
Enhancing bone radiology images classification through appropriate preprocessing: a deep learning and explainable artificial intelligence approach
Introduction
Deep learning techniques are now widely being implemented in the medical field, one of the more popular directions is using neural networks for medical image classification (1,2). There has been a large number of high-performance deep learning classification models for medical images, and since the emergence of the explainable artificial intelligence (XAI) concept, more and more researchers involved XAI in their work. For those works involving deep learning models and XAI techniques for medical image classification, the most straightforward approach is to achieve the best performance possible with the deep learning model, in the meantime to use XAI techniques to ensure that the model ‘learned’ the correct knowledge and made the correct classification based on that (3-5).
Current research regarding the use of deep learning for medical image recognition is thriving, many researchers focus on the design and structure of the model itself. For common image recognition, many models achieve remarkable performance (6,7). In their works, many of them performed basic standard data preprocessing steps. Yet for medical images, to say professionally, they too need ‘special treatment’ as the diseases they represent.
As we know, in common image classification tasks, an image being processed by the model will be recognized because of the object presented in the image. This means, every part of said object will support the correspondent prediction. There is a method of data augmentation process called “cropping”, used widely to increase data samples for better training and performance. Cropping is essentially segmenting an image into several partial images, and they are added under the same class as new data, thereby increasing the size of the dataset. For common images where we mentioned every part of the object supports the true label, cropping is a good way for data augmentation same as flipping, rotation, etc. But for medical images, the object is the body part in the image, the key indication of a disease is only lying on a part, even a very small fragment of this object, and the same indications are what help human radiologists to diagnose and determine the diseases from medical images. And now we can see that cropping is not a suitable data augmentation method for medical images, because once implemented, we risk having new images that do not contain key indications be classified as being ill. What we wish to point out is that medical images are different from common ones, not only because of the reason that we explained above, but also because we need to be cautious and precise when we develop methods for important medical use. Medical image data such as computed tomography (CT) scans, X-rays, and magnetic resonance imaging (MRI), currently often have high resolution, clear boundaries between body parts, also between background and object itself. We believe that it will be necessary and beneficial to process accordingly for higher accuracy and better performance.
On the other hand, with the thriving development of XAI thanks to researchers’ awakened awareness of accountability and reliability of their creations, XAI techniques have become popular among black box model research fields for validation and verification (8). Essentially even with extraordinary accuracy and performance, black box models might possibly learn the wrong knowledge and achieve through the wrong way, and XAI helps humans to interpret black box models, and thereby supervise, observe and verify.
Additionally, for critical fields such as radiology and medicine where human well beings are concerned, choosing a suitable XAI method in case-specific scenarios for the human users to appropriately understand black box models when they are involved in important decision-making processes is also a priority, as stated in the work proposed by Retzlaff et al. (9).
In this paper, we utilize one of the most trending XAI techniques—gradient-weighted class activation mapping (GradCAM), to illustrate the necessity of extending preprocessing methods for medical images specifically.
Related works
In the current research field, there have been tens of thousands of achievements regarding medical image recognition using deep learning models. Among these, lung disease is one of the most popular topics. Its trending reasons, besides the recent outbreak of coronavirus disease 2019 (COVID-19) and the immediate research demands that came with it, are because lung-related image data is extraordinarily abundant, far more than other diseases. Even before deep learning research started to thrive in the medical field, this abundance gave researchers rich materials and large space to develop. After the concept of computer-aided diagnosis (CAD) systems emerged, Hua et al. (10) proposed the combined design of convolutional neural network (CNN) and deep belief network (DBN) and illustrated satisfactory improvement in the model performance of lung cancer X-ray image classification. The Throax-Net proposed by Wang et al. (11) achieves classification of 14 lung diseases, containing two branches within the operation system—a classification branch and an attention branch—that together produce an output averaged and binarized by both operators. There is also the model proposed by Liang et al. (1), a combination design of dilated convolutional and residual structure, implemented on the classification of pediatric lung pneumonia. With a more specific range but still rich amount of chest X-ray image data, it achieves 96.7% recall and 92.7% F1-score. For the thriving research of COVID-19 diagnosis and classification, for example, the models proposed by Keles et al. (12), plainly called ‘Covid19-CNNet’ and ‘Covid19-ResNet’, achieved 97.61% and 94.28% accuracy respectively when performing multi-class classification on lung radiology image datasets with both normal cases, COVID-19 cases, and viral pneumonia cases.
As we can presume from the above works, many researchers in this field emphasize model design to achieve higher performance. Few of them pay close attention to the image data itself and the method of pre-processing before practice. For example, in the work of Salehinejad et al. (13), they utilize a generative adversarial network to generate new synthetic images for extended training material, which is essentially an upsampling technique for imbalanced image datasets. The principle is similar to image data augmentation, a sort of pre-processing method. Because of the already scarce medical image data, there is no strict downsampling for it. However, in the work of Shin et al. (14), they implemented a transfer learning technique to reduce the requirement of training material from CNNs so that even with a smaller amount of medical image data, a fair training effect can be achieved. In these two works, the researchers put effort into designing and calibrating sophisticated networks and techniques, thereby achieving innovative results. Statistically, this type of publication is still less than conventional research, and we seek a more effort- and time-friendly approach to improve the performance of processing medical images. To address this perspective, in this paper, we propose an extended preprocessing approach for medical image data.
Even though there is much deep learning-related research for medical applications, XAI has only been introduced into this field for less than a decade. The consideration of accountability and reliability of creation for critical use has risen, and XAI techniques provide the means to transfer trust and confidence from black-box computer systems to human users. Researchers started to use XAI as a validation and verification tool for the black-box models. For example, in the work of Majkowska et al. (15), they proposed deep learning models for chest radiograph interpretations, along with their validations and evaluations using XAI techniques. In this work, they used four CNNs for the detection of fractures, nodules/masses, pneumothorax, and opacity from chest X-ray images. The models showed equally decent performance as human experts, with slightly higher sensitivity. They used SmoothGrad (16), an XAI technique where one randomly adds noise into input images, calculates derivatives of feature maps, and removes the noise in the resulting saliency map. Based on the explanation produced by SmoothGrad, experts can assess them and provide valuable inputs regarding where the model might present errors. Dunnmon et al. (17) and Rajpurkar et al. (18) both used the classic CAM method to validate their deep learning models developed for interpreting chest radiographs. CheXNeXt (18) was developed to detect 14 different types of lung pathologies and achieved human-expert level performance on 11 types. CAM results showed that the model focused on meaningful region of interest (ROI) and produced correct detection.
Methods
Dataset
In the experiment, we use the musculoskeletal radiographs (MURA) dataset developed by Rajpurkar et al. (19), which is recognized as currently the largest public osteo dataset worldwide. MURA dataset contains a total of 40,895 images categorized according to the body parts, which includes: elbow, finger, forearm, hand, humerus, shoulder and wrist. Dataset is organized by order of patient, and data masking has been performed originally from source (Stanford University), therefore no patient information is contained, only random index is assigned to each patient. Each patient file contains at least one, at most three different angles of images for the same subject. Extraction of data follows the csv file provided by original developer. For our experiment, we have extracted the elbow and shoulder images, which respectively represents different kind of redundant image elements. The two datasets are stored and utilized independently for different runs of experiment.
The elbow dataset contains a total of 5,396 images categorized into two classes: normal and abnormal. According to official document, training set contains a total of 4,931 images, among which 2,006 are abnormal and 2,925 are normal, and within testing set there are 235 normal images and 230 abnormal images.
On the other hand, the shoulder dataset contains a total of 8,942 images categorized into two classes: normal and abnormal. According to official document, there are 4,211 normal images and 4,168 abnormal images in training set, 285 normal images and 278 abnormal images in testing set.
The data that support the findings of this study are openly available at https://github.com/ushashwat/MURA-Bone-Abnormality-Detection.
Preprocessing
The workflow of preprocessing independent images is illustrated in Figure 1, which includes the following four steps:
- Contrast enhancement (brightness included), since there are a faire portion of X-ray images that too dark, the model might not be able to distinguish clearly the features on the images.
- Background cropping. As mentioned before, images should be removed of the black background outside the image frame.
- Centered cropping. From our observation, all the images contain the upper arm and forearm (in the case of elbow dataset), along with the nearby areas that often contain the logos, while the abnormality areas are almost always at or around the joint area. Therefore, a centered cropping is performed to remove background areas containing logos as well as surrounding forearm and upper arm areas including muscles and skin tissues.
- Final step is subject extraction. This step is to extract only the main subject.
We considered a precise manual processing of removing complete background along with irrelevant logos and other components from the images, but with the amount of almost 14 thousand images, the workload would be overwhelming. Therefore, we utilized automated functions by Python coding to achieve the removing process automatically.
By using the functions provided by OpenCV (20) packages in Python, we are able to preprocess the images following the workflow illustrated in Figure 1 for all 14 thousand images in approximately 7 minutes with low computational cost, using a loop function packing all the operations performed for each image. Using OpenCV functions, we first enhance the contrast to level 2, and enhanced brightness to 60. Then by setting a threshold distinguishing black and white pixels in the images, backgrounds are cropped. The centered cropping is performed by customizing cropping size based on the boarders of the images. Finally, the subject extraction is achieved by again using thresholding function. The first three steps help to perform and more precise subject extraction.
Our preprocessing method has the advantages of automaticity, low time consumption/human labor/computational cost/possible financial cost, and easy manipulation, also produces valuable data samples, suitable for researchers with high volume image data and limited resources.
Most images are successfully processed and essential parts are extracted as shown in Figure 2 below (original image left, processed image right), small percentage of images with low resolution are being cut out of essential parts, but these images are unable to provide too much valuable information for the model initially, therefore we determine that this automated process greatly reduces experimental cost and the small error has low negative influence towards the results, so we choose this process over manual process in our experiment.
As an example of XAI explanation shown in Figure 3, we can observe that in unprocessed images with background and irrelevant parts (image on the right), the explanation ROI would appear on the Figure 3 its background or even on the opposite side of the true ROI, also on the logo (label) of the right or left hand for the image (as shown in the image on the left inside the red frame), which we clearly understand is irrelevant to the outcome. As we are using the elbow dataset, they are categorized by left- and right-hand side; therefore, images are tagged with the logo “L” or “R”. Even though they are irrelevant to the classification (for diagnosing purposes only, not considering other surgery purposes), there is a high possibility that the ratio of left- and right-side abnormal cases is imbalanced in the datasets, which could lead to potentially biased training for the model. If, for example, there are more left-side abnormal elbow cases than right-side, the model could recognize that right-side images are less likely to suffer illness than left-side. To achieve accurate training, removing the irrelevant indicators is necessary.
Finally, an important step of rebalancing is conducted for both datasets. As we can see, the ratio of normal and abnormal classes is slightly imbalanced for both the training set and testing set, where normal images outnumber abnormal ones. Since the objective of a CAD system is always to find and identify abnormal cases and factors, more material to train the model to identify abnormal cases is preferred. To avoid the effect of this imbalance, we manually discarded 919 normal cases from the training set and 5 normal cases from the testing set, and achieved the ideal balance ratio. Therefore, for our experiment, we have 4,012 training images (2,006 in each class) and 460 testing images (230 in each class).
CNN models
For our experiment, we selected five classic CNN models included in TensorFlow Keras applications, which are known for their excellent performance in image classification. For models belonging to the same branch, we selected one representative from each family. They are: VGG16 (21), ResNet50 (22), EfficientNet (23), MobileNet (24), and DenseNet201 (25). Details for each model are shown in Table 1. For descriptions and concept definitions, please refer to the referenced literature.
Table 1
| Model | Parameters | Layer | Pretrain |
|---|---|---|---|
| VGG16 | 138 million | 16 | ImageNet |
| ResNet50 | 25,636,712 | 50 | ImageNet |
| MobileNet | 4.2 million | 28 | ImageNet |
| EfficientNetB0 | 5,330,564 | 237 | ImageNet |
| DenseNet201 | 20,242,984 | 201 | ImageNet |
Parameter configuration
To ensure a same experimental environment for every selected model, we use the exact same parameter settings for all models. We conducted parameter screening to search for a suitable parameter configuration. The screening process is done for the following three parameters: batch size, optimizer and initial learning rate. For batch size, we chose between 16 and 32; for optimizer, we chose between stochastic gradient descent (SGD) and Adam; for initial learning rate, we chose between 0.01 and 0.001. Forming a total of eight combinations of parameters, we use them on all models mentioned in Table 1 to conduct independent test runs using the elbow dataset. For the test runs using each of the eight parameter combinations, five accuracies will be produced, and a variance value of these accuracies is calculated for each combination. We selected the combination that possesses the lowest variance. The final configuration is shown in (Table 2).
Table 2
| Parameters | Setting |
|---|---|
| Input size | 128×128 |
| Batch size | 16 |
| Optimizer | SGD |
| Initial learning rate | 0.01 |
| Learning rate reduction | 0.1 |
| Loss function | Categorical |
| Activation function | Relu |
| Epoch | 10 |
All models are pre-trained with ImageNet, as various research has shown that ImageNet pre-trained models generally have significantly superior performance over untrained ones. SGD, stochastic gradient descent.
All models are pre-trained with ImageNet, as various research has shown that ImageNet pre-trained models generally have significantly superior performance over untrained ones.
XAI method
The XAI method used for our experiment is GradCAM, proposed by Selvaraju et al. (26), on the concept of “generalization of CAM”, namely the base concept “class activation mapping” by Zhou et al. (27). Since the CAM is initially implemented under the premise of having global average pooling (GAP) layer instead of the last fully connected layer, it requires the neural network structure to be changed and the model might need retraining. In the original CAM method, feature maps are processed by GAP and produce correspondent weighted feature vectors with the same length as the channels of the feature maps. CAM is the linear sum of weights of all layers’ feature maps towards the predicted class. At the final softmax layer, its size is generally different from input, therefore the CAM result also needs to be oversampled to the same size as the input image, then overlay it on original image for a final observational result. And GradCAM works in similar mechanism without the requirement of GAP, therefore it is applicable for different structures of neural networks. We call GradCAM as a “class-oriented method”, because in its process, a “class of interest” is initially given for the target image and assigned a gradient value 1, which will then be backpropagate to rectify feature maps and form a rough area of interest for the “class of interest”. Therefore, GradCAM produces a heatmap of ROI with vague boundaries. Since GradCAM produces heatmaps based on the model structure itself, it is known to produce stable explanations for same data sample on same models.
As all CNN models are black box models, users cannot determine whether the model makes its prediction based on the correct knowledge judging only from its prediction results. Therefore, the necessity of XAI emerges. As mentioned in the Related Works Section, many researchers now value the reliability of the predictions made by black box models especially for medical-used ones since it is crucial that doctors are getting reliable aid and results from black box models, therefore they now use XAI methods to validate their results, and this has often been the sole purpose of using XAI in their research. In our work, the XAI technique is not only used to evaluate whether the models make reliable predictions, but also, through using XAI, we discovered that CNN models trained with unprocessed image datasets would focus on the wrong features (background, logos) to make predictions, and thereby inspires us to propose the effective, efficient, low cost and easily operated preprocessing method for specifically medical images.
Experimental setup
Our experiment is a two-dimensional process. Vertically, experiments will be conducted on elbow dataset and shoulder dataset using the same five CNN models. Horizontally, for each dataset, experiments will be conducted twice, for un-processed background and processed background respectively, evaluation is performed as previously mentioned, in both XAI method and statistical method. Our experiment process includes the following main steps:
- Individually putting the preprocessed two datasets with background and irrelevant part through selected CNNs and produced trained models.
- Select testing image samples and put them through the correspondent trained models.
- Using XAI methods, produce correspondent ROI for testing samples.
- Remove background and irrelevant part from both datasets and input into unweighted CNNs, produced trained models.
- Input the same testing image samples into new trained models.
- Using XAI methods, produce correspondent new ROIs for testing samples.
Same steps performed for both dataset in both forms (processed and un-processed).
According to our multiple experiments, most times the performance trends to stable after epoch No. 10, therefore the epoch is set to 10.
Results
Results will be presented by the order of the two datasets respectively, containing statistical analysis and visual analysis.
For both elbow and shoulder dataset, respectively five test samples will be selected. For a just and fair experimental comparison, only as training material that the images will be processed, the samples being tested afterwards are all unprocessed images, to precisely test the improvement made by training with processed material.
For clearer description and reading, the models trained by raw images will be referred to as ‘Model A’, and the models trained by processed images will be called ‘Model B’.
Quantitative results: elbow dataset
Quantitative performance for the elbow dataset is shown in Tables 3,4.
Table 3
| Model | VGG16 | ResNet50 | MobileNet | EfficientNetB0 | DenseNet201 |
|---|---|---|---|---|---|
| A | 0.72 | 0.71 | 0.74 | 0.63 | 0.78 |
| B | 0.78 | 0.74 | 0.77 | 0.79 | 0.80 |
| Aug | 0.06 | 0.03 | 0.03 | 0.16 | 0.02 |
Model A, the models trained by raw images; Model B, the models trained by processed images; Aug, performance augmentation (from Model A to Model B).
Table 4
| Model | VGG16 | ResNet50 | MobileNet | EfficientNetB0 | DenseNet201 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | TN | TP | TN | TP | TN | TP | TN | TP | TN | |||||
| A | 71 | 73 | 71 | 73 | 70 | 78 | 47 | 79 | 66 | 90 | ||||
| B | 74 | 81 | 64 | 84 | 78 | 79 | 72 | 85 | 71 | 90 | ||||
Model A, the models trained by raw images; Model B, the models trained by processed images. TP, true positive; TN, true negative.
Figures 4,5 present a comparison of validation accuracy and validation loss for ResNet50.
Figure 6 presents a comparison of validation accuracy for EfficientNetB0. Figure 7 presents a comparison of validation loss for EfficientNetB0.
Figure 8 shows the confusion matrix of DenseNet21 trained with raw images and processed images, respectively; the confusion matrix values are normalized.
Visual results: elbow dataset
Five testing samples are randomly selected from the original MURA dataset elbow part before preprocessing, and they are never included or involved in any training and testing process.
Regarding the testing samples, they are raw images without any preprocessing (sample for the testing samples from shoulder dataset to be used in the next part of experiment). We intentionally use the raw images to experiment Models A and B, to demonstrate that, after being trained by the “effective training materials” mentioned above, the models are capable of predicting new images based on the correct attention areas where the illness occurs, even when the new images come black background and logos, or other irrelevant features.
The original images are shown in Figure 9.
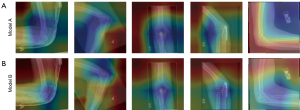
Figures 10,11 present the GradCAM explanations for the 5 testing samples from the models trained with raw images (Model A corresponding to Figure 10A and Figure 11A), and from the models trained with processed images (Model B, corresponding to Figure 10B and Figure 11B). Order of the testing samples (Nos. 1 to 5) is the same as shown in Figure 9. Comparison will not be conducted between models since performance differences between models are not our objective. Only the differences in explanations between the same model using different training materials will be compared. Therefore, we do not need all five selected models’ results. Judging from general performance, we determine that models VGG16 and DenseNet201 are suitable for the elbow dataset. Hence, the explanations produced from these two models will be presented and compared.

Quantitative results: shoulder dataset
Table 5 presents the accuracy comparisons for selected models between being trained with raw images and processed images, Table 6 presents the comparisons of true positive (TP) and true negative (TN) rates.
Table 5
| Model | VGG16 | ResNet50 | MobileNet | EfficientNetB0 | DenseNet201 |
|---|---|---|---|---|---|
| A | 0.61 | 0.70 | 0.66 | 0.75 | 0.66 |
| B | 0.68 | 0.71 | 0.73 | 0.79 | 0.75 |
| Aug | 0.07 | 0.01 | 0.07 | 0.04 | 0.09 |
Model A, the models trained by raw images; Model B, the models trained by processed images; Aug, performance augmentation (from Model A to Model B).
Table 6
| Model | VGG16 | ResNet50 | MobileNet | EfficientNetB0 | DenseNet201 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | TN | TP | TN | TP | TN | TP | TN | TP | TN | |||||
| A | 56 | 66 | 69 | 70 | 64 | 71 | 75 | 76 | 36 | 95 | ||||
| B | 67 | 69 | 70 | 71 | 65 | 81 | 76 | 81 | 74 | 76 | ||||
Model A, the models trained by raw images; Model B, the models trained by processed images. TP, true positive; TN, true negative.
Figures 12,13 illustrate the accuracy and loss of MobileNet trained with raw and processed images.
Figures 14,15 respectively present the accuracy and loss comparison from Densenet_201. Figure 16 presents the comparison of confusion matrix from EfficientNetB0.
Visual results: shoulder dataset
Five testing samples are selected from the original MURA dataset shoulder part, all testing samples have never been included or involved in any training and testing process. Original images are as shown in Figure 17.
Judging from general performance, we determined that MobileNet and ResNet50 are the two models suitable for the shoulder dataset. Figures 18,19 contain the explanation images produced using GradCAM for the testing samples (ordered as shown in Figure 17 from Nos. 1 to 5) processed by these two models but trained with respectively raw images (Model A, corresponding to Figure 18A and Figure 19A) and processed images (Model B, corresponding to Figure 18B and Figure 19B).


Discussion
This section presents the corresponding discussions of the respected experimental results. For better reading and comprehension, the discussions will be presented in the same order as the “Results” section.
Quantitative results: elbow dataset
As we can observe from Table 3, in general, the performance of all selected models is enhanced when trained with images processed to remove background and irrelevant components. Specifically, for the models EfficientNetB0 and ResNet50, validation accuracy is improved by 10% and 16%, respectively. For the other models, improvements range from 2% to 6%. Table 4 presents the comparisons of true prediction rates from selected models trained with different materials. We can see that their capability of producing true predictions is improved, with the improvement being slightly more in TP predictions than in TF predictions, but the models are generally more accurate in distinguishing negative cases.
We can also observe from Figure 4 that the model accuracy values improved during the training process; curve A started from a very low value of 0.027 and raised, while curve B, although relatively unstable, appears to have a higher average accuracy. From Figure 5, we can see that the validation loss curve for Model B fluctuates approximately between 0.25 and 1.25, whereas for Model A, the loss reached as high as 4.34 at epoch 4. Therefore, the average validation loss is improved for Model B compared to Model A.
From Figure 6, one can observe that for Model A, the validation accuracy fluctuates vividly, while for Model B, it increases more stably. Model evaluation results showed that the mean accuracy value is improved for Model B. From Figure 7, we can observe that Model A’s loss reached 1.163, while Model B’s loss values are all less than 0.475. Therefore, we can see that the overall loss values of Model B are improved compared to Model A, and the loss curve of Model B is comparatively more stable.
From Figure 8, we can observe that, the TN rate is actually improved by 0.003 in actual digits. We can see that besides the decent TN rate, the TP rate is improved by 0.05. This indicates that preprocessing the elbow dataset helps the model reduce false negative predictions. For all selected models, generally speaking, using processed image data will lead to better results.
Visual results: elbow dataset
As we observe from Figure 10, for the randomly selected testing samples, VGG16 presents a decent performance with fair accuracy and relies on roughly the right areas. For samples Nos. 3 and 4, the indications lay almost on top of the right area, but with training on processed images, we can see that the area subtly adjusted to more precisely match the exact right position. On test samples Nos. 2 and 5, the explanations show attention to the background and the logos “L” and “R”, which suggests that the model might sometimes be confused by these elements. With training on processed images, attention drifted away from the logos, and the ROI area became narrower and more concentrated. For sample No. 1, which is a clear case of an upper arm bone fracture on the lower forearm bone, we can see that the ROI presented by Model A is almost completely opposite. With training on processed images, the ROI drifted closer to the right area, although not center. For sample No. 5, the ROI somehow located on the bone fragment that is lost away from the bone tissue due to the fracture. With training on processed images, the attention still does not shift, only becoming narrower and more concentrated on the fragments as opposed to the fractured femur; therefore, this improvement is only technical but not strictly medical.
From the explanation provided by DenseNet201 as shown in Figure 11, we can see more clearly that the background and logos could possibly affect the training process. Significantly, for sample No. 5, the attention area is solely on the unrelated muscle area, background, and logo of the image, with the forearm bone showing mild interest (yellow area). It is clear that Model A does not perceive anything abnormal in this image and processes the different elements as a whole. After being trained with processed images, in Figure 11B for sample No. 5, the presented ROI roughly covers both the bone fragment and the fractured femur head. In sample No. 1, Model A produces a wider and less precise explanation. We can see that the explanation produced by Model B has a narrower cover area but not on the correct area, which is a similar case with sample No. 5 in the VGG16 model. We presume that the model recognized other indications that do not meet the expert’s standard, and the training is not yet sufficient to help the model recognize these types of abnormalities. Observing column 4, we can see that for sample No. 4, the model trained with raw images produced an explanation concentrated on the inner-side muscle instead of the joint, but the model trained with processed images focused on the correct area of the abnormal joint, which is a good case demonstrating effectiveness.
Quantitative results: shoulder dataset
We can see on Tables 5,6 that the overall accuracy for all selected models are improved, here the DenseNet201 improved most from 0.66 to 0.75 of 9%.
From Table 6, we can see that almost all rates of producing correct prediction are improved, overall seeing that the improvement on TN cases are slightly more than on TP case. For DenseNet201 who also obtained most accuracy improvement similar to elbow dataset, after being trained with processed images, the has significant improvement on the capability of recognizing positive images comparing to being trained with raw images, although the recognition rate for negative samples has dropped.
From Figures 12,13, we can observe that when trained with raw images, the curves of both validation accuracy and validation loss bounce vividly indicating instability of prediction. For Model B trained with processed image, the curve of validation accuracy maintains relatively stable, although loss curve trends upward but mean loss value is still lower than the one of Model A.
Figures 14,15 respectively showed that with raw images, Densenet201 started low at the accuracy of 0.183, both accuracy and loss curves are unstable, highest loss reaches 2.67. On the other hand, when trained with processed images, validation accuracy curve maintains relatively stable starting from 0.708, loss curve maintains between 0.6 and 1, which is more than 2 times lower than the loss of model A. From Figure 16, we can see that with true predictions number improved, False predictions reduces. TP rate is improved from 0.75 to 0.76, TN rate is improved from 0.76 to 0.81, indicating that the preprocessing benefits the model more on reducing false positive predictions.
Visual results: shoulder dataset
We can observe from Figure 18A that trained with raw images, MobileNet produces comparatively vague parameters with lower saturation ROIs, though they all seem to cover approximately the key areas. Lighter blue color indicates the model does not sufficiently rely on these areas as much as other models do (deep, saturated blue color as in other models), but they still are the positive indications for the MobileNet. On the two samples with logos (Nos. 1 and 3), it still showed mild interest on the logos (yellow area). After being trained with processed images, light blue color become deeper indicating the model relies on the ROIs more to make its predictions, and the ROIs for all five samples appears to become narrower and more concentrated, for sample No. 1 although the ROI area is narrower and more concentrated, the rib cage area is still considered an indication for abnormality and even more centered than the explanation of Model A, we could surmise that during both training course the rib cage consistently attracts the model’s attention regardless of the other irrelevant parts being processed out. For samples Nos. 2, 3 and 4, the positions of the ROIs shifted slightly to more centered areas around the bone tissues, sample No. 5 appears to be centered on muscle tissue.
From Figure 19A, we can see that, ResNet50 produces more concentrated areas of interest with clear edges than MobileNet, and surprisingly on sample No. 3, it presents opposing interest individually the logo area rather than only ‘not interest’ (connecting with background area as a whole irrelevant part as seen in Figure 19A sample No. 1), which proves that model does process them separately and has different level of affect from different elements. Also on sample No. 1, the area of interest includes rib bones, which is obviously irrelevant to shoulder join abnormality, therefore although it includes the right indicating area, performance can still be improved, similar case on sample No. 3 whose ROI also covered rib bone area. After being trained with processed images, on sample No. 1 it shows similar situation that we see on the above MobileNet explanation, where we can see that both models, whether trained with raw or processed image, agree that the rib bone area of the patient that provided sample image No. 1, contains elements supporting the abnormality. Which is an interesting phenomena that the two models might possibly see patterns that does not meet human eyes and standards but actually connect to the abnormality. On sample No. 2 and five all presented ROIs centered on approximately the same location but with more concentrated coverage, on the other hand sample Nos. 3 and 4’s ROIs shifted slightly their positions, on sample No. 3 the area is no longer centered on the rib bone, and same for sample No. 4 no longer covers the muscle tissue under the armpit.
Conclusions
In many current studies on developing deep learning models for medical image recognition, it is observed that they often perform basic standard preprocessing on the images, which is appropriate for all image data in general. However, we hold the opinion that due to the special nature of medical images, an extended preprocessing process is necessary. This is especially true when modifying the model’s structure, design, and calibrating parameters, which can sometimes be exhaustive with less reward. The extended preprocessing method proposed in this paper for medical image data is easily implemented, time and effort-friendly, and effective for performance improvement. Additionally, our proposal through a deep learning and XAI approach highlights that for the development of CAD systems specifically for medical images, special factors and aspects must be considered, such as the background and “irrelevant parts”, both of which are emphasized in this paper. As evidenced by the experimental results and XAI visualization, they significantly influenced the outcome.
Corresponding with what was mentioned in the Introduction section about the “special treatment for medical image data”, in our experiment, the data augmentation process excluded the step of cropping. This method, which crops one image into several and adds them as new data under the same class, is considered inappropriate for processing medical images. Our consideration includes the following two points: the datasets contain a fair amount of data as they are, thus, we do not need a data augmentation process to increase the training material, thereby avoiding the risk of compromising dataset integrity. Additionally, within the abnormal images, areas that indicate abnormality are only partial but not general, which is different from object detection where all parts of the object support the true classification result. If cropping or data augmentation is performed, even the cropped images from abnormal samples that do not contain an abnormal factor will still be labeled as abnormal, possibly confusing the model and negatively affecting model performance.
XAI has been a crucial component in our findings. Thanks to the explanations provided by using XAI, we discovered that models trained with unprocessed raw images with irrelevant features (background, logos, etc.) indeed could be biased towards these features, hence our proposing the appropriate, efficient, low-cost and easily operated preprocessing method for the medical image training materials. We firmly believe in the necessity of XAI, which is an important bridge that connects human intelligence and AI. For the black box models developed for important purposes such as radiology and medicine, doctors would be benefited from them on improving efficiency, receiving aid on diagnosis, yet all information coming from the models are for references, the ultimate determination lays with human ourselves. As stated in the work proposed by Sorantin (28) quote “For legal reasons, humans remain in control and are responsible for decision-making—even when AI is doing/supporting the decision”.
Future work
Therefore, for future research, we believe that an explanation for black box models can serve more purpose than just proving that the model operates as expected. By implementing XAI techniques, we can not only validate the model’s classification results but also identify removable elements beforehand, thereby improving the effect of data preprocessing. Our future research direction points towards combining medical image segmentation and classification. Medical image segmentation techniques are currently very mature and accurate, enabling the pinpointing of exact areas with clear boundaries, which could possibly further improve the preprocessing of medical images. For example, for femoral head necrosis, the method of observation is through radiology images of the patients’ articulatio coxae. Since femoral head necrosis develops exclusively on the femur head bone, yet radiology images capture the whole hip compartment, which contains what we call “irrelevant parts” for this disease such as the spinal cord, upper thigh bone, and partial rib bones. These irrelevant parts could possibly distract an AI model. Therefore, if our future research could further narrow down the precise area corresponding to specific diseases as a preprocessing method, model performance is hopefully to be improved, and the model would be more trustworthy for medical applications.
On the other hand, on the note mentioned in Introduction Section where importance of selecting appropriate XAI method for case-specific scenarios (28), we believe new XAI approaches designed exclusively for radiology is in order, and to that aim, the “Counterfactual Explanation” proposed in the work of Del Ser et al. (29) would be an interesting solution with great potential. In their work, the concepts “Causability” and “Plausibility” can be important base factors when developing new XAI techniques, and thereby produce advanced, credible XAI methods, providing further confidence and trustworthiness between human and AI.
Acknowledgments
None.
Footnote
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1745/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed 2020;187:104964. [Crossref] [PubMed]
- Gharaibeh M, Almahmoud M, Ali MZ, Al-Badarneh A, El-Heis M, Abualigah L, Altalhi M, Alaiad A, Gandomi AH. Early Diagnosis of Alzheimer’s Disease Using Cerebral Catheter Angiogram Neuroimaging: A Novel Model Based on Deep Learning Approaches. Big Data Cogn. Comput 2022;6:2.
- Lei Y, Tian Y, Shan H, Zhang J, Wang G, Kalra MK. Shape and margin-aware lung nodule classification in low-dose CT images via soft activation mapping. Med Image Anal 2020;60:101628. [Crossref] [PubMed]
- Yang C, Rangarajan A, Ranka S. Visual Explanations From Deep 3D Convolutional Neural Networks for Alzheimer's Disease Classification. AMIA Annu Symp Proc 2018;2018:1571-80. [PubMed]
- Palatnik de Sousa I, Maria Bernardes Rebuzzi Vellasco M, Costa da Silva E. Local Interpretable Model-Agnostic Explanations for Classification of Lymph Node Metastases. Sensors (Basel) 2019;19:2969. [Crossref] [PubMed]
- Liu L, Feng W, Chen C, Liu M, Qu Y, Yang J. Classification of breast cancer histology images using MSMV-PFENet. Sci Rep 2022;12:17447. [Crossref] [PubMed]
- Ma L, Su X, Ma L, Gao X, Sun M. Deep learning for classification and localization of early gastric cancer in endoscopic images. Biomedical Signal Processing and Control 2023;79:104200. [Crossref]
- Shen X, Luo J, Tang X, Chen B, Qin Y, Zhou Y, Xiao J. Deep Learning Approach for Diagnosing Early Osteonecrosis of the Femoral Head Based on Magnetic Resonance Imaging. J Arthroplasty 2023;38:2044-50. [Crossref] [PubMed]
- Retzlaff CO, Angerschmid A, Saranti A, Schneeberger D, Röttger R, Müller H, Holzinger A. Post-Hoc vs Ante-Hoc Explanations: XAI Design Guidelines for Data Scientists. Cognitive Systems Research 2024;86:101243. [Crossref]
- Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ. Computer-aided classification of lung nodules on computed tomography images via deep learning technique. Onco Targets Ther 2015;8:2015-22. [PubMed]
- Wang H, Jia H, Lu L, Xia Y. Thorax-Net: An Attention Regularized Deep Neural Network for Classification of Thoracic Diseases on Chest Radiography. IEEE J Biomed Health Inform 2020;24:475-85. [Crossref] [PubMed]
- Keles A, Keles MB, Keles A. COV19-CNNet and COV19-ResNet: Diagnostic Inference Engines for Early Detection of COVID-19. Cognit Comput 2021; Epub ahead of print. [Crossref] [PubMed]
- Salehinejad H, Valaee S, Dowdell T, Colak E, Barfett J. Generalization of deep neural networks for chest pathology classification in x-rays using generative adversarial networks. 2018 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP); 2018:990-4.
- Shin HC, Roth HR, Gao M, Lu L, Xu Z, Nogues I, Yao J, Mollura D, Summers RM. Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning. IEEE Trans Med Imaging 2016;35:1285-98. [Crossref] [PubMed]
- Majkowska A, Mittal S, Steiner DF, Reicher JJ, McKinney SM, Duggan GE, Eswaran K, Cameron Chen PH, Liu Y, Kalidindi SR, Ding A, Corrado GS, Tse D, Shetty S. Chest Radiograph Interpretation with Deep Learning Models: Assessment with Radiologist-adjudicated Reference Standards and Population-adjusted Evaluation. Radiology 2020;294:421-31. [Crossref] [PubMed]
- Smilkov D, Thorat N, Kim B, Viégas F, Wattenberg M. SmoothGrad: removing noise by adding noise. arXiv 2019 arXiv:1706.03825.
- Dunnmon JA, Yi D, Langlotz CP, Ré C, Rubin DL, Lungren MP. Assessment of Convolutional Neural Networks for Automated Classification of Chest Radiographs. Radiology 2019;290:537-44. [Crossref] [PubMed]
- Rajpurkar P, Irvin J, Ball RL, Zhu K, Yang B, Mehta H, et al. Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists. PLoS Med 2018;15:e1002686. [Crossref] [PubMed]
- Rajpurkar P, Irvin J, Bagul A, Ding D, Duan T, Mehta H, Yang B, Zhu K, Laird D, Ball RL, Langlotz C, Shpanskaya K, Lungren MP, Ng AY. MURA Dataset: Towards Radiologist-Level Abnormality Detection in Musculoskeletal Radiographs. arXiv 2017. arXiv:1712.06957.
- Bradski G. The OpenCV Library. Dr. Dobb’s Journal of Software Tools 2000;120:122-5.
- Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 3rd International Conference on Learning Representations (ICLR 2015). Computational and Biological Learning Society; 2015.
- He K, Zhang X, Ren S, Sun J, Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016:770-8.
- Tan M, Le Q. EfficientNet: Rethinking Model Scaling for Convolutional Neural Networks. Proceedings of the 36th International Conference on Machine Learning Proceedings of Machine Learning Research 2019;97:6105-14.
- Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017. arXiv:1704.04861.
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:2261-9.
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. 2017 IEEE International Conference on Computer Vision (ICCV); 2017:618-26.
- Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A. Learning Deep Features for Discriminative Localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016:2921-9.
- Sorantin E, Grasser MG, Hemmelmayr A, Tschauner S, Hrzic F, Weiss V, Lacekova J, Holzinger A. The augmented radiologist: artificial intelligence in the practice of radiology. Pediatr Radiol 2022;52:2074-86. [Crossref] [PubMed]
- Del Ser J, Barredo-Arrieta A, Díaz-Rodríguez N, Herrera F, Saranti A, Holzinger A. On Generating Trustworthy Counterfactual Explanations. Information Sciences 2024;655:119898. [Crossref]

















