
Searching for genetic determinants for left ventricular non-compaction
Introduction
Left ventricular non-compaction (LVNC) is a pathology classified by the American Heart Association (AHA) as a genetically determined primary cardiomyopathy (1). The European Society of Cardiology (ESC) classified it in 2008 as unclassified cardiomyopathy (2), but since 2023 have classified LVNC as the other traits and syndromes associated with cardiomyopathy phenotypes (3). There are several criteria for the diagnosis of cardiomyopathy based primarily on imaging tests—transthoracic echocardiogram (TTE) and cardiac magnetic resonance (CMR). However, none of them are precise enough and often lead to the over-recognition of this disease. The criteria most commonly used in TTE are Jenni et al. (4), Chin et al. (5), Stöllberger et al. (6); in CMR: Petersen et al. (7) and Jacquier et al. (8). Unfortunately, they are not correlated with genetic studies. In some cases, they have been established on small groups of patients and their effectiveness is unsatisfactory. Unfortunately, they are not correlated with genetic studies. In some cases, they have been established on small groups of patients and their effectiveness is unsatisfactory. Recent studies suggest that many LVNC criteria lack significant clinical correlation, which affects their utility in clinical practice (9,10). The long-term survival is overall lower in patients with LVNC compared with the general population. However, it depends, among other things, on systolic dysfunction and the location of non-compaction (11). The risk of death increases with the degree of cardiovascular failure. It has been shown that patients with this diagnosis are at a high risk of serious complications. They often require heart transplantation and are at risk of sudden cardiac death (SCD), and therefore require close clinical monitoring (12). Recently, guidelines from the European Society of Cardiology relating to the prevention of SCD have been published. It has been stated that the implantation of a cardioverter-defibrillator in patients with LVNC in primary prevention of NCC has a high-class IIa recommendations (13).
The dominant hypothesis is that LVNC is a distinct and genetically induced cardiomyopathy (14). The most frequently mentioned genes potentially related to this cardiomyopathy are the sarcomere-encoding genes β-myosin heavy chain (MYH7), α-cardiac actin (ACTC1), cardiac troponin T (TNNT2), myosin binding protein-C (MYBPC3), LIM—domain binding protein 3 (LBD3) and the X-linked gene taffazin (TAZ) (15).
Genetic testing in LVNC is strongly guided by a comprehensive clinical evaluation of the patient and their family. Isolated LVNC, with no left ventricle (LV) dysfunction and detected incidentally on CMR will have a very low genetic testing yield compared to LVNC associated with other cardiomyopathies and LV dysfunction, syndromic features, and/or a strong family history where the genetic testing yield will be significantly higher. On the other hand, the absence of clinical symptoms at the time of detection of hypertrabeculation meeting LVNC criteria in imaging studies does not exclude that in the future such symptoms may appear in some people.
Therefore, it is worth finding a good method to distinguish patients who will meet LVNC criteria who are potentially at risk for symptoms such as arrhythmia, systolic dysfunction or SCD from those who do not. It can also not be excluded that hypertrabeculation meeting the LVNC criteria will only predispose people to clinical symptoms when the body will be burdened with other additional factors such as severe infections, extensive operations, immunotherapy, etc. Genetic testing could then be used to search for such potentially at-risk people, provided that there is a strong correlation between genetic changes and the degree of LV trabeculation.
The lack of a gold standard and good method for differentiating patients with LVNC from patients without LVNC also implies potential discrepancies and ambiguous results for both its definitive diagnosis and prognostic stratification. The aim of this study is to present the results of genetic testing of selected regions in genomic DNA isolated from the peripheral blood of participants distinguished based on the Petersen’s criteria. We would like to check if there are any correlations between the most often described genes and their regions and Petersen’s phenotypes. The results obtained could contribute to the possible exclusion or confirmation of the usefulness of analysing specific DNA segments in the context of LVNC.
Methods
Study groups selection
After obtaining the Bioethics Committee’s (Resolution No 18/2017 from Świętokrzyska Medical Chamber) approval, we retrospectively analyzed the result of CMR studies in Świętokrzyskie Cancer Center Hospital performed between January 2013 and September 2017. A number of 23 patients who met Petersen’s criteria agreed to participate in the research and take blood samples for genetic testing. All of these 23 patients were referred for CMR to look for causes of arrhythmia or left ventricular systolic failure or due to positive family history for LVNC, or suspected LVNC in the TTE study. None of them had any co-existing heart diseases, such as ischemic heart disease, myocarditis, hypertension, heart tumors, other congenital heart defects, etc. None of the patients meeting LVNC criteria were also pregnant or athletes.
Next, in the period between October 2017 and September 2019, in the study, we prospectively included 24 volunteers who did not meet Petersen’s criteria. Petersen’s criteria were complied with ratio of non-compacted to compacted myocardium (NC/C) ≥2.3. The cohort characteristic is presented in Table 1.
Table 1
| Participants (n=47) | Age (years) | Sex | ||||
|---|---|---|---|---|---|---|
| Mean (SD) | Median (Q1, Q3) | Range | Male, n (%) | Female, n (%) | ||
| NC/C <2.3 (n=23) | 41.57 (11.30) | 44.00 (34.00–50.00) | 44.00 | 11 (47.82) | 12 (52.17) | |
| NC/C ≥2.3 (n=24) | 42.33 (16.22) | 44.00 (32.25–57.50) | 55.00 | 14 (58.33) | 10 (41.66) | |
NC/C, non-compacted to compacted myocardium; SD, standard deviation; NC/C <2.3, participants not meeting the Petersen’s criteria; NC/C ≥2.3, participants meeting the Petersen’s criteria.
There were no family connections between the subjects. A total of 47 blood samples were collected from participants (22 females and 25 males, 11–66 years old). The distribution of sex and age had no influence on the results of statistical analyses. Patients with post-infarction lesions, heart tumors, congenital heart defects, pregnant women and athletes were not included in the research.
All participants, and in some cases their legal guardians, obtained information about the project, gave their informed consent to participate in the project and could withdraw from participation at any time. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Magnetic resonance imaging
All CMR exams were performed at the 1.5-T system (Gyroscan NT, Philips Healthcare, Best, The Netherlands) at Świętokrzyskie Cancer Center in Kielce from 2013 to 2019. In all subjects contiguous stack of short-axis (SA) two-dimensional steady-state free precession (SSFP) cine images, encompassing the LV from apex to base, was obtained. Next, three long-axis views in SSFP cine images were acquired: two-chamber (2CH), three-chamber (3CH), and four-chamber (4CH) ones. Imaging parameters included: repetition time (TR) 2.7 or 3.4 ms, time to echo (TE) 1.4 or 1.7 ms, and flip angle of 60°. Slice thickness was 8.0 mm (no interslice gap), and field-of-view (FOV) 300 or 320 mm.
Additionally, T2-weighted images in short tau inversion recovery (STIR) sequence, T1-weighted images in turbo spin echo (TSE) sequence before and after gadolinium-based contrast agents administration, and late gadolinium enhancement (LGE) images were obtained only in patients who met Petersen’s criteria. These images were not analyzed in this study.
Images were transferred to a dedicated workstation (Extended MR WorkSpace 2.6.3.5, Philips Healthcare). Measurements of the compacted and non-compacted layers of the myocardium were manually performed according to Petersen (7). The NC/C in diastole was calculated for each of the three long-axis views, and only the maximal ratio was then used for analysis. Petersen’s criteria were complied with NC/C ≥2.3. The apex (segment 17) was excluded from the measurements. All CMR examinations were assessed by the single reviewer.
Polymerase chain reaction (PCR) design
The automatically isolated DNA by MagCore® Genomic DNA Whole Blood Kit (RBC Bioscience Corp.) were used for the amplification of 5 different regions of selected genes. All primers (synthesized by the oligo.pl company) were designed in this study using BLAST software (The National Center for Biotechnology Information—NCBI) and verified by OligoAnalyzer™ Tool (Integrated DNA Technologies, Inc., Coralville, IA, USA). The sequences of the primers, the size of PCR products, and the gene regions with accession numbers are presented in Table 2. The reaction mixture was prepared with cDNA (approx. 20 ng) in a 25 µL reaction mixture containing a 12.5 µL DreamTaq™ Green DNA Polymerase Master Mix (2x) (Thermo Fisher Scientific™, MA, USA) and 10 pmol of each primer and refilled with MiliQ water. DNA amplification was carried out in MasterCycler X50 (Eppendorf, SE, Hamburg, Germany). Cycling conditions were as follows: denaturation at 95 ℃ for 3 minutes followed by amplification for 30 cycles at 95 ℃ for 1 minute, 60 ℃ for 1 minute and 72 ℃ for 1 minute, followed by a final extension at 72 ℃ for 8 minutes. After electrophoresis on 2% agarose gel with the addition of the Syngen GreenDNA Gel Stain (Syngen Biotech Sp. z o.o., Wrocław) the PCR products were visualized under UV and subsequently sequenced by outsourcing (Macrogen Europe BV, Amsterdam, The Netherlands). The sequences of the PCR products were compared to the reference sequences available from GeneBank using the BLAST platform (Table 2). The significance of detected SNVs and molecular consequence was determined based on data deposited in NCBI’s ClinVar. The Population frequencies (ƒ) was provided in reference to gnomAD Exomes resource available in VarSome (https://varsome.com/) and Sorting Intolerant From Tolerant (SIFT) (https://sift.bii.a-star.edu.sg) database. The Population frequencies (ƒ) represent the frequency of the alternate allele in different sub-populations (in total, in Europe, in our study), ƒ is the number of occurrences of the alternative allele (including homozygotes and heterozygotes, available online: https://cdn.amegroups.cn/static/public/qims-24-470-1.xlsx) divided by the allele number in the sub-population. The results were analyzed in relation to the latest reference Homo sapiens genome GRCh38.p14. In order to verify and analyze the molecular consequences of identified SNVs and their functions, we performed SIFT and Combined Annotation-Dependent Depletion (CADD) scores analysis. We display scores above 30 as ‘likely deleterious’ and scores below as ‘likely benign’. Variants with scores over 30 are predicted to be the 0.1% most deleterious possible substitutions in the human genome (16).
Table 2
| Name | Primer’s sequence (5’-3’) | PCR product [bp] | Gene (assembly) | Location | Amplification region | |
|---|---|---|---|---|---|---|
| From | To | |||||
| ACTC-F | AGCAAAGATGAGAAGACCTCAGAA | 801 | 3’UTR ACTC1 (NG_007553.1) | NC_000015.10 | 34,788,268 | 34,789,047 |
| ACTC-R | TGGAATGTGCTAGACAGGAACT | |||||
| TNNT2-F | TTAGAGCTTGGCAGACACCG | 779 | TNNT2 (NG_007556.1) | NC_000001.11 | 201,358,962 | 201,359,680 |
| TNNT2-R | CTCCAGCGTTGCTCTTTGTC | |||||
| LDB3-F | GGGGAACAGACACAGAACGG | 724 | LDB3 (NG_008876.1) | NC_000010.11 | 86,681,206 | 86,681,811 |
| LDB3-R | CCACCTGTGGAGAGCTGTATG | |||||
| MYH7-F | TTAGAGCTGGAAGAATCCTTGA | 718 | MYH7 (NG_007884) | NC_000014.9 | 23,413,604 | 23,414,200 |
| MYH7-R | GTTCTTTGGACTCTTTCCTGG | |||||
| TAZ-F | CTTTTCCTTGCAGGAGATGGC | 810 | TAZ (NG_009634.2) | NC_000023.11 | 154,419,551 | 154,420,210 |
| TAZ-R | TCCATAGTGCTGAGTGGGAGA | |||||
PCR, polymerase chain reaction; F, forward; R, reverse; bp, base pairs.
Data analysis
The age distribution was normal (P=0.17, Shapiro-Wilk test), the correlation between the study groups was analyzed using a two-tailed, paired t-test, where P<0.05 meant statistically significant. who met Petersen’s criteria (17,18).
GraphPad Prism, version 9 (San Diego, CA, USA) was used for the analyses and derivation of figures.
Results
General analysis
The total of 47 DNA samples were analyzed based on the selected regions of five genes: 3’UTR of ACTC1, between 16th and 17th exons of TNNT2, surrounding exon 5th of LDB3, between 6th and 8th exons of TAZ, between 38th and 39th exons of MYH7. Sequence failed for 5 samples (lack of sequencing quality despite 3 repetitions): 1 for ACTC1, 2 for TNNT2, and 2 for LDB3. In total, 248 substitutions in exons and introns were identified for all analyzed samples: ACTC1: 77/46 samples, TNNT2: 4/45 samples, LDB3: 56/44 samples, TAZ: 0/47 samples, MYH7: 111/47 samples. The values of NC/C: ≥2.3 (n=24) and <2.3 (n=23) were used to differentiate participants into two groups. The median number of substitutions was 5 in both groups of participants with the slight tendency of higher number of substitutions in the NC/C ≥2.3 group (Figure 1).
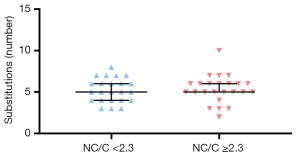
No statistically significant difference (unpaired t-test two-tailed, P>0.05) was detected between the mentioned groups. There were also no statistically significant differences [two-way analysis of variance (ANOVA) test] in terms of gender and the Petersen’s criteria (data not shown). Furthermore, no significant difference in either downward or upward trends in the number of substitutions in relation to the increasing NC/C value was observed (Figure 2), even in the case of individual SNV analysis.
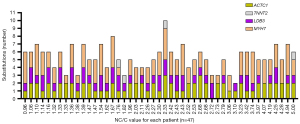
We identified insertions or deletions not deposited in any database so far (data not shown). They were in intron sites and did not have statistical significance, so we omitted them from the analyses, because would require more detailed verification by another method, e.g., next-generation sequencing (NGS).
Specific analysis
As many as 14 different single nucleotide variants have been identified (Table 3, Figure 3).
Table 3
| Gene | SNV | Clinical significance | Molecular consequence | ƒ in total (in Europe) |
ƒ in our study | N | Presence | Absence | P value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NC/C≥2.3 | NC/C<2.3 | NC/C≥2.3 | NC/C<2.3 | |||||||||
| ACTC1 (3’) | rs8037241 | Benign | Non coding | 0.21 (0.199) | 0.086* | 46 | 6 | 2 | 17 | 21 | 0.24 | |
| ACTC1 (3’) | rs533021 | Benign | Non coding | Not found (0,67) | 0.66 | 46 | 23 | 21 | 0 | 2 | 0.48 | |
| ACTC1 (3’) | rs604689 | Benign | Non coding | Not found (0,599) | 0.52 | 46 | 12 | 13 | 11 | 10 | >0.99 | |
| TNNT2 (int) | rs45576635 | Not reported | Non coding | 0.028 (0.0314)* | 0.033* | 45 | 1 | 1 | 22 | 21 | 0.51 | |
| TNNT2 (e17) | rs3729998 | Benign/Likely benign | Missense | 0.0249 (0.0289)* | 0.011* | 45 | 0 | 1 | 23 | 21 | 0.74 | |
| TNNT2 (e17) | rs3730244 | Benign | Missense | 0.023 (0.027)* | 0.011* | 45 | 1 | 0 | 22 | 22 | 0.41 | |
| LDB3 (int) | rs2248643 | Benign | Non coding | 0.744 (0.704) | 0.68 | 44 | 19 | 20 | 3 | 2 | 0.56 | |
| LDB3 (int) | rs2675686 | Benign | Non coding | 0.229 (0.194) | 0.193 | 44 | 7 | 9 | 15 | 13 | 0.24 | |
| LDB3 (e5) | rs35507268 | Benign | Missense | 0.006 (0.007)* | 0.011* | 45 | 0 | 1 | 23 | 21 | 0.48 | |
| MYH7 (int) | rs3729833 | Benign | Non coding | 0.186 (0.162) | 0.148 | 47 | 8 | 5 | 16 | 18 | >0.99 | |
| MYH7 (int) | rs770065133 | Not reported | Non coding | Not found | 0.38 | 47 | 19 | 17 | 5 | 6 | 0.51 | |
| MYH7 (e39) | rs727503240 | Uncertain significance | Missense | 0.000036 (0.000042)* | 0.010* | 47 | 1 | 0 | 23 | 23 | 0.74 | |
| MYH7 (e39) | rs397516254 | Likely pathogenic | Missense | Not found | 0.42 | 47 | 19 | 21 | 5 | 2 | 0.41 | |
| MYH7 (int) | rs2331979 | Benign | Non coding | 0.422 (0.369) | 0.23 | 47 | 12 | 9 | 12 | 14 | 0.56 | |
P value was calculated by Fisher exact test, two-sided, with statistical significance P<0.05. *, rare variants according to gnomAD allele frequency <0.1%; SNV, single nucleotide variant deposited in NCBI database; clinical significance and molecular consequence, in reference to data deposited in ClinVar of NCBI platform; ƒ, Population frequencies in reference to gnomAD Exomes resource available in VarSome and SIFT database, ƒ calculated in our study (including homozygotes and heterozygotes); 3’, 3’UTR end; int, intron, e, exon; n, total number of participants with positive result of sequencing; presence, number of participants with identified SNV; absence, number of participants without SNV; NC/C, ratio of non-compacted to compacted myocardium; NC/C <2.3, participants not meeting the Petersen’s criteria; NC/C ≥2.3, participants meeting the Petersen’s criteria. NCBI, National Center for Biotechnology Information; SIFT, Scale-Invariant Feature Transform.
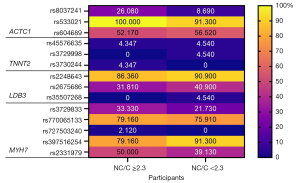
They were previously published in Gene Bank (NCBI) and other sources (VarSome, gnomAD). The regions of ACTC1 and MYH7 were the most numerous in mutations. TNNT2 had unique, rarely occurring mutations. The most common genetic variants were rs533021 (3’UTR of ACTC1), rs2248643 (LDB3) and rs397516254 (MYH7). They were present in an average of 76% of all participants. The percentage of the participants with the particular SNVs distinguished into two group based on the Petersen criteria (NC/C ≥2.3/<2.3) was analyzed (Figure 3). The rs8037241 (3’UTR of ACTC1) occurred on average 3 times more often in the group of participants meeting Petersen’s criteria. The remaining SNVs showed on average a few to 10% difference in occurrence between groups. An inverse association between the number of SNVs and the meeting the Petersen’s criteria was demonstrated for studied LDB3 region and rs397516254 in exon 39 of the MYH7 gene. This indicates the tendency to reduce the risk of cardiomyopathy. The occurrence of all identified SNVs were analyzed statistically. SNVs were four times more common in non-coding regions than in coding regions (exons) [Chi-square test, P<0.0001; odds ratio (OR) =4.124; relative ratio (RR) =2.577]. However, no significant differences were found between the study groups with respect to individual SNVs (Table 3). Using external databases (gnomAD, VarSome, ClinVar, SIFT), we analyzed the frequency of individual SNVs in our population in relation to the frequency of occurrence worldwide and in Europe (Table 3). The rare variants were estimated <0.1% according to gnomAD allele frequency. In most cases, the frequency of the tested variants in our study group was similar compared to the European and/or global population. However, in two cases of MYH7 gene the frequency was not determined yet, what in case of rs397516254 is particularly important due to its likely pathogenic for gene expression and high frequency in our study group. Most of the detected SNVs in exons cause a missense change but were nevertheless defined as benign except for rs397516254 as we mentioned above. The second one—rs727503240, present also in 39th exon of MYH7 also seems to be of interest due to its undetermined significance according to data from the ClinVar database, but it is very rare in the human population. Due to the lack of complete information or in order to verify the available data regarding the molecular sequences of the occurrence of the tested variants, we performed SIFT and CADD score analysis (Table 4). These above-mentioned variants in exon 39th of MYH7 gene have been termed likely deleterious, as well as rs35507268 in exon 5th of LDB3 gene. The status of the remaining variants was determined to be likely benign.
Table 4
| Gene | CADD | SIFT | |||
|---|---|---|---|---|---|
| Score | Classification | Score | Qualitative prediction | ||
| ACTC1 | |||||
| rs8037241 | 3.229 | Likely benign | NA | NA | |
| rs533021 | 2.748 | Likely benign | NA | NA | |
| rs604689 | 2.798 | Likely benign | NA | NA | |
| TNNT2 | |||||
| rs45576635 | 1.624 | Likely benign | NA | NA | |
| rs3729998 | 7.966 | Likely benign | Nd | NA | |
| rs3730244 | 0.039 | Likely benign | Nd | NA | |
| LDB3 | |||||
| rs2248643 | 1.098 | Likely benign | NA | NA | |
| rs2675686 | 0.161 | Likely benign | NA | NA | |
| rs35507268 | 21 | Likely benign | 0.051–0.25 | Deleterious* | |
| MYH7 | |||||
| rs3729833 | 7.916 | Likely benign | NA | NA | |
| rs770065133 | 21.0 | Likely benign | NA | NA | |
| rs727503240 | 35 | Likely deleterious | Nd | NA | |
| rs397516254 | 46 | Likely deleterious | Nd | NA | |
| rs2331979 | 0.101 | Likely benign | NA | NA | |
*, low confidence to tolerated. CADD, Combined Annotation-Dependent Depletion; SIFT, Sorting Intolerant From Tolerant; SNV, single nucleotide variant; ND, no data; NA, not applicable.
The rare mutations were also analyzed for co-occurrence: rs8037241 (3’UTR of ACTC1), rs3729998 (TNNT2e.12), and rs727503240 (MYH7e.39). They did not reveal the correlation between the participants distinguished based on the Petersen’s criteria (Table 5).
Table 5
| SNV | Positive | Negative | P value | |||
|---|---|---|---|---|---|---|
| NC/C ≥2.3 | NC/C <2.3 | NC/C ≥2.3 | NC/C <2.3 | |||
| rs8037241 (ACTC1) | 7 | 2 | 17 | 21 | 0.136 | |
| rs3730244 (TNNT2e.17) | ||||||
| rs727503240 (MYH7e.39) | ||||||
P value was calculated by Fisher exact test, two-sided, with statistical significance P<0.05. SNV, single nucleotide variant assigned as “rs” number published in NCBI database. Positive, participants with at least one SNV; negative, participants without any SNVs. NC/C, ratio of non-compacted to compacted myocardium; NCBI, National Center for Biotechnology Information.
However, the presence of at least one of these SNVs may increases the risk of cardiomyopathy 4.3 times. We want to emphasize that only 8.6% of participant which did not met Petersen’s criteria and 29% of participants meet the Petersen criteria (NC/C ≥2.3) had at least one of the listed SNVs.
Discussion
LVNC is a pathology that still evokes numerous controversies and discussions regarding the criteria for its diagnosis, presentation, prognosis, and even classification into the appropriate group of diseases. Originally in the Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of Cardiomyopathies published in 1996, LVNC was recognized as an unclassified cardiomyopathy (19). The European Society of Cardiology (ESC) classified it in 2008 as unclassified cardiomyopathy also (2), but since 2023 have classified LVNC as the other traits and syndromes associated with cardiomyopathy phenotypes (3). However, the American Heart Association has classified LVNC as primary genetic cardiomyopathy since 2006 (1).
Existing proposals of criteria for the diagnosis of this pathology in ultrasound (4-6,20,21), CMR (7,8,22-27), and CT (28,29), as well as discussions in this field for many years, show that there is still no perfect and unambiguous criterion, nor is there still a gold standard for confirming/excluding this pathology. In addition, they are not correlated with genetic studies. Recent studies suggest that many LVNC criteria lack significant clinical correlation, which affects their utility in clinical practice (9,10). Presumably, Petersen’s criteria are gaining popularity and are still widely recognized for their simplicity of application and no requirement for specialized software. In our study, we decided to use Petersen’s criteria to isolate a group of patients with LVNC. The vast majority of publications on patients with LVNC use the Petersen’s criteria as an inclusion criteria, both in diagnostic and clinical researches. Currently, there are no official guidelines on whose criteria to use, although some proposals seem to be more appropriate, they have not yet gained widespread clinical application or been widely used in research work. Because LVNC is a rare disease and it is difficult to collect large material in one center, using a commonly used diagnostic criterion, smaller studies can be included in meta-analyses of many studies.
In addition to classification and diagnostic criteria, there are also discrepancies concerning the naming itself. There are proponents of the name LVNC, but there are also many voices that the appropriate myocardial structure, which is the subject of this analysis, should be called “excessive trabeculation”, or “hypertrabeculation” (30,31). However, due to the widespread recognition of the acronym LVNC, the authors decided to use this name.
As the years pass and more reports appear, there are also increasing ambiguities about which genes and mutations determine the formation of LVNC. The 2011 consensus between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) on the genetic diagnosis of people with cardiomyopathy and channelopathies concluded that genetic changes in MYH7, ACTC1, TNNT2, MYBPC3, LBD3, and TAZ are important for the LVNC development (15). In the following years, there have been more reports of finding new mutations in patients with LVNC, which tend to obscure the genotype of patients meeting LVNC criteria in imaging studies. So far, about 190 genes in which mutations may be associated with LVNC have been described, and in each of them, several to several dozen different loci have been discovered (32). Thus, the number of potentially relevant mutations in LVNC may reach several hundred and close to 1,000. For example, 70 such loci are described in the MYH7 gene, in ACTC1—7, in TNNT2—8, in LDB3—2, and in TAZ—at least 23 (32-34). This is complicated by the fact that many mutations in LVNC patients are also present in other cardiomyopathies (14,32-35). On the other hand, it is not uncommon for two phenotypes of cardiomyopathy to coexist in one patient, e.g., DCM/LVNC or LVNC/HCM (36,37). Thus, in the context of the continuing lack of unambiguous diagnostic criteria for LVNC in imaging studies and the multitude of potential mutations, genetic tests do not currently have indications supported by strong observational evidence and the authors’ consensus in patients with this pathology but are only indicated in some cases as potentially useful. Genetic testing should not be performed in case of isolated LVNC with normal LV function, no associated syndromic features, and no family history. The role of genes in other cardiomyopathies—DCM, HCM, restrictive cardiomyopathy (RCM) and ARVC—is better understood, and genetic tests are more useful in diagnostic, therapeutic, and prediction processes (38,39).
We decided to analyze the frequency of single nucleotide variants in selected regions of the MYH7, ACTC1, TNNT2, LDB3, and TAZ genes and their correlation with NC/C morphology in order to contribute to the selection of the most important mutations for LVNC and the rejection of the less important ones. The regions for PCR amplification and sequencing were selected based on literature data, choosing those genes that appear to be the best candidates for differentiating LVNC cardiomyopathy from others, and/or from healthy subjects. Within each gene, those regions were selected, which are characterized by the presence of specific SNVs published by other authors as correlating with LVNC or other cardiomyopathies. Looking for these SNVs, we found another one. Our studies did not show a significant association between single nucleotide variants in selected regions of individual genes and the degree of trabeculation of the LV of the heart. However, some of our observations may be interesting and provide additional information to those published so far.
We confirmed the known association that mutations occur significantly more often in non-coding regions than in coding regions. However, some of them may indicate some correlation with the phenotype which meet Petersen’s criteria, probably due to their position in the regulatory regions. The most interesting seems to be MYH7. There is a very large number of reports of a potential pathological role of SNVs in MYH7 among LVNC patients (40,41). The MYH7 is the largest of the analyzed genes (it contains 40 exons) and is characterized by high genetic variability. In our study, the PCR amplicon of MYH7 flanked part of the intron before the 38th exon to the intron behind the 39th exon. That region indicates 28 variants reported in GeneBank (NCBI), mainly in the 39th exon. However, we did not detect any SNVs in the 38th exon but two in the 39th exon, of which one missense mutation (rs727503240 present in only one patient with NC/C ≥2.3) in ClinVar is defined as uncertain significance. However, according to CADD score analysis it was classified as likely deleterious, what can suggest its influence on the gene expression. It should be mentioned that its frequency is rare both in Europe and around the world, which may additionally suggest its important role for MYH7 translation. Walsh et al. (42) made similar observations. Further, we observed that second important missense variant in the 39th exon of MYH7 - rs397516254 was 12% more common in participant which not meeting the Petersen criteria, what could suggest its protective potential. However, both ClinVar database and the CADD score analysis indicates likely pathogenic status, and only two healthy participants more revealed this SNV. It should be emphasized that this genetic variant is quite common in our study group, but the frequency in the world is unknown. We want to emphasize that it is worth further looking at this SNV to assess its association with LVNC and perhaps clinical diagnosis in healthy patients requires more detailed analyses. This conclusion is in line with the others (43), which determined this variant to be likely pathogenic (ClinVar, NCBI) but for primary dilated cardiomyopathy development. This should be certainly explored further in future research.
The region behind ACTC1 (3’UTR of ACTC1) is also associated with cardiomyopathy (44-47). Based on literature data, we chose this region for analysis. Nevertheless the width of the amplified region, we identified SNVs which are classified as benign (ClinVar) and confirmed by CADD score analysis. They frequency is also relatively high, these variants are observed in people with different NC/C values, suggesting no association with LVNC. An interesting, benign rs533021 was detected in all individuals who complied with the Petersen’s criteria, but also in 91% of patients with NC/C <2.3. Li et al. [2013] also detected this variant but in patients with other cardiomyopathy (DCM) as well as in healthy subjects but at a lower frequency than in our study (48). Another rs604689 in 3’UTR of ACTC1 detected in each of the subjects’ groups of our research in about 50% of cases is also known as benign in DCM and HCM (49). Our studies did not show a significant relationship between this SNV and LVNC, either.
The next studied region of TNNT2 flanked the 16th and 17th exons. As with ACTC1, the GeneBank database (NCBI) contains many variants of unconfirmed clinical significance, while 17 SNVs of confirmed clinical significance (Cited Variations—dbSNP 2. 0 Build 156 v2 all data based on Homo sapiens Annotation Release 110), most described as benign or uncertain significance, and only some as pathogenic (rs111377893) or likely pathogenic (rs376923877, rs727504247, rs141121678, rs367785431, rs397516484, rs121964857) are mostly present on 17th exon. Of these, we detected only 3 SNVs individually in 4 patients from different cohorts: rs45576635, rs3729998, rs3730244. Even those present in exons, despite missense changes and their rare occurrence in the human population, do not indicate a pathogenic potential. Similarly, a rare occurrence of mutations in the TNNT2 gene was demonstrated by Kassem et al. [2013] in a study of the Egyptian population and Santos et al. [2012] in research on the Portuguese population with HCM (50,51). Rani et al. [2014] in a study on a large group of Indians population with DCM and a healthy control group found 16 SNVs, but there were only rs45576635, which was present in our study as well (52).
The amplified LDB3 region included the intron region of about 200 nucleotides ahead and 7 nucleotides behind the 5th exon. In this region, GeneBank (NCBI) has submitted 19 variants of verified clinical significance (Cited Variations—dbSNP 2. 0 Build 156 v2 all data based on Homo sapiens Annotation Release 110), mainly in the mentioned exon. Most of them are benign or likely benign, or pathogenic (53). We detected only one—rs35507268—in one symptomatic participant who did not meet the Petersen’s criteria. The ClinVar (NCBI) database describes it as benign or likely benign with 0.7% European population frequency (54). Against, SIFT score analysis indicates its pathogenic potential, CADD score analysis did not confirm this. It seems that it is worth further looking at this variant, because of the ambiguous picture. We also detected two other variants in the intron (rs2248643, rs2675686) without confirmed clinical significance in PubMed (NCBI). It should be noted that we did not detect two known SNVs (rs45487699, rs121908337), which were submitted as pathogenic for LVNCs (55).
The last amplified TAZ region encompassed almost the entire 6th exon, followed by the intron, the 7th exon, the intron, and the 8th exon with the subsequent intron. This area, as well as the whole gene, is characterized by relatively low genetic variability compared to the other analyzed gene. In our results, we did not find any variant in the studied region. Nevertheless, there are many reports and official positions of scientific societies confirming the importance of this gene in heart disease, including LVNC (15,35,38,56).
An important role in the search for patients potentially at risk of cardiomyopathy can be played by a specially prepared genetic test including an appropriate set of selected genetic loci. Although we did not find a significant correlation between the co-occurrence of individual mutations with LVNC, it is worth noting that the presence of one of the four mutations in the range rs8037241 (ACTC1 3’UTR), rs3729998 (TNNT2e. 12), and rs727503240 (MYH7e. 39) increases the risk of LVNC more than 4 times. To summarize further, three missense SNVs seem to be particularly interesting due to their occurrence in the MYH7 (rs727503240, rs397516254) and LDB3 (rs35507268) exons, their pathogenic potential, and especially rs397516254 – due to its high frequency in our study but unknown in the world.
The limitation of our studies is the relatively small size of the subjects studied. It is difficult to gather an impressively large group of patients because LVNC is a rare disease. However, our work has the potential to be used in collective work and meta-analyses as is the case for rare diseases. The small number of groups of patients with LVNC is evidenced in the systematic review and meta-analysis by van Waning et al. (35) for instance, who analyzed geno- and phenotypes of 561 LVNC patients distributed in 172 studies (on average just over 3 patients per publication).
The patients included in our studies were in a very wide age range and also had different clinical symptoms, but they all met the same Petersen criteria. Our goal was not to correlate genotype with clinical symptoms in individual age groups, but to try to find a correlation between genotype and the degree of left ventricular trabeculation. Whether increased left ventricular trabeculation has any specific relevance to clinical aspects in patients of different ages should be the focus of further work. In our opinion, first of all, it is necessary to systematically analyze and correlate the appropriate changes in the genome with the myocardial phenotype in imaging studies to find a uniform reference point for further work. In our work, we analyzed only certain sections of selected genes, but this may be the first step in finding significant changes in the genome in the case of selection of individuals with hypertrabeculation first, then determining the possible clinical significance of this phenotype and potentially final definition and diagnosis of LVNC.
Conclusions
There are many studies that analyze both true LVNC and patients with mild hypertrabeculation that is not clinically relevant. Firstly, we would like to eliminate as much noise that can be found in the literature on LVNC on genetics and clinical observations based on non-imperfect criteria and find a strong reference point for further proposals. We based our study on comparing a group of patients with completely healthy individuals without any clinical symptoms or increased trabeculation. Analyzing the same areas of the genome in healthy people and people suspected of having LVNC could shed new light on this issue.
To our knowledge, no studies have been published comparing the prevalence of selected SNVs in a group of healthy subjects and in a group meeting the Petersen criteria for LVNC.
Among both completely healthy individuals who did not meet the Petersen criteria for LVNC as well as those with symptoms who met these criteria, we found a similar incidence of SNVs in the ACTC1, TNNT2, LDB3 and MYH7 genes segments analyzed.
Further studies are also required to confirm or exclude “potentially protective” SNV in the 39th exon of MYH7 (rs397516254) and the role of co-occurrence of individual SNVs in rs8037241 (ACTC1 3’UTR), rs3729998 (TNNT2e. 12), and rs727503240 (MYH7e. 39) for the increase of the risk of LVNC.
Acknowledgments
Funding: This work was supported by the program of
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-470/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Approval was granted by the Bioethics Committee of Świętokrzyska Medical Chamber (Resolution No. 18/2017). All participants, and in some cases, their legal guardians, obtained information about the project, gave their informed consent to participate in the project and could withdraw from participation at any time.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, Moss AJ, Seidman CE, Young JBAmerican Heart Association. Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation 2006;113:1807-16. [Crossref] [PubMed]
- Elliott P, Andersson B, Arbustini E, Bilinska Z, Cecchi F, Charron P, Dubourg O, Kühl U, Maisch B, McKenna WJ, Monserrat L, Pankuweit S, Rapezzi C, Seferovic P, Tavazzi L, Keren A. Classification of the cardiomyopathies: a position statement from the European Society Of Cardiology Working Group on Myocardial and Pericardial Diseases. Eur Heart J 2008;29:270-6. [Crossref] [PubMed]
- Arbelo E, Protonotarios A, Gimeno JR, Arbustini E, Barriales-Villa R, Basso C, Bezzina CR, Biagini E, Blom NA, de Boer RA, De Winter T, Elliott PM, Flather M, Garcia-Pavia P, Haugaa KH, Ingles J, Jurcut RO, Klaassen S, Limongelli G, Loeys B, Mogensen J, Olivotto I, Pantazis A, Sharma S, Van Tintelen JP, Ware JS, Kaski JPESC Scientific Document Group. 2023 ESC Guidelines for the management of cardiomyopathies. Eur Heart J 2023;44:3503-626.
- Jenni R, Oechslin E, Schneider J, Attenhofer Jost C, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart 2001;86:666-71. [Crossref] [PubMed]
- Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium. A study of eight cases. Circulation 1990;82:507-13. [Crossref] [PubMed]
- Stöllberger C, Finsterer J, Blazek G. Left ventricular hypertrabeculation/noncompaction and association with additional cardiac abnormalities and neuromuscular disorders. Am J Cardiol 2002;90:899-902. [Crossref] [PubMed]
- Petersen SE, Selvanayagam JB, Wiesmann F, Robson MD, Francis JM, Anderson RH, Watkins H, Neubauer S. Left ventricular non-compaction: insights from cardiovascular magnetic resonance imaging. J Am Coll Cardiol 2005;46:101-5. [Crossref] [PubMed]
- Jacquier A, Thuny F, Jop B, Giorgi R, Cohen F, Gaubert JY, Vidal V, Bartoli JM, Habib G, Moulin G. Measurement of trabeculated left ventricular mass using cardiac magnetic resonance imaging in the diagnosis of left ventricular non-compaction. Eur Heart J 2010;31:1098-104. [Crossref] [PubMed]
- Ivanov A, Dabiesingh DS, Bhumireddy GP, Mohamed A, Asfour A, Briggs WM, Ho J, Khan SA, Grossman A, Klem I, Sacchi TJ, Heitner JF. Prevalence and Prognostic Significance of Left Ventricular Noncompaction in Patients Referred for Cardiac Magnetic Resonance Imaging. Circ Cardiovasc Imaging 2017;10:e006174. [Crossref] [PubMed]
- Zemrak F, Raisi-Estabragh Z, Khanji MY, Mohiddin SA, Bruder O, Wagner A, Lombardi M, Schwitter J, van Rossum AC, Pilz G, Nothnagel D, Steen H, Nagel E, Prasad SK, Deluigi CC, Dill T, Frank H, Schneider S, Mahrholdt H, Petersen SE. Left Ventricular Hypertrabeculation Is Not Associated With Cardiovascular Morbity or Mortality: Insights From the Eurocmr Registry. Front Cardiovasc Med 2020;7:158. [Crossref] [PubMed]
- Vaidya VR, Lyle M, Miranda WR, Farwati M, Isath A, Patlolla SH, Hodge DO, Asirvatham SJ, Kapa S, Deshmukh AJ, Foley TA, Michelena HI, Connolly HM, Melduni RM. Long-Term Survival of Patients With Left Ventricular Noncompaction. J Am Heart Assoc 2021;10:e015563. [Crossref] [PubMed]
- Habib G, Charron P, Eicher JC, Giorgi R, Donal E, Laperche T, Boulmier D, Pascal C, Logeart D, Jondeau G, Cohen-Solal A. Working Groups 'Heart Failure and Cardiomyopathies' and 'Echocardiography' of the French Society of Cardiology. Isolated left ventricular non-compaction in adults: clinical and echocardiographic features in 105 patients. Results from a French registry. Eur J Heart Fail 2011;13:177-85. [Crossref] [PubMed]
- Zeppenfeld K, Tfelt-Hansen J, de Riva M, Winkel BG, Behr ER, Blom NA, et al. 2022 ESC Guidelines for the management of patients with ventricular arrhythmias and the prevention of sudden cardiac death. Eur Heart J 2022;43:3997-4126. [Crossref] [PubMed]
- Jefferies JL. Left Ventricular Noncompaction Cardiomyopathy: New Clues in a Not So New Disease? J Am Heart Assoc 2021;10:e018815. [Crossref] [PubMed]
- Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, Camm AJ, Ellinor PT, Gollob M, Hamilton R, Hershberger RE, Judge DP, Le Marec H, McKenna WJ, Schulze-Bahr E, Semsarian C, Towbin JA, Watkins H, Wilde A, Wolpert C, Zipes DPHeart Rhythm Society (HRS). European Heart Rhythm Association (EHRA). HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Europace 2011;13:1077-109. Erratum in: Europace 2012;14:277.
- Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet 2014;46:310-5. [Crossref] [PubMed]
- Pagano M, Gauvreau K. Principles of Biostatistics. 2nd edition. Chapman and Hall/CRC; 2020.
- Deeks J, Higgins J. Statistical Algorithms in Review Manager 5. Statistical Algorithms in Review Manager 5. Published online January 1, 2010.
- Richardson P, McKenna W, Bristow M, Maisch B, Mautner B, O'Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I, Martin I, Nordet P. Report of the 1995 World Health Organization/International Society and Federation of Cardiology Task Force on the Definition and Classification of cardiomyopathies. Circulation 1996;93:841-2. [Crossref] [PubMed]
- Tarando F, Coisne D, Galli E, Rousseau C, Viera F, Bosseau C, Habib G, Lederlin M, Schnell F, Donal E. Left ventricular non-compaction and idiopathic dilated cardiomyopathy: the significant diagnostic value of longitudinal strain. Int J Cardiovasc Imaging 2017;33:83-95. [Crossref] [PubMed]
- Cortés M, Oliva MR, Orejas M, Navas MA, Rábago RM, Martínez ME, Taibo M, Palfy J, Rey M, Farré J. Usefulness of speckle myocardial imaging modalities for differential diagnosis of left ventricular non-compaction of the myocardium. Int J Cardiol 2016;223:813-8. [Crossref] [PubMed]
- Grothoff M, Pachowsky M, Hoffmann J, Posch M, Klaassen S, Lehmkuhl L, Gutberlet M. Value of cardiovascular MR in diagnosing left ventricular non-compaction cardiomyopathy and in discriminating between other cardiomyopathies. Eur Radiol 2012;22:2699-709. [Crossref] [PubMed]
- Stacey RB, Andersen MM, St Clair M, Hundley WG, Thohan V. Comparison of systolic and diastolic criteria for isolated LV noncompaction in CMR. JACC Cardiovasc Imaging 2013;6:931-40. [Crossref] [PubMed]
- Hautvast GL, Breeuwer M, Lobregt S, Gerritsen F. Automatic exclusion of papillary muscles and trabeculae from blood volume measurements in cine cardiac magnetic resonance images. Computers in Cardiology 2006;33:57-60.
- Kubik M, Dąbrowska-Kugacka A, Dorniak K, Kutniewska-Kubik M, Daniłowicz-Szymanowicz L, Lewicka E, Szurowska E, Raczak G. Influence of observer-dependency on left ventricular hypertrabeculation mass measurement and its relationship with left ventricular volume and ejection fraction - comparison between manual and semiautomatic CMR image analysis methods. PLoS One 2020;15:e0230134. [Crossref] [PubMed]
- Captur G, Muthurangu V, Cook C, Flett AS, Wilson R, Barison A, Sado DM, Anderson S, McKenna WJ, Mohun TJ, Elliott PM, Moon JC. Quantification of left ventricular trabeculae using fractal analysis. J Cardiovasc Magn Reson 2013;15:36. [Crossref] [PubMed]
- Choi Y, Kim SM, Lee SC, Chang SA, Jang SY, Choe YH. Quantification of left ventricular trabeculae using cardiovascular magnetic resonance for the diagnosis of left ventricular non-compaction: evaluation of trabecular volume and refined semi-quantitative criteria. J Cardiovasc Magn Reson 2016;18:24. [Crossref] [PubMed]
- Melendez-Ramirez G, Castillo-Castellon F, Espinola-Zavaleta N, Meave A, Kimura-Hayama ET. Left ventricular noncompaction: a proposal of new diagnostic criteria by multidetector computed tomography. J Cardiovasc Comput Tomogr 2012;6:346-54. [Crossref] [PubMed]
- Sidhu MS, Uthamalingam S, Ahmed W, Engel LC, Vorasettakarnkij Y, Lee AM, Hoffmann U, Brady T, Abbara S, Ghoshhajra BB. Defining left ventricular noncompaction using cardiac computed tomography. J Thorac Imaging 2014;29:60-6. [Crossref] [PubMed]
- Petersen SE, Jensen B, Aung N, Friedrich MG, McMahon CJ, Mohiddin SA, Pignatelli RH, Ricci F, Anderson RH, Bluemke DA. Excessive Trabeculation of the Left Ventricle: JACC: Cardiovascular Imaging Expert Panel Paper. JACC Cardiovasc Imaging 2023;16:408-25. [Crossref] [PubMed]
- Wang J, Han Y, Chen Y. Discourage LVNC or Revise the Criteria of LVNC? JACC Cardiovasc Imaging 2023;16:868. [Crossref] [PubMed]
- Rojanasopondist P, Nesheiwat L, Piombo S, Porter GA Jr, Ren M, Phoon CKL. Genetic Basis of Left Ventricular Noncompaction. Circ Genom Precis Med 2022;15:e003517. [Crossref] [PubMed]
- Arbustini E, Favalli V, Narula N, Serio A, Grasso M. Left Ventricular Noncompaction: A Distinct Genetic Cardiomyopathy?. J Am Coll Cardiol 2016;68:949-66. Correction appears in J Am Coll Cardiol 2016;68:1821.
- Miszalski-Jamka K, Jefferies JL, Mazur W, Głowacki J, Hu J, Lazar M, Gibbs RA, Liczko J, Kłyś J, Venner E, Muzny DM, Rycaj J, Białkowski J, Kluczewska E, Kalarus Z, Jhangiani S, Al-Khalidi H, Kukulski T, Lupski JR, Craigen WJ, Bainbridge MN. Novel Genetic Triggers and Genotype-Phenotype Correlations in Patients with Left Ventricular Non-Compaction. Circ Cardiovasc Genet 2017;10:e001763. [Crossref] [PubMed]
- van Waning JI, Moesker J, Heijsman D, Boersma E, Majoor-Krakauer D. Systematic Review of Genotype-Phenotype Correlations in Noncompaction Cardiomyopathy. J Am Heart Assoc 2019;8:e012993. [Crossref] [PubMed]
- Jaouadi H, El Louali F, Wanert C, Cano A, Ovaert C, Zaffran S. Dilated-Left Ventricular Non-Compaction Cardiomyopathy in a Pediatric Case with SPEG Compound Heterozygous Variants. Int J Mol Sci 2022;23:5205. [Crossref] [PubMed]
- Kochańska S, Spałek M, Wróbel G, Korzeluch W, Michałowska I, Kuder T, Wożakowska-Kapłon B. Coexistence of hypertrophic cardiomyopathy and left ventricular non-compaction cardiomyopathy-a description of two cases. Quant Imaging Med Surg 2022;12:4016-21. [Crossref] [PubMed]
- Wilde AAM, Semsarian C, Márquez MF, Shamloo AS, Ackerman MJ, Ashley EA, Sternick EB, Barajas-Martinez H, Behr ER, Bezzina CR, Breckpot J, Charron P, Chockalingam P, Crotti L, Gollob MH, Lubitz S, Makita N, Ohno S, Ortiz-Genga M, Sacilotto L, Schulze-Bahr E, Shimizu W, Sotoodehnia N, Tadros R, Ware JS, Winlaw DS, Kaufman ES, Aiba T, Bollmann A, Choi J-I, Dalal A, Darrieux F, Giudicessi J, Guerchicoff M, Hong K, Krahn AD, MacIntyre C, Mackall JA, Mont L, Napolitano C, Ochoa JP, Peichl P, Pereira AC, Schwartz PJ, Skinner J, Stellbrink C, Tfelt-Hansen J, Deneke T. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Europace 2022;24:1307-67. [Crossref] [PubMed]
- Vogiatzi G, Lazaros G, Oikonomou E, Lazarou E, Vavuranakis E, Tousoulis D. Role of genetic testing in cardiomyopathies: A primer for cardiologists. World J Cardiol 2022;14:29-39. [Crossref] [PubMed]
- Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, Cook S, Dillon MW, Garcia J, Haverfield E, Jongbloed JDH, Macaya D, Manrai A, Orland K, Richard G, Spoonamore K, Thomas M, Thomson K, Vincent LM, Walsh R, Watkins H, Whiffin N, Ingles J, van Tintelen JP, Semsarian C, Ware JS, Hershberger R, Funke B. Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genet Med 2018;20:351-9. [Crossref] [PubMed]
- Miura F, Shimada J, Kitagawa Y, Otani K, Sato T, Toki T, Takahashi T, Yonesaka S, Mizukami H, Ito E. MYH7 mutation identified by next-generation sequencing in three infant siblings with bi-ventricular noncompaction presenting with restrictive hemodynamics: A report of three siblings with a severe phenotype and poor prognosis. J Cardiol Cases 2019;19:140-3. [Crossref] [PubMed]
- Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, Minikel EVExome Aggregation Consortium. MacArthur DG, Farrall M, Cook SA, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med 2017;19:192-203. [Crossref] [PubMed]
- Lakdawala NK, Funke BH, Baxter S, Cirino AL, Roberts AE, Judge DP, Johnson N, Mendelsohn NJ, Morel C, Care M, Chung WK, Jones C, Psychogios A, Duffy E, Rehm HL, White E, Seidman JG, Seidman CE, Ho CY. Genetic testing for dilated cardiomyopathy in clinical practice. J Card Fail 2012;18:296-303. [Crossref] [PubMed]
- Toledano-Delgado FJ, Jimenez-Alcantara I, Cobo-Molinos J, Carrasco-Avalos F, Mazuelos F, Urbano-Moral JA. Description of a Novel Cardiac Phenotype Associated With a Missense Variant in the Cardiac α-Actin (ACTC1) Gene. Circ Genom Precis Med 2023;16:e003963. [Crossref] [PubMed]
- Teng GZ, Dawson JF. The Dark Side of Actin: Cardiac actin variants highlight the role of allostery in disease development. Arch Biochem Biophys 2020;695:108624. [Crossref] [PubMed]
- Frank D, Yusuf Rangrez A, Friedrich C, Dittmann S, Stallmeyer B, Yadav P, Bernt A, Schulze-Bahr E, Borlepawar A, Zimmermann WH, Peischard S, Seebohm G, Linke WA, Baba HA, Krüger M, Unger A, Usinger P, Frey N, Schulze-Bahr E. Cardiac α-Actin (ACTC1) Gene Mutation Causes Atrial-Septal Defects Associated With Late-Onset Dilated Cardiomyopathy. Circ Genom Precis Med 2019;12:e002491. [Crossref] [PubMed]
- Rangrez AY, Kilian L, Stiebeling K, Dittmann S, Yadav P, Schulze-Bahr E, Frey N, Frank D. Data on the role of cardiac α-actin (ACTC1) gene mutations on SRF-signaling. Data Brief 2020;28:105071. [Crossref] [PubMed]
- Li X, Luo R, Mo X, Jiang R, Kong H, Hua W, Wu X. Polymorphism of ZBTB17 gene is associated with idiopathic dilated cardiomyopathy: a case control study in a Han Chinese population. Eur J Med Res 2013;18:10. [Crossref] [PubMed]
- The National Center for Biotechnology Information, ClinVar, Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/315647/?oq=rs604689[External+allele+ID]&m=NM_005159.4(ACTC1):c.*2090A%3EG Accessed 30 May 2023.
- Kassem HSh, Azer RS, Saber-Ayad M, Moharem-Elgamal S, Magdy G, Elguindy A, Cecchi F, Olivotto I, Yacoub MH. Early results of sarcomeric gene screening from the Egyptian National BA-HCM Program. J Cardiovasc Transl Res 2013;6:65-80. Erratum in: J Cardiovasc Transl Res 2013;6:663. [Crossref] [PubMed]
- Santos S, Marques V, Pires M, Silveira L, Oliveira H, Lança V, Brito D, Madeira H, Esteves JF, Freitas A, Carreira IM, Gaspar IM, Monteiro C, Fernandes AR. High resolution melting: improvements in the genetic diagnosis of hypertrophic cardiomyopathy in a Portuguese cohort. BMC Med Genet 2012;13:17. [Crossref] [PubMed]
- Rani DS, Dhandapany PS, Nallari P, Narasimhan C, Thangaraj K. A novel arginine to tryptophan (R144W) mutation in troponin T (cTnT) gene in an indian multigenerational family with dilated cardiomyopathy (FDCM). PLoS One 2014;9:e101451. [Crossref] [PubMed]
- Norton N, Robertson PD, Rieder MJ, Züchner S, Rampersaud E, Martin E, Li D, Nickerson DA, Hershberger RE. National Heart, Lung and Blood Institute GO Exome Sequencing Project. Evaluating pathogenicity of rare variants from dilated cardiomyopathy in the exome era. Circ Cardiovasc Genet 2012;5:167-74. [Crossref] [PubMed]
- Delio M, Patel K, Maslov A, Marion RW, McDonald TV, Cadoff EM, Golden A, Greally JM, Vijg J, Morrow B, Montagna C. Development of a Targeted Multi-Disorder High-Throughput Sequencing Assay for the Effective Identification of Disease-Causing Variants. PLoS One 2015;10:e0133742. [Crossref] [PubMed]
- Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol 2003;42:2014-27. [Crossref] [PubMed]
- Hertz CL, Christiansen SL, Larsen MK, Dahl M, Ferrero-Miliani L, Weeke PE, Pedersen O, Hansen T, Grarup N, Ottesen GL, Frank-Hansen R, Banner J, Morling N. Genetic investigations of sudden unexpected deaths in infancy using next-generation sequencing of 100 genes associated with cardiac diseases. Eur J Hum Genet 2016;24:817-22. [Crossref] [PubMed]


