Bibliometric analysis of research on the application of deep learning to ophthalmology
Introduction
Deep learning has become a popular area in the field of machine learning and artificial intelligence (AI) research (1), and has attracted significant attention in scientific investigations. It is widely used in many fields, such as computer vision, natural language processing, and data mining. Recently, deep learning has revolutionized the diagnosis and prediction of ocular diseases, especially fundus diseases (2,3). Numerous studies have achieved excellent results in applying deep learning to the management of eye diseases by training and validating algorithms for diabetic retinopathy (DR) (4,5), age-related macular degeneration (6,7), glaucoma (8), myopia (9), and retinopathy of prematurity (10,11). In various diagnostic tasks, deep learning models, such as those studied by Gulshan et al. (12) and Ting et al. (13), have achieved levels of accuracy comparable to medical experts. The application of deep learning techniques to three major tasks in ophthalmology (i.e., segmentation, classification, and prediction) has greatly enhanced the efficiency and accuracy of disease diagnosis and management.
This study was the first to conduct a bibliometric analysis of deep learning applied to ophthalmology research. It aimed to identify publishing trends in deep learning ophthalmology research via an analysis of ophthalmology-related literature, and to identify influential journals, institutions, authors, and countries. Additionally, it sought to explore international collaboration networks, research hotspots, and emerging topics. This study also aimed to provide a comprehensive bibliometric perspective on the application of deep learning to ophthalmology, providing readers with a quick overview of the field and its advancements.
Methods
Inclusion and exclusion criteria
Articles were included in the meta-analysis if they met the following criteria: (I) included the following search terms: Topic = (“deep learning”) and Topic = (“ocular” or “Ophthalmol” or “Cornea” or “glaucoma” or “retinopathy” or “retinal” or “cataract” or “choroidal neovascularization” or “strabismus” or “refractive error” or “myopia” or “macula” or “fundus” or “optic nerve” or “optic disc” or “optic cup”); (II) were published between 2015 and 2024; and (III) had an “article” document type. All the retrieved articles were manually checked, and any irrelevant articles were excluded.
Data collection
The Web of Science (WOS) is a multidisciplinary literature database that contains a number of multi-seeded repositories, including the Science Citation Index Expanded, Social Sciences Citation Index, Arts and Humanities Citation Index, and Conference Proceedings Citation Index. It contains more than one billion searchable citations across more than 250 disciplines, and it also has great influence and authority worldwide. For this study, the literature data were collected on a specific day (September 11, 2024). The retrieved articles had to meet the inclusion and exclusion criteria. In total, 3,428 publications were manually reviewed, 373 irrelevant articles were then excluded, and the bibliometric data of 3,055 publications were downloaded in plain-text format from the WOS Core Collection (WOSCC). The following information was collected from the articles: journal, country, institution, author, keyword, and reference.
Analytical tools and visualization maps
Visualization of similarities (VOS)-viewer is a widely used bibliometric analysis software that facilitates various types of qualitative studies. In this study, VOS-viewer (version 1.6.18) (14) was used to analyze the co-authorship network of institutions and authors. A visual map was created for the co-occurrence analysis of high-frequency author keywords. A descriptive analysis was conducted of the publication year, journal, country, institution, and citation text. The bibliometric approach was used in this study to identify the topic and purpose of each study. It revealed the relationships between authors and institutions, highlighting hot topics and trends by examining high-frequency words in the literature to examine the knowledge base of the field.
Results
Publishing trends
From 2015 to 2024, a total of 3,428 articles were retrieved from the WOSCC. After strictly applying the inclusion and exclusion criteria, 3055 articles remained. The first article on deep learning applied to ophthalmic research was published in 2015 (15). The number of publications gradually increased from 2015 to 2018, but in this period, fewer than 100 articles were published per year. However, from 2019 to 2023, there was a rapid surge in publications, and in 2023, 725 articles were published. This represented a significant 6.47-fold increase compared to the total number of publications from 2015 to 2018. In total, 92 countries conducted deep learning research in ophthalmology. The global distribution of deep learning research is depicted in Figure 1. Among these countries, China (n=1,187) had the highest number of publications, followed by the United States (n=673), India (n=425), United Kingdom (n=249), and South Korea (n=236).
High-impact institutions, authors, and cooperations
A total of 13,383 authors affiliated with 3,691 institutions worldwide contributed to the field of deep learning in ophthalmic research. Of the 3,691 institutions included in the study, 166 high-producing institutions were identified with a set threshold of 10. Similarly, of the 13,383 authors, 133 high-producing authors were identified with a set threshold of 10. As Table 1 shows, Sun Yat-sen University emerged as the leading institution with the highest number of published articles (n=109). The Chinese Academy of Sciences and Capital Medical University also made notable contributions. Among the institutions, the Chinese Academy of Sciences had the highest number of citations. Cheng and Bogunovic were the most prolific authors, having published the most articles (n=35), followed by Schmidt-Erfurth. While Raman was the most cited author in the field.
Table 1
| Rank | Institutions | Authors | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Content | TP | TC | Connections | Content | TP | TC | Connections | ||
| 1 | Sun Yat-sen University | 109 | 3,424 | 75 | Ching-Yu Cheng | 35 | 2,115 | 44 | |
| 2 | Chinese Academy of Sciences | 100 | 4,497 | 65 | Hrvoje Bogunovic | 35 | 1,407 | 11 | |
| 3 | Capital Medical University | 89 | 3,752 | 70 | Ursula Schmidt-Erfurth | 33 | 976 | 5 | |
| 4 | Singapore National Eye Center | 80 | 3,930 | 74 | Tien Yin Wong | 32 | 2,473 | 43 | |
| 5 | Shanghai Jiao Tong University | 74 | 3,210 | 58 | Haotian Lin | 30 | 935 | 30 | |
| 6 | National University Singapore | 70 | 3,478 | 66 | Carol Y. Cheung | 30 | 831 | 46 | |
| 7 | Wenzhou Medical University | 64 | 702 | 41 | Juan Ye | 30 | 420 | 4 | |
| 8 | Jinan University | 61 | 598 | 43 | Jiang Liu | 29 | 2,615 | 9 | |
| 9 | Medical University Vienna | 60 | 1,862 | 41 | Yitian Zhao | 27 | 1,848 | 13 | |
| 10 | Zhejiang University | 57 | 812 | 30 | Mingguang He | 27 | 1,071 | 18 | |
| 11 | Chinese University Hong Kong | 55 | 3,566 | 73 | Pearse A. Keane | 25 | 2,468 | 27 | |
| 12 | Stanford University | 54 | 3,211 | 45 | Aaron Y. Lee | 25 | 719 | 22 | |
| 13 | Seoul National University | 54 | 1,136 | 23 | Rajiv Raman | 23 | 4,353 | 25 | |
| 14 | Duke Nus Medical School | 50 | 1,692 | 49 | Kai Jin | 21 | 388 | 3 | |
| 15 | University College London | 46 | 2,680 | 52 | Weihua Yang | 21 | 185 | 2 | |
| 16 | Duke University | 46 | 1,521 | 24 | Emily Y. Chew | 20 | 551 | 19 | |
| 17 | Sungkyunkwan University | 45 | 847 | 35 | Tin Aung | 19 | 1,849 | 23 | |
| 18 | Johns Hopkins University | 43 | 1,916 | 38 | Daniel S. W. Ting | 19 | 988 | 41 | |
| 19 | Moorfields Eye Hospital | 43 | 1,338 | 46 | Tyler Hyungtaek Rim | 19 | 702 | 30 | |
| 20 | University Washington | 42 | 950 | 47 | Yih-Chung Tham | 19 | 608 | 37 | |
TP, total publications; TC, total citations; connections, cooperation between institutions or authors.
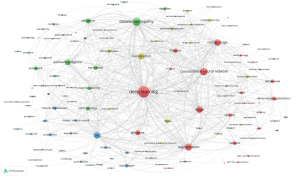
To examine the global cooperations, a co-authorship analysis of the 166 high-producing institutions was performed (Figure 2). Of these 166 high-producing institutions, the largest shared network comprised 164 institutions, which were further divided into eight distinct clusters. Sun Yat-sen University, Capital Medical University, Singapore National Eye Center, Medical University Vienna, Stanford University, Duke University, Zhejiang University, and Moorfields Eye Hospital were the core units of each cluster. The red cluster, which comprised 32 institutions, was the largest, and Sun Yat-sen University and Chinese Academy of Sciences were prominent nodes in this cluster. Table 1 details the cooperation connections between institutions. Sun Yat-sen University was the high-producing institution with the most connections in the network, with 75 connections, followed by Singapore National Eye Centre, with 74 connections, and Chinese University Hong Kong, with 73 connections. Further, 114 authors formed a collaborative network, resulting in 10 clusters. Cheung has collaborated with 46 highly productive authors, Cheng has collaborated with 44 highly productive authors, and Wong has collaborated with 43 highly productive authors. The co-authorship network of authors is shown in Figure 3.
Co-occurrence analysis of keywords
A total of 6,330 keywords were selected from the 3,055 retrieved articles. The threshold for high-frequency author keywords was set at 20, resulting in 120 keywords that met the criteria (Table S1). A co-occurrence analysis was conducted of these 120 high-frequency authorship keywords, and a network diagram was created to visualize the high-frequency keywords. As Figure 4 shows, the keywords with the highest frequency positioned at the center of the network diagram were deep learning (n=1,530), DR (n=797), convolutional neural network (CNN) (n=658), optical coherence tomography (OCT) (n=567), and segmentation (n=561). The following four clusters were identified based on keywords: (I) deep learning for the segmentation and feature extraction of ophthalmic images; (II) deep learning for the automatic detection and classification of ophthalmic images; (III) application of deep learning to ophthalmic imaging techniques; and (IV) deep learning for the diagnosis and management of ophthalmic diseases.
Highly cited references
Scientific research is based on previous research, and the analysis of references can help us to understand the knowledge base of a particular field of research. A total of 3,055 publications cited 60,147 references. The top 20 references with the highest number of citations are shown in Table 2, and the number of citations for the top 20 was distributed between 166 and 749.
Table 2
| Rank | Year | First author | Title | Source | Citations | DOI |
|---|---|---|---|---|---|---|
| 1 | 2015 | Olaf Ronneberger | U-Net: convolutional networks for biomedical image segmentation | Lecture Notes in Computer Science | 749 | 10.1007/978-3-319-24574-4_28 |
| 2 | 2016 | Kaiming He | Deep residual learning for image recognition | IEEE Conference on Computer Vision and Pattern Recognition | 678 | 10.1109/cvpr.2016.90 |
| 3 | 2016 | Varun Gulshan | Development and validation of a deep-learning algorithm for detection of diabetic retinopathy in retinal fundus photographs | Jama-Journal of the American Medical Association | 631 | 10.1001/jama.2016.17216 |
| 4 | 2017 | Alex Krizhevsky | ImageNet classification with deep convolutional neural networks | Communications of the ACM | 405 | 10.1145/3065386 |
| 5 | 2017 | Daniel Shu Wei Ting | Development and validation of a deep learning system for diabetic retinopathy and related eye diseases using retinal images from multiethnic populations with diabetes | Jama-Journal of the American Medical Association | 351 | 10.1001/jama.2017.18152 |
| 6 | 2004 | J. Staal | Ridge-based vessel segmentation in color images of the retina | IEEE Transactions on Medical Imaging | 288 | 10.1109/tmi.2004.825627 |
| 7 | 2017 | Rishab Gargeya | Automated identification of diabetic retinopathy using deep learning | Ophthalmology | 261 | 10.1016/j.ophtha.2017.02.008 |
| 8 | 2016 | Christian Szegedy | Rethinking the inception architecture for computer vision | IEEE Conference on Computer Vision and Pattern Recognition | 253 | 10.1109/cvpr.2016.308 |
| 9 | 2018 | Daniel Shafiee Kermany | Identifying medical diagnoses and treatable diseases by image-based deep learning | Cell | 234 | 10.1016/j.cell.2018.02.010 |
| 10 | 2009 | Jia Deng | ImageNet: a large-scale hierarchical image database | IEEE Conference on Computer Vision and Pattern Recognition | 231 | 10.1109/cvprw.2009.5206848 |
| 11 | 2017 | Gao Huang | Densely connected convolutional networks | IEEE Conference on Computer Vision and Pattern Recognition | 226 | 10.1109/cvpr.2017.243 |
| 12 | 2018 | Jeffrey De Fauw | Clinically applicable deep learning for diagnosis and referral in retinal disease | Nature Medicine | 211 | 10.1038/s41591-018-0107-6 |
| 13 | 2014 | Etienne Decenciere | Feedback on a publicly distributed image database: the Messidor database | Image Analysis & Stereology | 197 | 10.5566/ias.1155 |
| 14 | 2015 | Jonathan Long | Fully convolutional networks for semantic segmentation | IEEE Conference on Computer Vision and Pattern Recognition | 192 | 10.1109/cvpr.2015.7298965 |
| 15 | 2016 | Michael David Abramoff | Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning | Investigative Ophthalmology & Visual Science | 190 | 10.1167/iovs.16-19964 |
| 16 | 2018 | Zhixi Li | Efficacy of a deep learning system for detecting glaucomatous optic neuropathy based on color fundus photographs | Ophthalmology | 189 | 10.1016/j.ophtha.2018.01.023 |
| 17 | 2000 | Alicia Hoover | Locating blood vessels in retinal images by piecewise threshold probing of a matched filter response | IEEE Transactions on Medical Imaging | 178 | 10.1109/42.845178 |
| 18 | 2015 | Diederik P. Kingma | Adam: a method for stochastic optimization | International Conference on Learning Representations | 167 | 10.48550/arxiv.1412.6980 |
| 19 | 2017 | Geert Litjens | A survey on deep learning in medical image analysis | Medical Image Analysis | 167 | 10.1016/j.media.2017.07.005 |
| 20 | 2015 | Olga Russakovsky | ImageNet large-scale visual recognition challenge | International Journal of Computer Vision | 166 | 10.1007/s11263-015-0816-y |
Discussion
This study analyzed 3,055 articles in the WOSCC to identify hotspots and trends in deep learning applied to ophthalmic research from 2015 to 2024. In this study, bibliometric techniques were used to elucidate the research trends and hotspots in deep learning applied to ophthalmic research to identify the key trends, influential contributors (journals, countries, institutions, authors), and collaborative networks in the field. International collaboration networks, research hotspots, and emerging themes were also investigated to inform further research on deep learning applied to ophthalmology.
Publishing trends
The research literature on deep learning in ophthalmology has shown consistent growth over the years since the initial publication in this area in 2015. There was a significant surge in publications from 2019 to 2023, with the volume surpassing six times that of the previous 4 years combined. In terms of publication distribution by countries/regions, China had the largest share, followed by the United States.
International cooperation
Deep learning, as an emerging and interdisciplinary field, demonstrates extensive international cooperation. According to the co-authorship analysis, the greatest shared network comprised 164 institutions (98.80%), and approximately 85% of the high-producing authors were interconnected in the co-authorship network, indicating significant collaborations among them. Collaborations among high-yield institutions and high-yield authors enable deep learning to be applied to ophthalmic research at a deeper level and on a broader platform. In addition, high-yield institutions may offer financial support or mentorship to high-yield authors, providing them with the resources and guidance necessary to succeed in their research.
Research hot spots and emerging topics
The application of deep learning to the medical field has yielded significant outcomes, particularly in ophthalmology. The integration of deep learning and diverse imaging data has facilitated comprehensive disease analysis, enhancing the diagnosis of ophthalmic disorders. This has expanded the potential of the medical model by offering new avenues for diagnosis and treatment.
Cluster 1: deep learning in the segmentation and feature extraction of ocular images
Deep learning models have recently been widely used in computer vision (16), and have been mainly used in retinal image segmentation and feature extraction in ophthalmology (17,18). In image processing, CNNs are the most commonly used method for feature extraction. CNNs can be trained to segment retinal blood vessels and optic discs in large medical image datasets, and the use of generative adversarial networks (GANs) and the U-net framework can further improve segmentation accuracy.
In the field of image processing, widely used deep learning-based features are extracted by CNNs, which can learn relevant features and patterns from large retinal image datasets through a supervised learning process. In the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC) held in 2012, Krizhevsky et al. (19) used a CNN trained by this technique to reduce the average error rate of a 1,000-level target classification test from 25% to 16%. In recent years, several typical CNN architectures and fully convolutional network architectures have been effectively used for retinal fluid segmentation in OCT images. Lu et al. (17) proposed a framework for multi-class fluid segmentation and detection in retinal OCT images, which achieved the highest score in the Medical Image Computing and Computer-Assisted Intervention RETOUCH challenge held in 2017. Padilla-Pantoja et al. (20) used a deep learning model of CNNs for semantic segmentation and feature extraction to identify macular edema, which had an accuracy, sensitivity, and specificity comparable to those of experts.
To meet some particular retinal fluid segmentation demands, many researchers have improved the traditional CNN architecture. Girish et al. (21) preprocessed selected regions in OCT images to enhance the semantics, and then segmented the intra-retinal-cyst regions using a CNN with a depth-separable convolutional filter, which not only helped in the generalization of the model but also avoided excessive barriers.
GANs were proposed by Goodfellow (22) as an unsupervised generative model. GANs can be used for a variety of applications in ophthalmology, including the segmentation of ocular structures such as the retina, optic nerve, and macula. Budai et al. (23) proposed a new vessel segmentation method that improves on the method proposed by Frangi, shortens the runtime, and segments low-resolution vessels. Deep learning-based models (24) that use slit-lamp anterior segmentation images to automatically segment and evaluate corneal neovascularization also provide a fast and efficient method for clinical disease assessment. GANs can also increase the training data by generating synthetic images with corresponding masks (25) to improve the performance of segmentation algorithms. In addition to segmentation, GANs have a role in image enhancement (26).
As another classical image segmentation framework, U-net was proposed by Ronneberger (27) in 2015. Lou et al. (28) proposed a U-net-based recursive residual CNN image analysis technique to measure the degree of inferior oblique overactivity through eye localization and segmentation, which provides an objective and accurate measurement for clinical practice and is expected to be applied to telemedicine. The ReLayNet, proposed by Roy et al. (29), performs the end-to-end segmentation of retinal layers and fluid masses. The framework has been extensively measured and validated against five state-of-the-art retinal layer segmentation methods (30-32). Conversely, the ReLayNet was shown to reliably segment retinal layers, even under highly pathologic conditions that severely affect the normal retinal layering structure. The use of the U-net in retinal image segmentation in ophthalmology can significantly reduce the amount of manual annotation required for training data, making it a more efficient and scalable method. In addition, the U-net is robust to changes in image quality and illumination, making it ideal for processing medical images.
Cluster 2: deep learning in automated detection and classification of ocular images
Deep learning algorithms have excelled in detecting and classifying retinal diseases (33-35) by using various image processing techniques, such as CNNs, transfer learning, attention mechanisms, and support vector machines. In 1998, the earliest classical CNN model for image classification was developed (36). The later AlexNet model (37) triggered the deep learning boom by significantly outperforming other methods in the ILSVRC held in 2012. Subsequent developments of the GoogLeNet model, the visual geometry group (VGG) model, the ResNet model, and the SENet model all did well in the ILSVRC.
Several studies have reported on the performance of CNN image classification in OCT (38-40). These studies combined CNN algorithms with OCT images to detect and characterize intraretinal fluid areas, and reported a performance comparable to that of retinal experts. For example, Li et al. (41) fine-tuned a VGG-16 network pre-trained on the ImageNet using a transfer learning method to classify diabetic macular edema (DME) and DR in OCT images, which showed a performance comparable to that of ophthalmologists. In another study, Schlegl (42) proposed and validated a robust and sensitive automated method that incorporated deep learning techniques to classify intraretinal cystic fluid, subretinal fluid, and non-fluid areas in OCT images. This powerful tool was found to be effective in detecting and quantifying various types of exudative macular fluid areas and exudative macular diseases.
In addition, some deep learning-based methods have been shown to be highly consistent with manual measurements in the diagnosis and analysis of eyelid diseases (43) and thyroid-associated ophthalmopathies (44), providing new tools for the diagnosis and treatment evaluation of more ophthalmopathies. Deep learning-based methods have also been used in computer-aided detection systems for DR. A trial conducted by He et al. (45) showed the high sensitivity and specificity of an AI-based system for the detection of DR and referable DR, demonstrating the feasibility of AI-based screening in community hospital outpatient clinics.
However, the application of deep learning to ophthalmology has its limitations. One of the challenges is the presence of artifacts in retinal images, which may affect the accuracy of automatic detection and classification. The interpretability of deep learning models is still a challenge that can affect their application in clinical decision making. The use of deep learning methods is expected to increase in the coming years as technology advances and more data becomes available.
Cluster 3: application of deep learning algorithms to ophthalmic imaging technology
Deep learning has been successfully applied to medical imaging, such as computed tomography and magnetic resonance imaging (46). In ophthalmology, it has been combined with fundus imaging techniques, including OCT and OCT angiography (OCTA), and fundus color photography, as well as Optomap Daytona scanning laser ophthalmoscopy and fundus fluorescein angiography (FFA), corneal topography (CT), and visual field (VF). OCT, the most commonly used ophthalmic examination instrument, has revolutionized the diagnosis and management of various ophthalmic diseases by analyzing OCT and OCTA images using deep learning technology (47-49). With the availability of large amounts of data and digitized visual images, deep learning algorithms can analyze these images and make informed decisions to assist in detecting and diagnosing ophthalmic diseases (50-52).
Lee et al. (53) showed the effectiveness of deep learning techniques in differentiating between the OCT images of normal and age-related macular degeneration patients by creating a database of OCT images linked to clinical endpoints in electronic medical records. By analyzing OCT images, deep learning algorithms can accurately identify DR for lesion detection and grading (4). Deep learning has also been used to develop automated diagnostic algorithms for ophthalmic diseases (54). Wang et al. (52) were the first to employ CNNs to diagnose and segment choroidal neovascularization in OCTA images.
Karri et al. (55) proposed a transfer learning-based OCT image classification method. They fine-tuned the GoogleNet model before training it on a small dataset. This study established a novel approach for training CNN models on small-scale datasets, and showed the potential of transfer learning in addressing the challenges posed by limited medical image data. A multi-view approach based on transfer learning (56) has also achieved robust and fully automated retinal layer segmentation in glaucoma patients with satisfactory results.
The combination of deep learning with other ophthalmic imaging technologies also shows the importance of deep learning technology. Campbell et al. (57) developed a computer-based image analysis system to analyze 77 fundus photographs for quantitative analysis of retinal vascular characteristics in retinopathy of prematurity. Yang et al. (58) combined deep learning with Optomap Daytona scanning laser ophthalmoscopy for refractive error prediction using ultra-widefield fundus photographs. Jin et al. (59,60) used FFA to detect and classify DR. In addition, their team automatically used FFA to detect non-perfused areas, microaneurysms, and leaks in DME. In addition, deep learning has also been applied to assist in the diagnosis of other ophthalmic diseases, and deep learning techniques combined with CT can be used for the detection and classification of conical corneas, and to assist optometrists in the fitting of keratoplasty lenses (61,62). Hussain et al. (63) developed a multimodal deep learning model that combines a patient’s OCT maps and VF maps with clinical data to predict glaucoma progression, which can help tailor treatment to the patient.
Cluster 4: deep learning in diagnosis and management of ocular diseases
The effectiveness of diagnosing and treating ocular illnesses is greatly enhanced by deep learning. In ophthalmology, deep learning is most often combined with clinical use in the diagnosis and management of DR. The increase in the prevalence of diabetes globally has led to an increase in the cost of DR screening in public health systems. Thus, there is a need to develop accurate and efficient diagnostic tools, as early detection and treatment are crucial in preventing vision loss (64). To optimize the screening process, Dai et al. (65) developed the DeepDR deep learning system. This system was validated on a local dataset and three external datasets, and had high accuracy with an area under the grading curve greater than 0.94 for different stages of DR. Similar deep learning methods for the automatic detection of DR have been developed by Gulshan et al. (12) and Ting et al. (3), both of which showed good sensitivity and specificity in evaluating fundus images.
Validation and accuracy are critical to the application of deep learning and machine learning algorithms in ophthalmology. Studies have shown that these algorithms can achieve high accuracy in detecting and assessing DR risk factors such as retinal damage and microaneurysms. The high agreement between human experts and these algorithms in DR diagnosis (12,13) also suggests that deep learning algorithms can be an important tool in ophthalmology.
Telemedicine has emerged as a promising approach for improving access to eye care in remote areas (66). Deep learning combined with DR screening can accurately detect and assess DR risk factors in retinal images, which can help alleviate the burden on ophthalmologists and improve access to eye care services (67-70). Li et al. (71) completed China’s first population-wide digital ophthalmology telescreening study applied to urban and rural areas, combining traditional and telemedicine system technologies, and conducted a cost-benefit analysis of age-related macular degeneration and DR telescreening, and found that the telescreen model-based on teleanalysis was effective and feasible. In addition to DR, deep learning and machine learning algorithms have shown encouraging results in the diagnosis and management of other ophthalmic diseases, such as open-angle glaucoma (72,73), myopia (58,74), retinopathy of prematurity (75-77), and keratitis. It is important to note that the diagnosis of glaucoma depends primarily on intraocular pressure, retinal nerve fiber thickness, optic nerve, and VF examination, and unlike DR, it is not a visual disease. Therefore, other examination results, such as OCT images, intraocular pressure, optic disc photographs, and VF data, need to be integrated into AI systems.
Although deep learning and machine learning algorithms have shown great potential in ophthalmology, their application still has limitations. For instance, these algorithms may be affected by variations in image quality and illumination, which may affect accuracy. Additionally, large datasets of high-quality images are needed to train the algorithms, which can be time-consuming and resource-intensive. More research needs to be conducted to improve the availability and quality of image data sets to improve the accuracy of the algorithms.
Limitations
This study had certain limitations that should be acknowledged. First, one limitation relates to the irregular spelling of authors’ names and institutions in publications, which might have introduced bias into the statistical results. The use of consistent identity documents (such as Open Researcher and Contributor Identifiers) could reduce this bias. Second, the literature included in this study was exclusively sourced from the WOS. The WOS holds a substantial amount of data; however, it is important to note that articles may exist that were not captured in the WOS, which might have resulted in deviations in the findings. Third, machine learning also belongs to AI; however, in this study, we mainly focused on the application of deep learning to ophthalmology. Finally, it would be remiss not to consider other potential biases. For example, well-connected groups frequently establish robust collaborative networks and publish articles together. Similarly, prominent authors are frequently invited to serve as co-authors to increase citations.
Conclusions
As deep learning technology continues to advance, it has great potential to contribute to the diagnosis and management of ophthalmic diseases. Through a visual data analysis, we learned about the current state of the application of deep learning to ophthalmology and its future development. Influential authors and institutions, collaborative networks, and emerging themes in the research provide valuable resources for subsequent researchers, as well as policymakers, to further advance the exploration of the field.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-1340/coif). C.P. reports that this work was supported by the Liaoning Provincial Science and Technology Medical-Industrial Intersection Joint Fund (No. 2022-YGJC-51) and the Shenyang Science and Technology Bureau Youth Talent Support Program (No. RC230675). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- De Fauw J, Ledsam JR, Romera-Paredes B, Nikolov S, Tomasev N, Blackwell S, et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med 2018;24:1342-50. [Crossref] [PubMed]
- Ting DSW, Peng L, Varadarajan AV, Keane PA, Burlina PM, Chiang MF, Schmetterer L, Pasquale LR, Bressler NM, Webster DR, Abramoff M, Wong TY. Deep learning in ophthalmology: The technical and clinical considerations. Prog Retin Eye Res 2019;72:100759. [Crossref] [PubMed]
- Wang Y, Yu M, Hu B, Jin X, Li Y, Zhang X, et al. Deep learning-based detection and stage grading for optimising diagnosis of diabetic retinopathy. Diabetes Metab Res Rev 2021;37:e3445. [Crossref] [PubMed]
- Gargeya R, Leng T. Automated Identification of Diabetic Retinopathy Using Deep Learning. Ophthalmology 2017;124:962-9. [Crossref] [PubMed]
- Grassmann F, Mengelkamp J, Brandl C, Harsch S, Zimmermann ME, Linkohr B, Peters A, Heid IM, Palm C, Weber BHF. A Deep Learning Algorithm for Prediction of Age-Related Eye Disease Study Severity Scale for Age-Related Macular Degeneration from Color Fundus Photography. Ophthalmology 2018;125:1410-20. [Crossref] [PubMed]
- Fang L, Cunefare D, Wang C, Guymer RH, Li S, Farsiu S. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed Opt Express 2017;8:2732-44. [Crossref] [PubMed]
- Phene S, Dunn RC, Hammel N, Liu Y, Krause J, Kitade N, Schaekermann M, Sayres R, Wu DJ, Bora A, Semturs C, Misra A, Huang AE, Spitze A, Medeiros FA, Maa AY, Gandhi M, Corrado GS, Peng L, Webster DR. Deep Learning and Glaucoma Specialists: The Relative Importance of Optic Disc Features to Predict Glaucoma Referral in Fundus Photographs. Ophthalmology 2019;126:1627-39. [Crossref] [PubMed]
- Xuan M, Wang W, Shi D, Tong J, Zhu Z, Jiang Y, Ge Z, Zhang J, Bulloch G, Peng G, Meng W, Li C, Xiong R, Yuan Y, He M. A Deep Learning-Based Fully Automated Program for Choroidal Structure Analysis Within the Region of Interest in Myopic Children. Transl Vis Sci Technol 2023;12:22. [Crossref] [PubMed]
- Luo Z, Ding X, Hou N, Wan J. A Deep-Learning-Based Collaborative Edge-Cloud Telemedicine System for Retinopathy of Prematurity. Sensors (Basel) 2022.
- Bao Y, Ming WK, Mou ZW, Kong QH, Li A, Yuan TF, Mi XS. Current Application of Digital Diagnosing Systems for Retinopathy of Prematurity. Comput Methods Programs Biomed 2021;200:105871. [Crossref] [PubMed]
- Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, Kim R, Raman R, Nelson PC, Mega JL, Webster DR. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016;316:2402-10. [Crossref] [PubMed]
- Ting DSW, Cheung CY, Lim G, Tan GSW, Quang ND, Gan A, et al. Development and Validation of a Deep Learning System for Diabetic Retinopathy and Related Eye Diseases Using Retinal Images From Multiethnic Populations With Diabetes. JAMA 2017;318:2211-23. [Crossref] [PubMed]
- van Eck NJ, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010;84:523-38. [Crossref] [PubMed]
- Gao X, Lin S, Wong TY. Automatic Feature Learning to Grade Nuclear Cataracts Based on Deep Learning. IEEE Trans Biomed Eng 2015;62:2693-701. [Crossref] [PubMed]
- Minaee S, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans Pattern Anal Mach Intell 2022;44:3523-42. [Crossref] [PubMed]
- Lu D, Heisler M, Lee S, Ding GW, Navajas E, Sarunic MV, Beg MF. Deep-learning based multiclass retinal fluid segmentation and detection in optical coherence tomography images using a fully convolutional neural network. Med Image Anal 2019;54:100-10. [Crossref] [PubMed]
- Staal J, Abràmoff MD, Niemeijer M, Viergever MA, van Ginneken B. Ridge-based vessel segmentation in color images of the retina. IEEE Trans Med Imaging 2004;23:501-9. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Communications of the ACM 2017;60:84-90.
- Padilla-Pantoja FD, Sanchez YD, Quijano-Nieto BA, Perdomo OJ, Gonzalez FA. Etiology of Macular Edema Defined by Deep Learning in Optical Coherence Tomography Scans. Transl Vis Sci Technol 2022;11:29. [Crossref] [PubMed]
- Girish GN, Saikumar B, Roychowdhury S, Kothari AR, Rajan J. Depthwise Separable Convolutional Neural Network Model for Intra-Retinal Cyst Segmentation. Annu Int Conf IEEE Eng Med Biol Soc 2019;2019:2027-31. [Crossref] [PubMed]
- Goodfellow IJ, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial networks. Communications of the ACM 2020;63:139-44.
- Budai A, Bock R, Maier A, Hornegger J, Michelson G. Robust vessel segmentation in fundus images. Int J Biomed Imaging 2013;2013:154860. [Crossref] [PubMed]
- Chu X, Wang X, Zhang C, Liu H, Li F, Li G, Zhao S. A deep learning-based model for automatic segmentation and evaluation of corneal neovascularization using slit-lamp anterior segment images. Quant Imaging Med Surg 2023;13:6778-88. [Crossref] [PubMed]
- Thambawita V, Salehi P, Sheshkal SA, Hicks SA, Hammer HL, Parasa S, Lange T, Halvorsen P, Riegler MA. SinGAN-Seg: Synthetic training data generation for medical image segmentation. PLoS One 2022;17:e0267976. [Crossref] [PubMed]
- Kebaili A, Lapuyade-Lahorgue J, Ruan S. Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. J Imaging 2023;9:81. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. arXiv:1505.04597. 2015. Available online: https://arxiv.org/abs/1505.04597
- Lou L, Huang X, Sun Y, Cao J, Wang Y, Zhang Q, Tang X, Ye J. Automated photographic analysis of inferior oblique overaction based on deep learning. Quant Imaging Med Surg 2023;13:329-38. [Crossref] [PubMed]
- Roy AG, Conjeti S, Karri SPK, Sheet D, Katouzian A, Wachinger C, Navab N. ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks. Biomed Opt Express 2017;8:3627-42. [Crossref] [PubMed]
- Karri SP, Chakraborthi D, Chatterjee J. Learning layer-specific edges for segmenting retinal layers with large deformations. Biomed Opt Express 2016;7:2888-901. [Crossref] [PubMed]
- Chiu SJ, Li XT, Nicholas P, Toth CA, Izatt JA, Farsiu S. Automatic segmentation of seven retinal layers in SDOCT images congruent with expert manual segmentation. Opt Express 2010;18:19413-28. [Crossref] [PubMed]
- Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640-51. [Crossref] [PubMed]
- Peng Y, Dharssi S, Chen Q, Keenan TD, Agrón E, Wong WT, Chew EY, Lu Z. DeepSeeNet: A Deep Learning Model for Automated Classification of Patient-based Age-related Macular Degeneration Severity from Color Fundus Photographs. Ophthalmology 2019;126:565-75. [Crossref] [PubMed]
- Ma F, Liu X, Wang S, Li S, Dai C, Meng J. CSANet: a lightweight channel and spatial attention neural network for grading diabetic retinopathy with optical coherence tomography angiography. Quant Imaging Med Surg 2024;14:1820-34. [Crossref] [PubMed]
- Rivas-Villar D, Hervella ÁS, Rouco J, Novo J. Joint keypoint detection and description network for color fundus image registration. Quant Imaging Med Surg 2023;13:4540-62. [Crossref] [PubMed]
- LeCun Y, Bottou L, Bengio Y, Haffner P. Gradient-based learning applied to document recognition. Proc IEEE 1998;86:2278-324.
- Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang ZH, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L. Imagenet large scale visual recognition challenge. Int J Comput Vis 2015;115:211-52.
- Chakravarthy U, Goldenberg D, Young G, Havilio M, Rafaeli O, Benyamini G, Loewenstein A. Automated Identification of Lesion Activity in Neovascular Age-Related Macular Degeneration. Ophthalmology 2016;123:1731-6. [Crossref] [PubMed]
- Samagaio G, Estévez A, Moura J, Novo J, Fernández MI, Ortega M. Automatic macular edema identification and characterization using OCT images. Comput Methods Programs Biomed 2018;163:47-63. [Crossref] [PubMed]
- Christopher M, Bowd C, Belghith A, Goldbaum MH, Weinreb RN, Fazio MA, Girkin CA, Liebmann JM, Zangwill LM. Deep Learning Approaches Predict Glaucomatous Visual Field Damage from OCT Optic Nerve Head En Face Images and Retinal Nerve Fiber Layer Thickness Maps. Ophthalmology 2020;127:346-56. [Crossref] [PubMed]
- Li F, Chen H, Liu Z, Zhang X, Wu Z. Fully automated detection of retinal disorders by image-based deep learning. Graefes Arch Clin Exp Ophthalmol 2019;257:495-505. [Crossref] [PubMed]
- Schlegl T, Waldstein SM, Bogunovic H, Endstraßer F, Sadeghipour A, Philip AM, Podkowinski D, Gerendas BS, Langs G, Schmidt-Erfurth U. Fully Automated Detection and Quantification of Macular Fluid in OCT Using Deep Learning. Ophthalmology 2018;125:549-58. [Crossref] [PubMed]
- Luo Y, Zhang J, Yang Y, Rao Y, Chen X, Shi T, Xu S, Jia R, Gao X. Deep learning-based fully automated differential diagnosis of eyelid basal cell and sebaceous carcinoma using whole slide images. Quant Imaging Med Surg 2022;12:4166-75. [Crossref] [PubMed]
- Shao J, Huang X, Gao T, Cao J, Wang Y, Zhang Q, Lou L, Ye J. Deep learning-based image analysis of eyelid morphology in thyroid-associated ophthalmopathy. Quant Imaging Med Surg 2023;13:1592-604. [Crossref] [PubMed]
- He J, Cao T, Xu F, Wang S, Tao H, Wu T, Sun L, Chen J. Artificial intelligence-based screening for diabetic retinopathy at community hospital. Eye (Lond) 2020;34:572-6. [Crossref] [PubMed]
- Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys 2019;29:102-27. [Crossref] [PubMed]
- Farsiu S, Chiu SJ, O'Connell RV, Folgar FA, Yuan E, Izatt JA, Toth CA. Age-Related Eye Disease Study 2 Ancillary Spectral Domain Optical Coherence Tomography Study Group. Quantitative classification of eyes with and without intermediate age-related macular degeneration using optical coherence tomography. Ophthalmology 2014;121:162-72. [Crossref] [PubMed]
- Kermany DS, Goldbaum M, Cai W, Valentim CCS, Liang H, Baxter SL, et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018;172:1122-1131.e9. [Crossref] [PubMed]
- Li M, Wan C. The use of deep learning technology for the detection of optic neuropathy. Quant Imaging Med Surg 2022;12:2129-43. [Crossref] [PubMed]
- Ng WY, Zhang S, Wang Z, Ong CJT, Gunasekeran DV, Lim GYS, Zheng F, Tan SCY, Tan GSW, Rim TH, Schmetterer L, Ting DSW. Updates in deep learning research in ophthalmology. Clin Sci (Lond) 2021;135:2357-76. [Crossref] [PubMed]
- Chan HP, Samala RK, Hadjiiski LM, Zhou C. Deep Learning in Medical Image Analysis. Adv Exp Med Biol 2020;1213:3-21. [Crossref] [PubMed]
- Wang J, Hormel TT, Gao L, Zang P, Guo Y, Wang X, Bailey ST, Jia Y. Automated diagnosis and segmentation of choroidal neovascularization in OCT angiography using deep learning. Biomed Opt Express 2020;11:927-44. [Crossref] [PubMed]
- Lee CS, Baughman DM, Lee AY. Deep learning is effective for the classification of OCT images of normal versus Age-related Macular Degeneration. Ophthalmol Retina 2017;1:322-7. [Crossref] [PubMed]
- Tsiknakis N, Theodoropoulos D, Manikis G, Ktistakis E, Boutsora O, Berto A, Scarpa F, Scarpa A, Fotiadis DI, Marias K. Deep learning for diabetic retinopathy detection and classification based on fundus images: A review. Comput Biol Med 2021;135:104599. [Crossref] [PubMed]
- Karri SP, Chakraborty D, Chatterjee J. Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration. Biomed Opt Express 2017;8:579-92. [Crossref] [PubMed]
- Gende M, de Moura J, Fernández-Vigo JI, Martínez-de-la-Casa JM, García-Feijóo J, Novo J, Ortega M. Robust multi-view approaches for retinal layer segmentation in glaucoma patients via transfer learning. Quant Imaging Med Surg 2023;13:2846-59. [Crossref] [PubMed]
- Campbell JP, Ataer-Cansizoglu E, Bolon-Canedo V, Bozkurt A, Erdogmus D, Kalpathy-Cramer J, Patel SN, Reynolds JD, Horowitz J, Hutcheson K, Shapiro M, Repka MX, Ferrone P, Drenser K, Martinez-Castellanos MA, Ostmo S, Jonas K, Chan RV, Chiang MF. Imaging and Informatics in ROP (i-ROP) Research Consortium. Expert Diagnosis of Plus Disease in Retinopathy of Prematurity From Computer-Based Image Analysis. JAMA Ophthalmol 2016;134:651-7. [Crossref] [PubMed]
- Yang D, Li M, Li W, Wang Y, Niu L, Shen Y, Zhang X, Fu B, Zhou X. Prediction of Refractive Error Based on Ultrawide Field Images With Deep Learning Models in Myopia Patients. Front Med (Lausanne) 2022;9:834281. [Crossref] [PubMed]
- Pan X, Jin K, Cao J, Liu Z, Wu J, You K, Lu Y, Xu Y, Su Z, Jiang J, Yao K, Ye J. Multi-label classification of retinal lesions in diabetic retinopathy for automatic analysis of fundus fluorescein angiography based on deep learning. Graefes Arch Clin Exp Ophthalmol 2020;258:779-85. [Crossref] [PubMed]
- Jin K, Pan X, You K, Wu J, Liu Z, Cao J, Lou L, Xu Y, Su Z, Yao K, Ye J. Automatic detection of non-perfusion areas in diabetic macular edema from fundus fluorescein angiography for decision making using deep learning. Sci Rep 2020;10:15138. [Crossref] [PubMed]
- Kamiya K, Ayatsuka Y, Kato Y, Shoji N, Mori Y, Miyata K. Diagnosability of Keratoconus Using Deep Learning With Placido Disk-Based Corneal Topography. Front Med (Lausanne) 2021;8:724902. [Crossref] [PubMed]
- Xu S, Yang X, Zhang S, Zheng X, Zheng F, Liu Y, Zhang H, Li L, Ye Q. Evaluation of the corneal topography based on deep learning. Front Med (Lausanne) 2023;10:1264659. [Crossref] [PubMed]
- Hussain S, Chua J, Wong D, Lo J, Kadziauskiene A, Asoklis R, Barbastathis G, Schmetterer L, Yong L. Predicting glaucoma progression using deep learning framework guided by generative algorithm. Sci Rep 2023;13:19960. [Crossref] [PubMed]
- Warpeha KM, Chakravarthy U. Molecular genetics of microvascular disease in diabetic retinopathy. Eye (Lond) 2003;17:305-11. [Crossref] [PubMed]
- Dai L, Wu L, Li H, Cai C, Wu Q, Kong H, Liu R, Wang X, Hou X, Liu Y, Long X, Wen Y, Lu L, Shen Y, Chen Y, Shen D, Yang X, Zou H, Sheng B, Jia W. A deep learning system for detecting diabetic retinopathy across the disease spectrum. Nat Commun 2021;12:3242. [Crossref] [PubMed]
- Jani PD, Forbes L, Choudhury A, Preisser JS, Viera AJ, Garg S. Evaluation of Diabetic Retinal Screening and Factors for Ophthalmology Referral in a Telemedicine Network. JAMA Ophthalmol 2017;135:706-14. [Crossref] [PubMed]
- Cheung CY, Tang F, Ting DSW, Tan GSW, Wong TY. Artificial Intelligence in Diabetic Eye Disease Screening. Asia Pac J Ophthalmol (Phila) 2019;8:158-64. [Crossref] [PubMed]
- Taylor CR, Merin LM, Salunga AM, Hepworth JT, Crutcher TD, O'Day DM, Pilon BA. Improving diabetic retinopathy screening ratios using telemedicine-based digital retinal imaging technology: the Vine Hill study. Diabetes Care 2007;30:574-8. [Crossref] [PubMed]
- Vujosevic S, Aldington SJ, Silva P, Hernández C, Scanlon P, Peto T, Simó R. Screening for diabetic retinopathy: new perspectives and challenges. Lancet Diabetes Endocrinol 2020;8:337-47. [Crossref] [PubMed]
- Daskivich LP, Vasquez C, Martinez C Jr, Tseng CH, Mangione CM. Implementation and Evaluation of a Large-Scale Teleretinal Diabetic Retinopathy Screening Program in the Los Angeles County Department of Health Services. JAMA Intern Med 2017;177:642-9. [Crossref] [PubMed]
- Li R, Yang Z, Zhang Y, Bai W, Du Y, Sun R, Tang J, Wang N, Liu H. Cost-effectiveness and cost-utility of traditional and telemedicine combined population-based age-related macular degeneration and diabetic retinopathy screening in rural and urban China. Lancet Reg Health West Pac 2022;23:100435. [Crossref] [PubMed]
- Girard MJA, Schmetterer L. Artificial intelligence and deep learning in glaucoma: Current state and future prospects. Prog Brain Res 2020;257:37-64. [Crossref] [PubMed]
- Wen JC, Lee CS, Keane PA, Xiao S, Rokem AS, Chen PP, Wu Y, Lee AY. Forecasting future Humphrey Visual Fields using deep learning. PLoS One 2019;14:e0214875. [Crossref] [PubMed]
- Yang Y, Li R, Lin D, Zhang X, Li W, Wang J, Guo C, Li J, Chen C, Zhu Y, Zhao L, Lin H. Automatic identification of myopia based on ocular appearance images using deep learning. Ann Transl Med 2020;8:705. [Crossref] [PubMed]
- Adekunle AN, Adkins A, Wang W, Kaplan HJ, de Castro JF, Lee SJ, Huie P, Palanker D, McCall M, Pardue MT. Integration of Perforated Subretinal Prostheses With Retinal Tissue. Transl Vis Sci Technol 2015;4:5. [Crossref] [PubMed]
- Ataer-Cansizoglu E, Kalpathy-Cramer J, You S, Keck K, Erdogmus D, Chiang MF. Analysis of underlying causes of inter-expert disagreement in retinopathy of prematurity diagnosis. Application of machine learning principles. Methods Inf Med 2015;54:93-102. [Crossref] [PubMed]
- Bolón-Canedo V, Ataer-Cansizoglu E, Erdogmus D, Kalpathy-Cramer J, Fontenla-Romero O, Alonso-Betanzos A, Chiang MF. Dealing with inter-expert variability in retinopathy of prematurity: A machine learning approach. Comput Methods Programs Biomed 2015;122:1-15. [Crossref] [PubMed]