malignancy Estimation. IEEE J Biomed Health Inform 2019;23:960-8. [Crossref] [PubMed]
- Masood A, Sheng B, Li P, Hou X, Wei X, Qin J, Feng D. Computer-Assisted Decision Support System in Pulmonary Cancer detection and stage classification on CT images. J Biomed Inform 2018;79:117-28. [Crossref] [PubMed]
- Surendar P. Diagnosis of lung cancer using hybrid deep neural network with adaptive sine cosine crow search algorithm. J Comput Sci 2021;53:101374.
- Zhu H, Han G, Peng Y, Zhang W, Lin C, Zhao H. Functional-realistic CT image super-resolution for early-stage pulmonary nodule detection. Future Generation Computer Systems 2021;115:475-85.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Proceedings of the 31st International Conference on Neural Information Processing Systems; 2017:6000-10.
- Huang J, Fang Y, Wu Y, Wu H, Gao Z, Li Y, Del Ser J, Xia J, Yang G. Swin transformer for fast MRI. Neurocomputing 2022;493:281-304.
- Wu Y, Schuster M, Chen Z, Le QV, Norouzi M, Macherey W, et al. Google's neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv:160908144. 2016.
- Zhang C, Zhang C, Zheng S, Qiao Y, Li C, Zhang M, Dam SK, Thwal CM, Tun YL, Huy LL, kim D, Bae SH, Lee LH, Yang Y, Shen HT, Kweon IS, Hong CS. A Complete Survey on Generative AI (AIGC): Is ChatGPT from GPT-4 to GPT-5 All You Need? arXiv preprint arXiv: 2303.11717. 2023.
- Zhang Q, Lu H, Sak H, Tripathi A, McDermott E, Koo S, Kumar S. Transformer transducer: A streamable speech recognition model with transformer encoders and rnn-t loss. ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2020:7829-33.
- Liu Z, Lin Y, Cao Y, Hu H, Wei Y, Zhang Z. Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada; 2021:9992-10002.
- Wang Y, Xie Y, Fan L, Hu G. STMG: Swin transformer for multi-label image recognition with graph convolution network. Neural Comput Applic 2022;34:10051-63.
- Zhao H, Zhang C, Zhu B, Ma Z, Zhang K. S3T: Self-Supervised Pre-Training with Swin Transformer For Music Classification. ICASSP 2022 - 2022 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Singapore, Singapore; 2022:606-10.
- Ayas S, Tunc Gormus E. SpectralSWIN: a spectral-swin transformer network for hyperspectral image classification. International Journal of Remote Sensing 2022;43:4025-44.
- Iqbal A, Sharif M. BTS-ST: Swin transformer network for segmentation and classification of multimodality breast cancer images. Knowledge-Based Systems 2023;267:110393.
- Gu Y, Chi J, Liu J, Yang L, Zhang B, Yu D, Zhao Y, Lu X. A survey of computer-aided diagnosis of lung nodules from CT scans using deep learning. Comput Biol Med 2021;137:104806. [Crossref] [PubMed]
- Dong X, Lei Y, Wang T, Thomas M, Tang L, Curran WJ, Liu T, Yang X. Automatic multiorgan segmentation in thorax CT images using U-net-GAN. Med Phys 2019;46:2157-68. [Crossref] [PubMed]
- Marques S, Schiavo F, Ferreira CA, Pedrosa J, Cunha A, Campilho A. A multi-task CNN approach for lung nodule malignancy classification and characterization. Expert Syst Appl 2021;184:115469.
- Armato SG 3rd, Zhao B, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 2011;38:915-31. [Crossref] [PubMed]
- Liu H, Cao H, Song E, Ma G, Xu X, Jin R, Liu C, Hung CC. Multi-model Ensemble Learning Architecture Based on 3D CNN for Lung Nodule Malignancy Suspiciousness Classification. J Digit Imaging 2020;33:1242-56. [Crossref] [PubMed]
- Liu L, Dou Q, Chen H, Qin J, Heng PA. Multi-Task Deep Model With Margin Ranking Loss for Lung Nodule Analysis. IEEE Trans Med Imaging 2020;39:718-28. [Crossref] [PubMed]
- Chen S, Ni D, Qin J, Lei B, Wang T, Cheng JZ. Bridging computational features toward multiple semantic features with multi-task regression: A study of CT pulmonary nodules. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, October 17-21, 2016, Proceedings, Part II 19. Springer; 2016.
- Jiang H, Gao F, Xu X, Huang F, Zhu S. Attentive and ensemble 3D dual path networks for pulmonary nodules classification. Neurocomputing 2020;398:422-30.
- Shen S, Han SX, Aberle DR, Bui AA, Hsu W. An Interpretable Deep Hierarchical Semantic Convolutional Neural Network for Lung Nodule Malignancy Classification. Expert Syst Appl 2019;128:84-95. [Crossref] [PubMed]
- Sun W, Zheng B, Qian W. Automatic feature learning using multichannel ROI based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med 2017;89:530-9. [Crossref] [PubMed]
- Hussein S, Cao K, Song Q, Bagci U. Risk stratification of lung nodules using 3D CNN-based multi-task learning. Information Processing in Medical Imaging: 25th International Conference, IPMI 2017, Boone, NC, USA, June 25-30, 2017, Proceedings 25. Springer; 2017.
- Hussein S, Gillies R, Cao K, Song Q, Bagci U. TumorNet: Lung nodule characterization using multi-view convolutional neural network with gaussian process. 2017 IEEE 14th International Symposium on Biomedical Imaging (ISBI 2017), Melbourne, VIC, Australia; 2017:1007-10.
- Buty M, Xu Z, Gao M, Bagci U, Wu A, Mollura DJ. Characterization of lung nodule malignancy using hybrid shape and appearance features. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2016: 19th International Conference, Athens, Greece, October 17-21, 2016, Proceedings, Part I 19. Springer; 2016.
- Han C, Kitamura Y, Kudo A, Ichinose A, Rundo L, Furukawa Y, Umemoto K, Li Y, Nakayama H. Synthesizing Diverse Lung Nodules Wherever Massively: 3D Multi-Conditional GAN-Based CT Image Augmentation for Object Detection. 2019 International Conference on 3D Vision (3DV), Quebec City, QC, Canada; 2019:729-37.
- Shi H, Lu J, Zhou Q. A novel data augmentation method using style-based GAN for robust pulmonary nodule segmentation. 2020 Chinese Control And Decision Conference (CCDC), Hefei, China; 2020:2486-91.
- Toda R, Teramoto A, Kondo M, Imaizumi K, Saito K, Fujita H. Lung cancer CT image generation from a free-form sketch using style-based pix2pix for data augmentation. Sci Rep 2022;12:12867. [Crossref] [PubMed]
- Punn NS, Agarwal S. Modality specific U-Net variants for biomedical image segmentation: a survey. Artif Intell Rev 2022;55:5845-89. [Crossref] [PubMed]







