
Deep learning for the detection of moyamoya angiopathy using T2-weighted images: a multicenter study
Introduction
Moyamoya angiopathy (MMA) is a progressive cerebrovascular disease with high prevalence in East Asian descendants (1,2), characterized by stenosis or occlusion of the distal internal carotid artery, proximal anterior cerebral artery and middle cerebral artery, followed by surrounding smog-like blood vessels (3). Transient ischemic attacks, cerebral infarction and intracerebral hemorrhage are common in MMA patients as the vessel stenosis progresses (3). The gold standard in the diagnosis of MMA is digital subtraction angiography (DSA). However, this procedure is invasive, expensive and requires the use of contrast media. Magnetic resonance angiography (MRA) and computed tomography angiography (CTA) are the non-invasive alternative methods for MMA diagnosis (4). However, vascular examination methods are not always included in certain cerebral MR examinations. MMA can be potentially missed in the initial MR examination without MRA. Flow voids on T2-weighted images (T2WI) have previously been assessed as the essential criteria in the diagnosis of MMA (5). But other anomalies on MR images tend to attract more attention and some newly employed radiologists may be not familiar to the features of MMA on T2WI. Therefore, it is necessary to recommend a method based on T2WI, which is one of the most classical sequences in cerebral MR examination.
Deep learning (DL) has been recently used in several radiologic applications, such as in the detection of cerebral aneurysms by CTA, MRA and DSA (6-9) and in segmentation and classification of primary bone tumors on radiographs (10). Three-dimensional (3D) coordinate attention residual network (3D CA-ResNet) can be used in cerebrovascular disease to detect the stenotic areas on MRA imaging in patients with MMA (11). Automatic diagnosis of MMA using DL can also be configured on different medical images with high specificity and sensitivity, such as DSA, T2WI and plain skull radiography (12-14), in order to timely recognize the re-hemorrhagic risk (13). However, existing literature shows training on a small sample size, and most of the works were not independently validated with multicenter data, which may result in decreased accuracy when applied to new, diverse datasets, affecting its overall reliability in clinical practice. In addition, DSA and plain skull radiography are not common imaging diagnostic methods in the clinical setting.
Shallow convolutional neural network (SCNN), LeNet-5 Convolutional Neural Network (LeNet), Visual Geometry Group Network (VGG), ResNet50, and Dense Convolutional Network (DenseNet) are classical DL models widely used in medical image classification, each with varying sensitivities and specificities (15-20). To detect MMA on a common imaging sequence, we selected prominent convolutional neural network (CNN) models (LeNet, VGG, ResNet, and DenseNet) frequently used in clinical research over recent years, aiming to identify the most reliable model. The layer numbers and structures of these models were implemented using Keras, and we documented all software libraries and packages used to ensure reproducibility, aligning with guidelines for artificial intelligence in medical imaging (21). In this study, multi-model analysis was performed using multi-center data to detect MMA from normal controls and patients with cerebrovascular disease except MMA. We present this article in accordance with the TRIPOD-AI reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1269/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Affiliated Drum Tower Hospital, Medical School of Nanjing University (No. 2021-026). All participating hospitals were informed and agreed to the study. Individual consent for this retrospective analysis was waived.
Subjects
A multicenter, retrospective study was performed aiming to detect MMA in patients from seven hospitals (site 1 to site 7) in China, based on axial two dimensional T2WI. The seven hospitals were in two provinces in eastern China, and included general and specialized hospitals, as well as adult and children’s hospitals. MMA was diagnosed on MRA or DSA according to the diagnostic guidelines (3) (Figure S1). MRA and T2WI sequences were obtained by an MR examination. The control group included patients with normal MRA and those with cerebrovascular disease except MMA, such as atherosclerosis, intracranial aneurysm, and arteriovenous malformation (AVM). The exclusion criteria were the following: (I) patients not subjected to MRA; (II) patients after craniotomy; (III) images with severe artifacts. The image parameters of T2WI and MRA were listed in Table S1.
Three continuous slices of T2WI at the central level of the basal cistern in each subject were acquired. All MR images from the seven sites were acquired in DICOM format and they were reviewed on RadiAnt software (version 2020.1.1). The age at the MR examination, manufacture and field strength of each image modality used were collected. The MRA score of each MMA patient was also assessed by an experienced neuroradiologist (M.W., with 11 years of neuroimaging experience) according to previous standards (4). Radiologist 1 and radiologist 2, with 3 and 12 years of neuroradiology experience respectively, each independently diagnosed the site 5 dataset using only three T2-weighted MRI slices, without knowledge of the final diagnosis.
Algorithm development methods
In this study, DL was based on Python (Version 3.8.11, http://www.python.org), using graphics processing unit (Pydicom: Version 2.1.2; and Cv2: version 4.5.2), and DL framework packages (Tensorflow: Version 2.4.0; and Keras: Version 2.4.0).
Data preprocessing in this study included the following aspects: (I) spatial standardization of images of each layer, using the method of zero value filling to standardize each image to 256×256 pixels; (II) after the standardization of the three-layer images, the image data of each subject formed a matrix of 256×256×3 dimensions as the input information of the DL network.
The CNNs used in this study included: SCNN, LeNet, VGG16, ResNet50 and DenseNet121 (22-24). See Appendix 1 for details.
Internal and external validation strategies
The training and validation strategy in this study was the following: 80% of MMA patients and control group from sites 1–4 were randomly selected as the training set, and the remaining 20% from sites 1–4 were used as the internal validation set. Independent data from sites 5–7 were used as an independent external validation set. Data augmentation in the training datasets was performed using the deflection angle, spatial displacement, scaling size, adding white noise and salt and pepper noise. The data were expanded from 1,436 to 14,360 groups of images. Data augmentation was not performed in both internal and external validation datasets. Finally, the optimal DL model was identified based on external validation results (Figure S2). The source code is publicly available (https://github.com/zhengjunjie1234/MRI_based_MMA_classification).
Statistical analysis
Statistical analysis was performed using SPSS (version 25.0, IBM SPSS Statistics, Armonk, NY, USA) and R (version 3.5.2, R Foundation for Statistical Computing, Vienna, Austria). Sensitivity, specificity, accuracy, the area under the receiver operating characteristic curve (AUC), and F1 score were calculated as values indicating the classification performance in the internal and external validation datasets, as well as the performance of the radiologists. Sensitivity, specificity, and accuracy were further calculated in the images taken from modalities of different manufacturers, field strength, age at the MR examination and MRA scores to assess the potential association of these factors to the optimal model performance. Chi-squared test or Fisher exact test was used to compare the difference of the five DL models in the external validation, among different manufacturers, field strength, age at the MR examination and MRA scores. Bonferroni correction was performed to evaluate the classifying accuracy of the models as well as the accuracy of the optimal model among different manufacturers and MRA scores. A value of P less than 0.05 was considered statistically significant.
Results
Data characteristics
A total of 1,038 patients with MMA (454 males, 46±11 years old), 1,211 normal controls (559 males, 44±14 years old) and 271 patients with cerebrovascular diseases except MMA (167 males, 53±12 years old) were included in this study. Demographics, MR manufacturers, field strength of all modalities, and MRA scores of MMA patients from each site were listed in Table 1.
Table 1
| Category | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 | Site 7 |
|---|---|---|---|---|---|---|---|
| MMA | 488 | 49 | 120 | 97 | 217 | 23 | 44 |
| Sex (male) | 193 | 22 | 59 | 41 | 102 | 13 | 24 |
| Age (years) | 49±11 | 56±12 | 60±10 | 46±15 | 46±15 | 59±12 | 7±3 |
| MRA score | |||||||
| ≤5 | 101 | 31 | 54 | 27 | 69 | 5 | 13 |
| 6–10 | 191 | 13 | 50 | 40 | 65 | 12 | 28 |
| ≥11 | 192 | 5 | 16 | 30 | 83 | 6 | 3 |
| MR scanner | |||||||
| GE | 1 | 0 | 0 | 50 | 57 | 0 | 0 |
| SIEMENS | 0 | 49 | 120 | 47 | 160 | 0 | 16 |
| PHILIPS | 264 | 0 | 0 | 0 | 0 | 7 | 28 |
| UMR | 219 | 0 | 0 | 0 | 0 | 16 | 0 |
| Field strength | |||||||
| 3.0 T | 466 | 49 | 111 | 66 | 109 | 16 | 28 |
| 1.5 T | 18 | 0 | 9 | 31 | 108 | 7 | 16 |
| Normal MRA | 578 | 40 | 73 | 149 | 292 | 29 | 50 |
| Sex (male) | 250 | 26 | 26 | 53 | 162 | 11 | 31 |
| Age (years) | 65±14 | 34±18 | 47±12 | 36±19 | 53±18 | 67±11 | 5±3 |
| MR scanner | |||||||
| GE | 84 | 0 | 0 | 100 | 64 | 0 | 0 |
| SIEMENS | 0 | 40 | 73 | 50 | 228 | 0 | 27 |
| PHILIPS | 420 | 0 | 0 | 0 | 0 | 9 | 23 |
| UMR | 74 | 0 | 0 | 0 | 0 | 20 | 0 |
| Field strength | |||||||
| 3.0 T | 308 | 40 | 70 | 99 | 187 | 20 | 23 |
| 1.5 T | 270 | 0 | 3 | 50 | 105 | 9 | 27 |
| Non-MMA cerebrovascular disease | 131 | 10 | 19 | 35 | 56 | 9 | 11 |
| Sex (male) | 71 | 5 | 10 | 29 | 39 | 4 | 9 |
| Age (years) | 61±16 | 54±14 | 58±10 | 64±14 | 58±18 | 72±12 | 8±3 |
| MR scanner | |||||||
| GE | 0 | 0 | 0 | 11 | 6 | 0 | 0 |
| SIEMENS | 0 | 10 | 19 | 58 | 50 | 0 | 1 |
| PHILIPS | 111 | 0 | 0 | 0 | 0 | 6 | 11 |
| UMR | 20 | 0 | 0 | 0 | 0 | 3 | 0 |
| Field strength | |||||||
| 3.0 T | 88 | 10 | 18 | 69 | 16 | 3 | 11 |
| 1.5 T | 43 | 0 | 1 | 0 | 40 | 6 | 1 |
Age was presented as mean ± standard deviation. The others were presented as numbers. Site 1, Nanjing Drum Tower Hospital, Affiliated Hospital of Medical School, Nanjing University; site 2, The Affiliated Sir Run Run Hospital of Nanjing Medical University; site 3, Xuyi People’s Hospital; site 4, the Affiliated Brain Hospital of Nanjing Medical University; site 5, Jinling Hospital, Affiliated Hospital of Medical School, Nanjing University; site 6, Lu’an Hospital of Anhui Medical University; site 7, Children’s Hospital of Nanjing Medical University. MMA, moyamoya angiopathy; MR, magnetic resonance; MRA, magnetic resonance angiography.
Model performance
The training set included 1,436 patients (608 MMA patients, 672 with normal MRA and 156 with cerebrovascular diseases except MMA), the internal validation set included 353 patients (146 MMA patients, 168 with normal MRA, 39 with cerebrovascular diseases except MMA) (Figure 1). The AUCs of SCNN, LeNet, VGG, ResNet and DenseNet in the internal validation set were 0.950, 0.936, 0.981, 0.985 and 0.974, respectively (Figure S3).
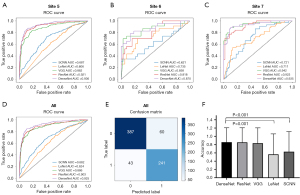
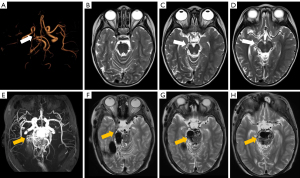
An independent external test datasets (sites 5–7) included 731 patients (284 MMA patients, 371 with normal MRA and 76 with cerebrovascular diseases except MMA), which was used to test the model. The results of the five models in the external datasets were listed in Figure 2 and Table S2. The F1 scores of SCNN, LeNet, VGG, ResNet and DenseNet in the external datasets were 0.626, 0.564, 0.833, 0.855 and 0.858, respectively. DenseNet showed the highest accuracy in the external validation data (0.859), which was significantly higher than that of SCNN (0.631, P<0.001) and LeNet (0.563, P=0.001), but not significantly higher than that of ResNet and VGG (0.855 and 0.834). The sensitivity and specificity of SCNN, LeNet, VGG, ResNet, DenseNet were 0.697 and 0.588, 0.732 and 0.456, 0.823 and 0.841, 0.782 and 0.901, 0.848 and 0.865, respectively. Forty-three MMA patients in the external datasets were missed while using DenseNet, and the rate of MRA that scored ≤5, 6–10 and ≥11 was 55.8%, 37.2% and 7%, respectively. Sixty patients were misdiagnosed as MMA in the external datasets, including 3 patients with AVM, 9 with atherosclerosis and 48 with normal MRA. Misdiagnosed patients from site 5 and site 7 were displayed in Figure S4 and Figure 3, respectively.
Comprehensive analysis
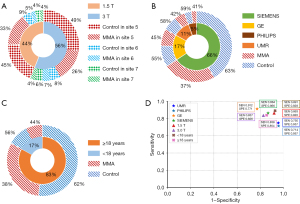
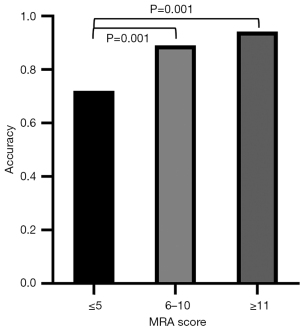
The MRA score was defined according to stenosis or occlusion of intracranial arteries, which may have an effect on collaterals at the skull base. The DenseNet model was used to explore whether the classification accuracy was affected by the MRA score. A total of 86 MMA patients had an MRA score ≤5, 145 MMA patients with score 6–10 and 53 MMA patients scored ≥11 in the external datasets. The accuracies of MRA with scores ≤5, 6–10 and ≥11 were 0.721, 0.890 and 0.943, respectively. The accuracy of MRA with score ≤5 was significantly lower than that of MRA with score 6–10 (χ2=10.734, P=0.001) and with score ≥11 (χ2=10.369, P=0.001). However, the accuracy of MRA with score 6–10 was not significantly different than that of the MRA with score ≥11 (χ2=1.292, P=0.256).
Different MR manufacturers and field strength may affect the accuracy of the model by producing images with different qualities. Images were acquired from four manufacturers and two field strengths in the external datasets. Images of 319 patients (47±23 years old, 155 males) were acquired under 1.5 T field strength and 412 patients (45±22 years old, 175 males) under 3.0 T field strength. The sensitivities and specificities of DenseNet were 0.863 and 0.920 for 1.5 T data and they were 0.837 and 0.826 for 3.0 T data, respectively. The accuracy of 1.5 T data was significantly higher than that of 3.0 T (0.883 vs. 0.886, χ2=6.559, P=0.01). In addition, the distribution of MRA score under 1.5T was significantly different from that under 3.0 T (χ2=12.852, P=0.002). The rate of MMA patients with MRA score ≥11 under 1.5T field strength was higher than that under 3.0 T (26.7% vs. 11.8%).
Images were acquired by UMR in 39 patients (65±23 years old, 21 males, 16 with MMA), by PHILIPS in 83 patients (22±26 years old, 44 males, 35 with MMA), by GE in 127 patients (48±18 years old, 72 males, 57 with MMA) and by SIEMENS in 482 patients (48±21 years old, 257 males, 176 with MMA). The sensitivities and specificities of DenseNet were 0.750 and 0.957 in UMR, 0.714 and 0.957 in PHILIPS, 0.912 and 0.771 in GE, 0.864 and 0.866 in SIEMENS, respectively. The accuracy among the four manufacturers was not significantly different (UMR: 0.829; PHILIPS: 0.855; GE: 0.850; SIEMENS: 0.865).
Age was also investigated as a potential factor affecting the classifying accuracy of the model. Two distribution peaks for age at the onset were found in patients with MMA: 5–9 and 45–49 years old (2). Hundred twenty-four patients (7±4 years old, 71 males, 55 with MMA) were less than 18 years old, 607 patients (54±16 years old, 323 males, 229 with MMA) were more than 18 years old in the external datasets. The sensitivities and specificities of DenseNet were 0.891 and 0.928 in patients with age less than 18 years old, and 0.838 and 0.854 in patients with age more than 18 years old. No significant difference was detected between the accuracies of DenseNet in patients with more than (0.848) and less than (0.911) 18 years old (χ2=0.013, P=0.908). The results of the comprehensive analysis were shown in Figures 4,5.
Radiologist performance
Radiologist 2 had similar diagnostic accuracy (0.862 vs. 0.857), but lower sensitivity (0.650 vs. 0.876) compared to DenseNet, while radiologist 1 showed lower accuracy (0.411 vs. 0.857) than DenseNet. Radiologist 2 demonstrated a trend similar to DenseNet, with accuracy increasing as MRA scores increased. DenseNet outperformed radiologist 2 in the 6–10 (0.924 vs. 0.638) and ≥11 (0.955 vs. 0.727) MRA score ranges. Radiologist 1 achieved high accuracy across different MRA score ranges, with no significant difference from DenseNet in the 6–10 (0.952 vs. 0.924) and ≥11 (0.932 vs. 0.955) ranges. The results were presented in Tables S3,S4.
Discussion
In this study, MR data were collected from seven hospitals with different acquisition parameters. A variety of imaging modalities and field strengths gave the model more generalization with less overfitting. DL models, SCNN, LeNet, VGG, ResNet and DenseNet, were used to classify the MMA patients included in the datasets, with DenseNet outperforming the other four models in terms of accuracy. In addition, the results of MMA patients with higher MRA scores had higher accuracy than those with lower MRA scores. However, MR manufacturers and age at the MR examination did not significantly affect the results.
The classification accuracy of DenseNet differed depending on different types of images. DenseNet model had lower accuracy in detecting cerebral hemorrhage on computed tomography (CT) images than ResNet (15). Five models, namely AlexNet, VGG, ResNet, DenseNet and Inception, have been used for colon cancer classification using public data. The results showed that ResNet had the highest accuracy, while VGG and DenseNet had slightly lower accuracies (25). Remedios et al. reported that DenseNet had the highest accuracy (0.830) in the classification of large vessel occlusion compared with ResNet, EfficientNet-B0, PhiNet and Inception Module-based network on CTA images (26). In this study, SCNN, LeNet, and VGG16 models were trained from scratch without pre-trained weights, while ResNet and DenseNet were initialized with pre-trained ImageNet weights, likely contributing to their superior performance. DenseNet emerged as the optimal model, achieving significantly higher accuracy than SCNN and LeNet. However, its accuracy was not significantly different from that of ResNet and VGG.
Classification of MMA was also previously performed based on DSA, MRA and cerebral oxygen saturation signals using DL and machine learning (27-29). However, these studies were carried out on a single site and MMA was diagnosed by DSA and MRA directly. Kim et al. used MMA patients from an independent site to validate the model based on plain skull radiography (12). Nevertheless, the accuracy was 0.759 using plain skull radiography images. MMA was difficult to diagnose directly based on only T2WI. As such, flow voids based on MR images have been used to diagnose MMA (5,30,31). Additionally, MMA diagnosis at the level of basal cistern showed the highest accuracy (0.928) using DL, as shown by a previous report (14). However, this study used a small sample and included only atherosclerosis patients as a control group. In the present study, external validation datasets came from three independent sites and the optimal model had an accuracy of 0.859. In addition, atherosclerosis, intracranial AVM and aneurysm were included in the control group.
In the comprehensive analysis, MRA score, manufacturers, field strength and age at the MR examination were included to explore potential factors that affect the optimal model. According to the results, the accuracy increased with the increase of the MRA score, which might be due to the number of collaterals at the skull base. Images with 1.5 T field strength had a higher accuracy than those with 3.0 T. However, the rate of MMA patients with MRA score ≥11 was higher in 1.5T data than in 3.0 T data. This might be the reason for the different accuracies in the two field strengths. The rate of MMA patients with MRA score ≤5 in the misdiagnosed patients was significantly higher than that with score ≥11. Sixty controls were also misdiagnosed as MMA. The reason might be collaterals of AVM and neovascular of atherosclerosis. Therefore, vascular examinations, such as DSA and MRA, are necessary to evaluate the intracranial vessels when patients with MMA are diagnosed.
There are several limitations in this study. First, only the manifestation of cerebrovascular disease on MRA was considered, but the influence of intracranial brain lesions, such as hydrocephalus, encephalitis and other lesions, on MMA classification was not considered. Although some patients with brain tumors were included in the control group, they were often combined with other imaging examinations so that MMA would not be missed. Second, MMA patients, patients with cerebrovascular disease except MMA and those with normal MRA were not distinguished from each other, and the sample size of patients with cerebrovascular disease except MMA was small. The inconsistent image characteristics of cerebrovascular lesions in patients without MMA, such as atherosclerosis, AVM, and other vascular lesions, could affect the results of the model classification. Moreover, the aim of the study was to detect MMA in clinical patients. Therefore, only binary classification was performed to evaluate the detection rate of MMA. The proportion of patients with cerebrovascular diseases without MMA was also lower than that of those with normal MRA, which may have bias to the result. However, it exists in the real clinical scenario, so few patients with cerebrovascular diseases without MMA were included in the control group of this study. Third, only three slices of T2WI were artificially included in the model classification instead of the whole sequence to improve the accuracy of MMA classification of the model. Future work will include the automatic detection and extraction of characteristic images in T2WI. Forth, since it is not possible to conduct a direct comparison between the model and radiologists under the same diagnostic conditions for MMA, the diagnostic accuracy of radiologists in this study may not accurately reflect their actual performance in real-world clinical settings.
Conclusions
In conclusion, DenseNet had higher sensitivity and accuracy in MMA detection than SCNN, LeNet, VGG and ResNet based on T2WI with different slice thickness and voxel size. No significant difference in classification results was detected between different MR manufacturers and age at the MR examination, but the classification accuracy of MMA with MRA score ≤5 was lower than that of MRA score more than 5. Therefore, the DenseNet model might be used to screen MMA patients in clinical practice to reduce the misdiagnosis rate of MMA patients.
Acknowledgments
None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD-AI reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-24-1269/rc
Funding: This work was supported by
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1269/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the Affiliated Drum Tower Hospital, Medical School of Nanjing University (No. 2021-026). All participating hospitals were informed and agreed to the study. Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Shang S, Zhou D, Ya J, Li S, Yang Q, Ding Y, Ji X, Meng R. Progress in moyamoya disease. Neurosurg Rev 2020;43:371-82. [Crossref] [PubMed]
- Kuroda S, Houkin K. Moyamoya disease: current concepts and future perspectives. Lancet Neurol 2008;7:1056-66. [Crossref] [PubMed]
- Hervé D, Kossorotoff M, Bresson D, Blauwblomme T, Carneiro M, Touze E, et al. French clinical practice guidelines for Moyamoya angiopathy. Rev Neurol (Paris) 2018;174:292-303. [Crossref] [PubMed]
- Houkin K, Nakayama N, Kuroda S, Nonaka T, Shonai T, Yoshimoto T. Novel magnetic resonance angiography stage grading for moyamoya disease. Cerebrovasc Dis 2005;20:347-54. [Crossref] [PubMed]
- Mikami T, Sugino T, Ohtaki S, Houkin K, Mikuni N. Diagnosis of moyamoya disease on magnetic resonance imaging: are flow voids in the basal ganglia an essential criterion for definitive diagnosis? J Stroke Cerebrovasc Dis 2013;22:862-8. [Crossref] [PubMed]
- Ueda D, Katayama Y, Yamamoto A, Ichinose T, Arima H, Watanabe Y, Walston SL, Tatekawa H, Takita H, Honjo T, Shimazaki A, Kabata D, Ichida T, Goto T, Miki Y. Deep Learning-based Angiogram Generation Model for Cerebral Angiography without Misregistration Artifacts. Radiology 2021;299:675-81. [Crossref] [PubMed]
- Shi Z, Miao C, Schoepf UJ, Savage RH, Dargis DM, Pan C, et al. A clinically applicable deep-learning model for detecting intracranial aneurysm in computed tomography angiography images. Nat Commun 2020;11:6090. [Crossref] [PubMed]
- Yang J, Xie M, Hu C, Alwalid O, Xu Y, Liu J, Jin T, Li C, Tu D, Liu X, Zhang C, Li C, Long X. Deep Learning for Detecting Cerebral Aneurysms with CT Angiography. Radiology 2021;298:155-63. [Crossref] [PubMed]
- Ueda D, Yamamoto A, Nishimori M, Shimono T, Doishita S, Shimazaki A, Katayama Y, Fukumoto S, Choppin A, Shimahara Y, Miki Y. Deep Learning for MR Angiography: Automated Detection of Cerebral Aneurysms. Radiology 2019;290:187-94. [Crossref] [PubMed]
- von Schacky CE, Wilhelm NJ, Schäfer VS, Leonhardt Y, Gassert FG, Foreman SC, Gassert FT, Jung M, Jungmann PM, Russe MF, Mogler C, Knebel C, von Eisenhart-Rothe R, Makowski MR, Woertler K, Burgkart R, Gersing AS. Multitask Deep Learning for Segmentation and Classification of Primary Bone Tumors on Radiographs. Radiology 2021;301:398-406. [Crossref] [PubMed]
- Zhang Z, Wang Y, Zhou S, Li Z, Peng Y, Gao S, Zhu G, Wu F, Wu B. The automatic evaluation of steno-occlusive changes in time-of-flight magnetic resonance angiography of moyamoya patients using a 3D coordinate attention residual network. Quant Imaging Med Surg 2023;13:1009-22. [Crossref] [PubMed]
- Kim T, Heo J, Jang DK, Sunwoo L, Kim J, Lee KJ, Kang SH, Park SJ, Kwon OK, Oh CW. Machine learning for detecting moyamoya disease in plain skull radiography using a convolutional neural network. EBioMedicine 2019;40:636-42. [Crossref] [PubMed]
- Lei Y, Zhang X, Ni W, Yang H, Su JB, Xu B, Chen L, Yu JH, Gu YX, Mao Y. Recognition of moyamoya disease and its hemorrhagic risk using deep learning algorithms: sourced from retrospective studies. Neural Regen Res 2021;16:830-5. [Crossref] [PubMed]
- Akiyama Y, Mikami T, Mikuni N. Deep Learning-Based Approach for the Diagnosis of Moyamoya Disease. J Stroke Cerebrovasc Dis 2020;29:105322. [Crossref] [PubMed]
- Zhou Q, Zhu W, Li F, Yuan M, Zheng L, Liu X. Transfer Learning of the ResNet-18 and DenseNet-121 Model Used to Diagnose Intracranial Hemorrhage in CT Scanning. Curr Pharm Des 2022;28:287-95. [Crossref] [PubMed]
- Bajić F, Orel O, Habijan M. A Multi-Purpose Shallow Convolutional Neural Network for Chart Images. Sensors (Basel) 2022;22:7695. [Crossref] [PubMed]
- Zhang Y, Liu YL, Nie K, Zhou J, Chen Z, Chen JH, Wang X, Kim B, Parajuli R, Mehta RS, Wang M, Su MY. Deep Learning-based Automatic Diagnosis of Breast Cancer on MRI Using Mask R-CNN for Detection Followed by ResNet50 for Classification. Acad Radiol 2023;30:S161-71. [Crossref] [PubMed]
- Li Y, Zhang Y, Zhang E, Chen Y, Wang Q, Liu K, Yu HJ, Yuan H, Lang N, Su MY. Differential diagnosis of benign and malignant vertebral fracture on CT using deep learning. Eur Radiol 2021;31:9612-9. [Crossref] [PubMed]
- Kumar V, Prabha C, Sharma P, Mittal N, Askar SS, Abouhawwash M. Unified deep learning models for enhanced lung cancer prediction with ResNet-50-101 and EfficientNet-B3 using DICOM images. BMC Med Imaging 2024;24:63. [Crossref] [PubMed]
- Sabottke CF, Spieler BM. The Effect of Image Resolution on Deep Learning in Radiography. Radiol Artif Intell 2020;2:e190015. [Crossref] [PubMed]
- Tejani AS, Klontzas ME, Gatti AA, Mongan JT, Moy L, Park SH, Kahn CE JrCLAIM 2024 Update Panel. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): 2024 Update. Radiol Artif Intell 2024;6:e240300. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 27-30 June 2016; Las Vegas, NV, USA. IEEE; 2016:770-8.
- Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 [Preprint]. 2014. Available online: https://doi.org/
10.48550 /arXiv.1409.1556 - Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; 21-26 July 2017; Honolulu, HI, USA. IEEE; 2017:4700-8.
- Ben Hamida A, Devanne M, Weber J, Truntzer C, Derangère V, Ghiringhelli F, Forestier G, Wemmert C. Deep learning for colon cancer histopathological images analysis. Comput Biol Med 2021;136:104730. [Crossref] [PubMed]
- Remedios LW, Lingam S, Remedios SW, Gao R, Clark SW, Davis LT, Landman BA. Comparison of convolutional neural networks for detecting large vessel occlusion on computed tomography angiography. Med Phys 2021;48:6060-8. [Crossref] [PubMed]
- Hu T, Lei Y, Su J, Yang H, Ni W, Gao C, Yu J, Wang Y, Gu Y. Learning spatiotemporal features of DSA using 3D CNN and BiConvGRU for ischemic moyamoya disease detection. Int J Neurosci 2023;133:512-22. [Crossref] [PubMed]
- Yin HL, Jiang Y, Huang WJ, Li SH, Lin GW. A Magnetic Resonance Angiography-Based Study Comparing Machine Learning and Clinical Evaluation: Screening Intracranial Regions Associated with the Hemorrhagic Stroke of Adult Moyamoya Disease. J Stroke Cerebrovasc Dis 2022;31:106382. [Crossref] [PubMed]
- Gao T, Zou C, Li J, Han C, Zhang H, Li Y, Tang X, Fan Y. Identification of moyamoya disease based on cerebral oxygen saturation signals using machine learning methods. J Biophotonics 2022;15:e202100388. [Crossref] [PubMed]
- Sawada T, Yamamoto A, Miki Y, Kikuta K, Okada T, Kanagaki M, Kasahara S, Miyamoto S, Takahashi JC, Fukuyama H, Togashi K. Diagnosis of moyamoya disease using 3-T MRI and MRA: value of cisternal moyamoya vessels. Neuroradiology 2012;54:1089-97. [Crossref] [PubMed]
- Mikami T, Kuribara T, Komatsu K, Kimura Y, Wanibuchi M, Houkin K, Mikuni N. Meandering flow void around the splenium in moyamoya disease. Neurol Res 2017;39:702-8. [Crossref] [PubMed]


