
A mutual reconstruction network model for few-shot classification of histological images: addressing interclass similarity and intraclass diversity
Introduction
Cancer has become a major cause of mortality worldwide, and the number of cancer-related deaths increases annually. Recent data show that over 19.3 million new cancer cases were diagnosed and reported in 2020 , leading to approximately 10 million deaths (1,2). Histologically, there are hundreds of histology image categories, adding complexity and challenge to classification (3). Whole slide image (WSI) scanners, which provide comprehensive views of histological slides, are crucial for the diagnosis and staging of cancer, thus informing treatment decisions. However, the primary diagnosis of cancer still predominantly relies on pathologists, a process that is time-consuming and depends heavily on experience. Additionally, the task of tissue classification varies across different cancer sites, and even within the same site, there may be different levels of category granularity. Hence, the automated classification of histological images is of immense value in cancer diagnosis. The concept of using computers for disease diagnosis was first introduced by Lusted in 1955 (4). Following this, Lodwick et al. pioneered the digitization of chest X-rays for developing a computer-aided diagnosis (CAD) system, which was later used to detect lung cancer (5).
The advent of deep learning methods has fundamentally changed this field. Researchers are now actively exploring different learning approaches, such as deep learning, for cancer diagnosis. For instance, Phankokkruad and colleagues used deep learning models including ResNet50V2, VGG16, and DenseNet201, along with transfer learning ensemble methods, to classify lung cancer (6). Mohalder et al. adopted deep learning-based approaches to predict colon cancer (7), while additionally Adu et al. proposed a new dual horizontal squash capsule network (DHS-CapsNet) for classifying lung and colon cancers (8). These studies demonstrate the potential and promising applications of deep learning in the field of cancer diagnosis.
Deep learning has advanced significantly in classification tasks, but its reliance on extensive, labeled data sets limits its generalizability when there is an insufficient amount of labeled data. Given the challenges associated with acquiring ample labeled data across a diversity of scenarios, this dependency restricts the advancement of deep learning technologies. Few-shot learning has emerged in recent years as a potent solution to this issue, yielding a plethora of new methods and research directions, primarily related to evaluating natural image-related datasets. Human beings possess the ability to comprehend new concepts with only a few samples. For instance, a child can recognize dogs after observing just a few images of them. Emulating this learning ability in machines to reduce the reliance on extensive annotated data has become a pivotal research direction. To address scenarios with limited samples, researchers have proposed few-shot learning, which involves using problem-solving approaches with only a small amount of annotated data. Few-shot learning initially emerged in the field of computer vision and subsequently gained traction in areas such as natural language processing. In medical image analysis tasks, both the acquisition and annotation of image data pose significant challenges. One of the reasons for this is that the collection of such images requires specialized equipment such as magnetic resonance imaging (MRI) and computed tomography (CT) scanners. Additionally, aside from the costly equipment, issues related to patient privacy also need to be considered. Another challenge arises due to the unique characteristics of certain diseases, which may impede the data collection process. For instance, some rare diseases have a limited number of patients, resulting in a scarcity of available images. In scenarios with extremely sparse data, conventional deep learning methods often struggle to perform effectively. Few-shot learning can be categorized into two main approaches: optimization-based methods (9,10) and metric-based methods (11-14). The concept of optimization-based methods was first introduced in model-agnostic meta-learning (MAML) (9), whose function is to train models on well-initialized parameters. Meta-learning long short-term memory (MetaLSTM) (15) represents an effective fine-tuning approach, while the meta-learning optimization network (MetaOptNet) (16) offers an end-to-end differentiated learning method. On the other hand, metric-based methods leverage predefined or online-trained metrics to learn deep representations, classifying new categories of images by measuring distances between support and query sets. Metric-based approaches have proven to be a superior choice for solving few-shot classification challenges. These can be further divided into three categories: global feature metric methods (11), local feature metric methods (13,14,17), and attention feature metric methods (12).
Due to the high cost and specialized expertise required for annotating histopathological images, there is a scarcity of labeled data in this domain, underscoring how critical few-shot learning is for the analysis of histopathological images. For the few-shot learning of histopathological images, there is a focus on classification tasks. In the domain of optimization-based methods, Chao and Belanger introduced an optimization-based learning approach, applying MAML to classify whole-genome doubling (WGD) in histopathological images (18). Another meta-learning-based method, MetaMed, was employed by Titoriya et al. for the classification of medical tumor images (19). Another team (20) applied the MAML method to classify histopathological images from different microscopes, as it should be noted that images scanned by different scanners may exhibit variable staining characteristics. Additionally, the MetaHistoSeg framework is worth mentioning; MAML was applied to a meta-dataset of histopathological image segmentation and was compared with a baseline of instance-based transfer learning (21). Moreover, in terms of metric-based methods, Shaikh et al. applied multiple-instance learning prototype networks to classify artifacts in histopathological images stained with hematoxylin and eosin (HE) or immunohistochemistry (22). Other authors (23) employed a deep Siamese network to address the knowledge transfer problem from specific to more general domains in histopathological image classification. The histology Siamese network model was first trained on a source domain dataset, Ds, which contains colon tissue, and was then fine-tuned on a target domain dataset, Dt, which contains colon, breast, and lung tissues. In other study, the performance of models trained with different loss functions in obtaining histopathological image embeddings was evaluated (24). The compared loss functions included triplet loss, multiclass-N-pair loss, and the proposed constellation loss. Prototype networks represent another example of metric learning methods. An intriguing extension of prototype networks using k-means was proposed that allows for the prediction of the category of unseen WSI based on a few examples (22). This method can also manage variations in image color and resolution caused by different microscopes and preprocessing steps in clinical settings.
Despite the significant clinical potential of the few-shot classification of histological images, current research methodologies do not fully meet the demands of clinical applications. For instance, the work by Chao et al. using MAML for few-shot classification of histological images achieved an area under the curve (AUC) of only 0.6944 (18). This limitation is partly due to the insufficient consideration of the fine-grained attributes of histological images leading to an ineffective capture of fine-grained information specific to certain tasks. In Figure 1, samples A–E belong to the tumor category of colorectal cancer yet exhibit significant differences in cell color and shape. However, sample F, representing the inflammatory category of colorectal cancer, shares similarities in color and cell morphology with sample E yet belongs to a different category. It is important to note that the preparation, fixation, and staining processes of histological images are subject to variations due to different operators, potentially leading to heterogeneity in image appearance. Consequently, the task of few-shot classification of histological images faces challenges of small interclass variance and large intraclass variance, presenting a complex scenario for accurate categorization. Several studies in traditional deep learning classification tasks for histological images have recognized this challenge and proposed models to address it. For instance, Jiang et al. introduced a fine-grained classification model named Breast TransFG Plus, based on the Transformer model, to classify breast HE-stained pathological images specifically for grading invasive ductal carcinoma of the breast (26). Liu et al. proposed a deep learning approach based on bilinear convolutional neural networks (BCNNs) for the fine-grained classification of breast cancer histopathological images (27). Li et al. sought to solve the challenging issue of small interclass variance and large intraclass variance in histological images by embedding prior knowledge into the feature extraction process (28). Several other studies have also focused on the fine-grained attributes of histological images (29-34).

In overcoming the challenges in the few-shot classification of histological images, it is crucial to discern and capture subtle and discriminative features. Current few-shot learning methods used for fine-grained natural images primarily focus on reconstructing query features from support features or employing attention mechanisms to manage interclass similarity. However, these solutions fail to effectively address the challenge of intraclass diversity (14,35-37). In response to this, we developed a mutual reconstruction network, designed to address small interclass variance and large intraclass variance simultaneously. This model initially uses a feature extractor to derive support and query features, which are then sequentially processed through a feature enhancement module (FEM) and a mutual reconstruction module for reconstruction. Finally, the task of few-shot classification of histological images is completed using a Euclidean metric module to calculate distances. This method was designed to overcome the issue of fine-grained features faced by the existing methods that manage the few-shot classification tasks of histological images, particularly for intraclass diversity and interclass similarity. By introducing the mutual reconstruction network, we sought to enhance the performance of few-shot classification tasks and provide new perspectives and methodologies for further research in the field of histological image processing.
The primary contributions of our research can be summarized as follows:
- We developed a mutual reconstruction network designed to address the challenges of small interclass variance and large intraclass variance in the few-shot classification tasks of histological images.
- We created a dedicated public dataset for the few-shot classification of histological images. For this dataset, our method demonstrated outstanding performance, achieving an accuracy of 91.12%. The mutual reconstruction network significantly outperformed other few-shot methods, and our findings offer new research directions and insights for few-shot classification tasks in histological imaging.
Methods
Problem statement
In the realm of few-shot classification, we aimed to train a model capable of leveraging knowledge acquired from a large, labeled example dataset, , often referred to as the source dataset. This model was trained to learn from a minimal number of examples in a new class set, , and successfully perform classification tasks. represents a dataset with a label space, , where the initial pretraining phase occurs. The subsequent phase, meta-testing, takes place on , which comprises a novel set of classes with a distinct label space, , characterized by a lack of overlap with . During the meta-testing phase, we employ episodic sampling, where a series of tasks are constructed from . Each task consists of two critical components: a support set and a query set. In these tasks, N categories are randomly selected, and then sample K examples from each chosen category are used to construct a support set of samples. The query set comprises M unlabeled query samples randomly drawn from these N categories within . Consequently, a series of N-way K-shot classification tasks is generated, where N is the number of selected categories, K is the number of examples chosen per category, and M is the number of samples in the query set. The objective of this approach is to test the model’s generalization capability through the examples in the support set and query set, enabling effective classification in the presence of new categories. The key to this strategy is the effective utilization of the knowledge from and its transfer to new categories in , which enhances classification performance. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Overview
The primary challenge in few-shot classification tasks of histological images lies in addressing intraclass diversity and interclass similarity, with the key to success being the ability of the model to learn subtle and discriminative features. The current approaches to this challenge primarily focus on resolving interclass similarity by reconstructing query features from support features. However, these methods often fall short of adequately addressing the challenge of intraclass diversity. In response, we devised an innovative solution: the mutual reconstruction network. This approach is designed to manage both intraclass diversity and interclass similarity simultaneously, thereby enhancing the performance in few-shot classification tasks for histological imaging. Our method takes into account the variations within categories and the similarities between categories in histological images, allowing for the more effective capture and utilization of feature information and thus more accurate classification.
Our proposed model, as illustrated in Figure 2, begins with the utilization of a feature extractor to process features from both support and query images. The choice of the feature extractor can range from traditional convolutional neural networks, such as Conv4, to more advanced residual networks, such as ResNet. Following this, the extracted features are fed into a FEM . This module employs a self-attention mechanism to self-reconstruct the extracted features, thereby accentuating the nuanced characteristics of each image category while minimizing the interference of irrelevant features. This step is crucial in better capturing and expressing the differences between images, ultimately enhancing classification performance. Subsequently, the enhanced features are introduced into a mutual reconstruction module . This module not only uses the enhanced features of the support set to reconstruct those of the query set but also employs the query set’s enhanced features to reconstruct the support set’s enhanced features. This bidirectional reconstruction strategy not only increases the interclass variance of features but also reduces intraclass variance, effectively addressing challenge of fine-grained features in histological image classification. Finally, a Euclidean metric module calculates the distances between the query features and reconstructed query features and those between the support features and reconstructed support features. Classification of the query samples is then conducted based on the weighted results of these distances.

FEM
The architecture of the FEM we have constructed is depicted in Figure 3. For the few-shot c-way k-shot classification task, we use a feature extractor to process features from samples , yielding feature representations , where h, w, and d represent the height, width, and number of channels of the features, respectively. Subsequently, these features are reorganized into local feature sequences with r spatial locations , where . In the next step, we combine the local feature sequence with spatially embedded position encoding , resulting in . Here, serves as the input to the FEM, with employing a sinusoidal position-encoding method (38). The output of the FEM module is obtained through self-attention computation as follows:
where , , and represent a set of learnable weight parameters, each with dimensions . Subsequently, is computed through consecutive operations of layer normalization (LN) and multilayer perceptron (MLP), in which serves as the output of the FEM.
Mutual reconstruction module
The structure of the intraclass-interclass dual awareness module is illustrated in Figure 4 and comprises two key steps. First, it involves the reconstruction of query features based on support features, addressing the challenge of interclass similarity in few-shot classification tasks of histological images. Second, it focuses on the reconstruction of the support features of a class based on query features, addressing the challenge of intraclass diversity in few-shot classification tasks of histological images.
Following the processing by the FEM, the enhanced support features for the class are obtained, denoted as , where , . Simultaneously, the enhanced query features are acquired, represented as , where . The subsequent step involves using the weight parameters , , and to reconstruct the enhanced support features and enhanced query features for each category. This process includes multiplying with the weight matrices , , and to obtain , , and , respectively, and similarly multiplying with , , and to obtain , , and , respectively, where , , and have dimensions . Finally, the derived , , and , , , and values are used to compute the reconstructed query feature from the support feature of the class and the reconstructed support feature from the query feature of the class. The computation formula is as follows:
Euclidean metric module
After the mutual reconstruction module processing, Euclidean metrics are used to compute the distances between query sample and support samples of the class, denoted as , as well as the distances from the support samples in the class to the query sample , represented as . This is followed by the weighted summation of these two distances to obtain the total distance . The computation formula is as follows:
In this equation, is a learnable weight parameter, initially set to 0.5 (39,40), and represents a learnable temperature factor.
Loss function
The distance is normalized to obtain . Based on , the loss function for the task can be calculated as follows:
In this context, is an indicator function, which takes the value of 1 when equals , and 0 otherwise.
Results
Dataset
To evaluate the effectiveness of our proposed method in few-shot classification tasks of histological images, we selected image samples from six distinct histological datasets, compiling them into a comprehensive few-shot histological image dataset. These six datasets include the Colorectal Cancer Tissue Phenotyping Dataset (CRC-TP) (25), Lung and Colon Cancer Histopathological Image Dataset (LC25000) (41), Breast Cancer Histopathological Image Classification Dataset (BreakHis) (42), National Center for Tumor Diseases and Colorectal Cancer Dataset (NCT-CRC) (43), The Cancer Genome Atlas (TCGA) dataset (44), and Malignant Lymphoma Classification Dataset (MLC) (45). Our amalgamated dataset encompassed a variety of tissue types and organs, comprising 34 tissue categories. We randomly selected 14,686 images for training, validation, and evaluation purposes. Table 1 displays the specific composition of the dataset.
Table 1
| Dataset | Number of tissue classes | Cancer type | Source |
|---|---|---|---|
| Training set | 8 | Breast | BreakHis |
| 2 | Stomach | TCGA | |
| 7 | Colorectum | CRC-TP | |
| 3 | Lymph nodes | MLC | |
| Validation set | 5 | Lung and colon | LC25000 |
| Evaluation set | 9 | Colorectum | NCT |
CRC-TP, Colorectal Cancer Tissue Phenotyping Dataset; LC25000, Lung and Colon Cancer Histopathological Image Dataset; BreakHis, Breast Cancer Histopathological Image Classification Dataset; NCT-CRC, National Center for Tumor Diseases and Colorectal Cancer Dataset; TCGA, The Cancer Genome Atlas; MLC, Malignant Lymphoma Classification Dataset.
In anticipation of future clinical applications, we opted to select cancer categories from the same anatomical site with similar appearances for the test set, rather than randomly choosing categories for the few-shot experiments. Our training set encompassed 20 categories, including those from the CRC-TP dataset (tumor, stroma, complex stroma muscle, debris, inflammatory, benign), the BreakHis dataset (adenosis, fibroadenoma, phyllodes tumor, tubular adenoma, carcinoma, lobular carcinoma, mucinous carcinoma, papillary carcinoma), TCGA dataset (microsatellite stable stomach tumor, mutated stomach tumor), and MLC dataset (chronic lymphocytic leukemia, follicular lymphoma, mantle cell lymphoma). The validation set comprised five categories from the LC25000 dataset (benign lung tissue, lung squamous cell carcinoma, lung adenocarcinoma, colon adenocarcinoma, and benign colonic tissue). Finally, the test set included nine categories from the NCT-CRC dataset (adipose, no tissue [background], debris, lymphocytes, mucus, smooth muscle, normal colon mucosa, cancer-associated stroma, colorectal adenocarcinoma epithelium).
Experimental setup
For the n-way k-shot setting, each episode encompasses n categories, with each category comprising k support images. We conducted model training on a training set consisting of 15 categories, each with 10 support images, and set up 15 query images for testing. The sizes of these images varied from 150×150 to 768×768 pixels, featuring a range of different aspect ratios. To facilitate model training, we preprocessed all images to a uniform size of 84×84 pixels. This adjustment was made due to considerations of future deployment on computationally constrained terminal medical devices and for facilitating comparison with existing research. In terms of data augmentation, we employed techniques such as cropping, horizontal flipping, and color jittering to enhance the training stability of the model (46-48). These operations contribute to increasing data diversity and enable the model to better adapt to various image variations.
Our experiments were conducted using the PyTorch framework on a GeForce RTX 3090 GPU (Nvidia Corp., Santa Clara, CA, USA) (49). We opted for Conv4 and ResNet12 as the image feature extractors and considered two primary factors. First, as we anticipated deploying the algorithm on terminal medical devices with limited computational resources in the future, we designed the feature extraction module to not excessively burden computational resources. Second, most research on few-shot fine-grained image classification employs these two network models as feature extractors. To facilitate better comparison with existing studies, we selected feature extractors similar to those used in previous research. These two backbone networks both accept images of size 84×84 as input; however, due to differences in their network structures, the size of the feature maps they generate varies. Specifically, Conv4 consists of four convolutional blocks, each comprising a convolutional layer with 64 channels, batch normalization, a rectified linear unit (ReLU) activation function, and a max pooling layer, resulting in feature maps 64×5×5 in size. ResNet12, on the other hand, includes four residual blocks, each containing three convolutional layers, generating feature maps 640×5×5 in size.
In our experiments, we used a stochastic gradient descent (SGD) optimizer with Nesterov momentum set to 0.9 and trained all Conv4 and ResNet12 models for 1,200 epochs. The initial learning rate was set to 0.1, with weight decay applied at a rate of 5e-4. The learning rate was decreased by a factor of 10% every 400 epochs. The best model was selected based on performance on the validation set, with evaluations conducted every 20 epochs. For all experiments, we report the average accuracy across 10,000 randomly generated tasks on the dataset, under the 5-way, 1-shot, 5-shot, and 10-shot settings, along with the 95% confidence intervals.
Evaluation metrics
For the few-shot multiclass tasks, we employed accuracy as the primary performance metric. Accuracy is defined as the proportion of correctly classified samples to the total number of samples. A higher accuracy indicates better performance, and the specific formula is as follows:
Where “Number of Correctly Classified Samples” refers to the count of samples correctly classified by the model, and “Total Number of Samples” is the total number of samples in the evaluation or test set. Accuracy ranges from 0% to 100%, with higher values indicating superior performance of the model in multiclass classification tasks. This metric was used to objectively assess the model’s performance and effectiveness in few-shot multiclass problems. The data in Tables 2-4 are presented as the mean ± standard deviation with 95% confidence intervals of 5-way classification accuracy for the few-shot histological image dataset.
Table 2
| Method | Backbone | 5-way 1-shot | 5-way 5-shot | 5-way 10-shot |
|---|---|---|---|---|
| MAML | Conv4 | 52.34%±1.23% | 59.13%±0.84% | 63.41%±0.77% |
| Resnet12 | 65.22%±0.65% | 67.69%±0.93% | 68.50%±0.63% | |
| Reptile | Conv4 | 55.78%±0.84% | 64.29%±0.74% | 69.11%±0.28% |
| Resnet12 | 68.90%±0.53% | 69.72%±0.78% | 74.14%±0.37% | |
| ProtoNet | Conv4 | 55.68%±0.43% | 76.66%±0.65% | 80.20%±0.21% |
| Resnet12 | 56.85%±0.68% | 72.96%±0.73% | 76.86%±0.89% | |
| DeepEMD | Resnet12 | 66.44%±0.26% | 78.10%±0.57% | 81.34%±0.69% |
| ReNet | Resnet12 | 68.27%±0.39% | 85.58%±0.20% | 89.07%±0.16% |
| FRN | Conv4 | 65.34%±1.10% | 80.82%±0.42% | 84.69%±0.22% |
| Resnet12 | 64.19%±0.87% | 81.89%±0.12% | 84.99%±0.43% | |
| Proposed | Conv4 | 71.87%±0.64% | 87.45%±0.31% | 90.42%±0.28% |
| Resnet | 75.37%±0.47% | 89.97%±0.31% | 92.09%±0.22% |
The data are presented as mean ± standard deviation on the 5-way classification accuracy for the few-shot histological image dataset. MAML, model-agnostic meta-learning; FRN, feature map reconstruction network.
Table 3
| Backbone | Method | 5-way 1-shot | 5-way 5-shot | 5-way 10-shot |
|---|---|---|---|---|
| Conv4 | Baseline (ProtoNet) | 55.68%±0.43% | 76.66%±0.65% | 80.20%±0.21% |
| FEM | 63.11%±0.79% | 80.14%±0.77% | 83.91%±0.29% | |
| MRM | 66.37%±0.19% | 82.79%±0.59% | 87.83%±0.34% | |
| FEM + MRM | 71.87%±0.64% | 87.45%±0.31% | 90.42%±0.28% | |
| ResNet12 | Baseline (ProtoNet) | 56.85%±0.68% | 72.96%±0.73% | 76.86%±0.89% |
| FEM | 63.14%±0.37% | 80.84%±0.71% | 83.91%±0.53% | |
| MRM | 70.20%±0.55% | 86.13%±0.42% | 89.97%±0.11% | |
| FEM + MRM | 75.37%±0.47% | 90.08%±0.31% | 92.09%±0.22% |
The data are presented as mean ± standard deviation on 5-way classification accuracy for the few-shot histological image dataset. FEM, feature enhancement module; MRM, mutual reconstruction module.
Table 4
| Backbone | Method | 1-shot | 5-shot | 10-shot |
|---|---|---|---|---|
| Conv4 | Baseline (ProtoNet) | 55.68%±0.43% | 76.66%±0.65% | 80.20%±0.21% |
| Our (Q→S) | 72.31%±0.35% | 84.77%±0.51% | 87.86%±0.53% | |
| Our (S→Q) | 63.76%±0.42% | 82.98%±0.71% | 86.59%±0.42% | |
| Our (mutual) | 71.87%±0.64% | 87.45%±0.31% | 90.42%±0.28% | |
| ResNet12 | Baseline (ProtoNet) | 56.85%±0.68% | 72.96%±0.73% | 76.86%±0.89% |
| Our (Q→S) | 75.72%±0.19% | 86.12%±0.44% | 89.21%±0.37% | |
| Our (S→Q) | 68.93%±0.39% | 85.21%±0.45% | 89.61%±0.39% | |
| Our (mutual) | 75.37%±0.47% | 90.08%±0.31% | 92.09%±0.22% |
The data are presented as mean ± standard on five-way classification accuracy for the few-shot histological image dataset. MRM, mutual reconstruction module.
Comparison with state-of-the-art methods
To rigorously evaluate the efficacy of our method in few-shot classification tasks of histological images, we designed a series of experiments based on a meticulously constructed few-shot histological image dataset. In this process, we selected several classic few-shot learning methods as benchmarks for comparison, including the optimization-based MAML and Reptile methods (9,10), the global feature metric-based ProtoNet method (11), the attention mechanism-based ReNet method (12), and the local feature metric-based DeepEMD and feature map reconstruction network (FRN) methods (13,14). The performance results of these models were obtained using publicly available code. Notably, the MAML, Reptile, and ProtoNet methods do not adequately address the challenges of fine-grained features in images. In contrast, the ReNet, DeepEMD, and FRN methods are emerging approaches for fine-grained few-shot classification tasks in natural images. These approaches were analyzed and compared with our proposed method.
We conducted accuracy tests for 5-way 1-shot, 5-way 5-shot, and 5-way 10-shot classification tasks using two different feature extractors, Conv4 and ResNet12. The detailed experimental results are presented in Table 2 and indicate that irrespective of the feature extractor used, our method possesses significant advantages in the 1-shot, 5-shot, and 10-shot experimental settings. Particularly noteworthy is the performance with ResNet12 as the feature extractor, where our method achieved a classification accuracy of up to 92.09% in the 5-way 10-shot experiment. This outcome emphatically validates the effectiveness of our method in handling few-shot classification tasks of histological images.
In this study, we aimed to address the challenge of fine-grained features present in few-shot classification tasks in histological imaging and compared our results with those of traditional few-shot methods such as MAML. Specifically, when using Conv4 as the backbone network, our method realized performance improvements of 19.53%, 28.32%, and 27.01% in the 1-shot, 5-shot, and 10-shot classification tasks, respectively. This significant enhancement in performance is attributed to our method’s effective capture and processing of fine-grained features.
For methods such as MAML, Reptile, and ProtoNet, which do not adequately consider the fine-grained features of histological images, the classification accuracy was generally poor. However, the DeepEMD and FRN methods, which adopt an innovative strategy of reconstructing query images to obtain discriminative features, demonstrated a degree of improvement in the few-shot classification tasks of histological images. Particularly noteworthy is the ReNet model, whose proposed dual-relationship modules use attention mechanisms to effectively capture subtle regions in histological images, which led to an excellent performance in this task, especially in the 10-shot classification tasks, with accuracies reaching up to 89.07%.
Regarding feature extractors, as illustrated in Figure 5, most models exhibited performance improvements when switching from Conv4 to ResNet12. However, it is notable that only the ProtoNet model showed a decrease in performance when switching to ResNet12 as the feature extractor, suggesting potential limitations in its handling of high-dimensional features. This finding provides valuable insights for future research, highlighting the crucial importance of the choice and optimization of feature extractors in the design of few-shot learning methods.
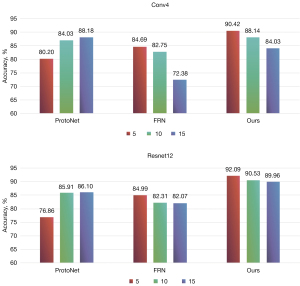
We also evaluated the performance of ProtoNet, FRN, and our newly proposed method when using different feature extractors (specifically Conv4 and ResNet12) and varied numbers of training set categories. As illustrated in Figure 5, our experimental results indicate that only ProtoNet exhibited a gradually improving performance as the number of categories increased in the training set. In contrast, our method and FRN performed best in 5-way tasks and had a declining performance as the number of categories in the training set increased. This trend may stem from ProtoNet’s use of a global feature metric-based approach, in which a greater amount of category information in each training round aids in prototype construction. On the other hand, the performance of the proposed method and FRN worsened with an increase number of training set categories, possibly due to overfitting. Notably, when the more complex ResNet12 feature extractor was employed, the performance decline was relatively less pronounced, with slightly better performance compared to that achieved with the Conv4 feature extractor. These results provide important insights into the problem of category induction in medical image processing and offer valuable guidance for model selection and training processes.
Ablation study
To further substantiate the efficacy and accuracy of our method and the design of its modules, we conducted ablation experiments on the few-shot histological image dataset using Conv4 and ResNet12 as feature extractors.
Initially, we assessed the effectiveness of the FEM and mutual reconstruction module in medical image processing. We assessed their impact on the performance of our method by individually removing these two modules, effectively reducing them to equivalent baseline models (5). The results presented in Table 3 clearly show that each module positively influenced the performance relative to the baseline model. In the 5-shot experimental setting, the independent use of either the FEM or the mutual reconstruction module led to performance improvements of 3.48% and 6.13%, respectively, compared to the baseline model. This indicates that both modules provided benefit in learning the microfeatures and discriminative features of histological images. Notably, when both the FEM and the mutual reconstruction module were used simultaneously, our method’s performance further improved, with an enhancement of 10.79% in the 5-shot setting, indicating the value of their complementarity.
Furthermore, we investigated the effectiveness of mutual reconstruction. To evaluate this, we modified our method in two ways: (I) by removing the reconstruction of support samples from query samples in mutual reconstruction (setting to 0 in Eq. [8]), denoted as “our (Q→S)”, and (II) by removing the reconstruction of query samples from support samples (setting to 1 in Eq. [8]), denoted as “our (S→Q)”. As shown in Table 4, both unidirectional and bidirectional reconstruction methods significantly achieved a performance surpassing that of the baseline model, demonstrating the effectiveness of reconstruction.
However, outside the 1-shot tasks, we observed that both unidirectional reconstruction methods underperformed compared to the bidirectional reconstruction method. The reason our (Q→S) method performed better in the 1-shot experiment is that each category only contained one sample, and hence there was no challenge of intraclass diversity. Therefore, addressing only the challenge posed by interclass similarity is most effective in this scenario. These results imply that the mutual reconstruction module not only addresses the challenge of interclass similarity in few-shot classification tasks of histological images but also successfully overcomes the complexity of intraclass diversity. Thus, our design choice of employing a bidirectional reconstruction method is both rational and effective.
Discussion
Few-shot learning enables deep learning models to achieve good generalization performance and efficiency with limited data. Its mechanism allows models to effectively learn from a small number of samples, mimicking the process of natural human learning. Few-shot learning holds significant clinical relevance and is poised to become one of the key clinical applications in medical image analysis for several reasons. Firs, due to privacy concerns, the high costs associated with data collection, and the intricate process of expert annotation, medical datasets are often limited in scale. Few-shot learning enables models to achieve robust generalization with minimal labeled examples, even in scenarios with scarce data, facilitating the development of effective medical imaging solutions. Second, few-shot learning alleviates the burden of manual annotation, requiring only a small number of labeled examples for each new task or medical condition. This functionality streamlines the annotation process and supports clinicians in completing time-consuming tasks. Additionally, few-shot learning demonstrates particular value in addressing rare medical conditions where traditional deep learning methods may struggle in conditions of sparse data. By leveraging knowledge from more common diseases, few-shot learning allows models to adapt to new instances with limited examples of both novel and rare cases. Moreover, the field of medicine constantly encounters new diseases, conditions, and imaging modalities. Few-shot learning enables medical imaging models to quickly adapt and learn from a few examples of these new tasks, facilitating their seamless integration into clinical practice. Despite the high clinical applicability of few-shot learning, it has not yet emerged as the optimal clinical solution, primarily due to constraints imposed by limited training data and specific challenges in medical image analysis. Further research is needed to overcome these challenges. To this end, we developed corresponding solutions to address the dual challenges of interclass similarity and intraclass diversity in few-shot classification tasks of histological images. More specifically, we adopted a bidirectional reconstruction strategy in the mutual reconstruction module.
The results of the ablation study indicate that both reconstruction tasks complemented each other. By reconstructing query features from support features, we could increase interclass variance, addressing the issue of inter-class similarity. In the ablation study, under the 1-shot setting in which the impact of intraclass diversity was not considered, we found that when the reconstruction of support samples was removed from query samples, our (Q→S) method performed best. This further corroborates the notion that reconstructing query features from support features can increase interclass variance and address the issue of interclass similarity, a conclusion also supported by several other studies (14,35,37). Furthermore, we addressed the issue of high intraclass variance through the reconstruction of support samples from query samples. The results from the ablation study show that in the sole reconstruction of support samples from query samples, our (S→Q) provided significant improvement relative to the baseline model, especially when the number of samples per category increased, even surpassing our (Q→S) in the 10-shot setting. This result further confirms that reconstructing support features from query features can reduce intraclass variance and address the issue of intra-class diversity. Although both unidirectional reconstruction methods, our (S→Q) and our (Q→S) significantly outperformed the baseline model and showed excellent performance in different experimental settings, almost all experimental results indicate that bidirectional reconstruction outperforms unidirectional reconstruction. Therefore, the combined strategy of bidirectional reconstruction and the reconstruction query features from support features, and vice versa, best fits future clinical application needs.
Considering the limitations of our study, we must explore methods that can enhance model performance, especially given the potential future increase in the variety of cancer categories in the source dataset. Moreover, further validation is needed to assess our model’s generalization and fitting capabilities across a broader range and different distribution conditions of data. The diversity and broader distribution of the source dataset data may significantly impact the results of our study, an issue we have not yet thoroughly examined.
In the comparison with other methods, our study not only revealed the potential of the mutual reconstruction network model to complete few-shot classification tasks of histological images but also provides new insights into managing fine-grained features in this context. We recognize the need for further research and exploration to fully realize the potential of this method, especially when faced with more complex source datasets and a broader range of clinical application scenarios.
In future research, we plan to introduce more powerful feature extractors and to preprocess images to larger dimensions during training and testing to mitigate the risk of significant information loss and thus further enhance our model’s fitting capability. Figure 5 shows the impact of using different feature extractors and varying numbers of categories (way numbers) on model performance during training. The results indicate a gradual decline in performance with an increasing number of categories in the training set. However, the decline in performance was somewhat milder with the more complex ResNet12 feature extractor than with the Conv4 feature extractor. We speculate that both feature extractors may have been subject to overfitting. In the future, with an increasing number of cancer categories in the source dataset and a broader data distribution, we plan to train our model using larger way numbers and more complex feature extractors (such as ResNet50 or Vision Transformer) to further enhance the performance of our proposed model. Additionally, we will continue to explore the potential applications of few-shot learning in the field of histological image processing. This series of explorations and improvements will help us better address complex medical image classification problems, providing more promising solutions for clinical practice.
Conclusions
To address the complex challenges of interclass similarity and intraclass diversity in few-shot classification tasks of histological images, we developed a novel mutual reconstruction network model. This model innovatively employs an FEM and a mutual reconstruction module for the bidirectional reconstruction of both support and query images. Rigorous ablation studies confirmed the synergistic effect of these modules. Our findings demonstrate that reconstructed query features using support features effectively increase inter-lass variance and that reconstructed support features using query features decrease intraclass variance. This dual reconstruction approach adeptly resolves the challenges of granularity encountered in histological image classification. When compared with existing few-shot methods, our model achieved commendable results. In a 5-way 10-shot experimental setup, the model attained an impressive accuracy of 92.09%. This achievement represents a substantial contribution to the field of automated classification of histological images, particularly in clinical applications.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-24-253/coif). All authors report that this work was supported by the Department of Natural Resources of Guangdong Province (grant No. GDNRC[2023]47), the National Key R&D Program of China (grant Nos. 2021ZD0112501 and 2021ZD0112502), the National Natural Science Foundation of China (grant Nos. U22A2098, 62172185, and 61876069), the Jilin Province Capital Construction Fund Industry Technology Research and Development Project (grant No. 2022C0471), the Changchun Key Scientific and Technological Research and Development Project (grant No. 21ZGN30), and the Jilin Province Natural Science Foundation (grant No. 20200201036JC). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov 2022;12:31-46. [Crossref] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer 2021; Epub ahead of print. [Crossref] [PubMed]
- Srinidhi CL, Ciga O, Martel AL. Deep neural network models for computational histopathology: A survey. Med Image Anal 2021;67:101813. [Crossref] [PubMed]
- LUSTED LB. Medical electronics. N Engl J Med 1955;252:580-5. [Crossref] [PubMed]
- Lodwick GS, Keats TE, Dorst JP. The coding of roentgen images for computer analysis as applied to lung cancer. Radiology 1963;81:185-200. [Crossref] [PubMed]
- Phankokkruad M. Ensemble transfer learning for lung cancer detection. DSIT 2021: 2021 4th International Conference on Data Science and Information Technology, 2021:438-42.
- Mohalder RD, Talukder KH. Deep learning based colorectal cancer (CRC) tumors prediction. 2021 12th International Conference on Computing Communication and Networking Technologies (ICCCNT), Kharagpur, India, 2021:01-06.
- Adu K, Yu Y, Cai J, Owusu-Agyemang K, Twumasi BA, Wang X. DHS-CapsNet: Dual horizontal squash capsule networks for lung and colon cancer classification from whole slide histopathological images. Int J Imaging Syst Technol 2021;31:2075-92. [Crossref]
- Finn C, Abbeel P, Levine S. Model-agnostic meta-learning for fast adaptation of deep networks. Proceedings of the 34th International Conference on Machine Learning, 2017:1126-35 .
- Nichol A, Schulman J. Reptile: a scalable metalearning algorithm. arXiv preprint arXiv: 180302999, 2018;2:4.
- Snell J, Swersky K, Zemel R. Prototypical networks for few-shot learning. Part of Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017;30.
- Kang D, Kwon H, Min J, Cho M. Relational embedding for few-shot classification. Proceedings of the IEEE/CVF International Conference on Computer Vision, 2021:8822-33.
- Zhang C, Cai Y, Lin G, Shen C. Deepemd: Few-shot image classification with differentiable earth mover’s distance and structured classifiers. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 2020:12200-10.
- Wertheimer D, Tang L, Hariharan B. Few-shot classification with feature map reconstruction networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2021:8012-21.
- Ravi S, Larochelle H. Optimization as a model for few-shot learning. International Conference on Learning Representations, 2016.
- Lee K, Maji S, Ravichandran A, Soatto S. Meta-Learning With Differentiable Convex Optimization. EProceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2019:10657-65.
- Li W, Wang L, Xu J, Huo J, Gao Y, Luo J. Revisiting local descriptor based image-to-class measure for few-shot learning. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2019:7260-8.
- Chao S, Belanger D. Generalizing Few-Shot Classification of Whole-Genome Doubling Across Cancer Types. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) Workshops, 2021:3382-92.
- Titoriya AK, Singh MP. Few-Shot Learning on Histopathology Image Classification. 2022 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 2022:251-6.
- Fagerblom F, Stacke K, Molin J. Combatting out-of-distribution errors using model-agnostic meta-learning for digital pathology. Medical Imaging 2021;11603:186-92. [Crossref]
- Yuan Z, Esteva A, Xu R. Metahistoseg: a python framework for meta learning in histopathology image segmentation. In: Engelhardt S, Oksuz I, Zhu D, Yuan Y, Mukhopadhyay A, Heller N, Huang SX, Nguyen H, Sznitman R, Xue Y. Deep Generative Models, and Data Augmentation, Labelling, and Imperfections. DGM4MICCAI DALI 2021 2021. Lecture Notes in Computer Science, Springer, 2021;13003:268-75.
- Shaikh NN, Wasag K, Nie Y. Artifact Identification in Digital Histopathology Images Using Few-Shot Learning. 2022 IEEE 19th International Symposium on Biomedical Imaging (ISBI), Kolkata, India, 2022:1-4.
- Medela A, Picon A, Saratxaga CL, Belar O, Cabezón V, Cicchi R, Bilbao R, Glover B. Few shot learning in histopathological images: reducing the need of labeled data on biological datasets. 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 2019:1860-4.
- Medela A, Picon A. Constellation Loss: Improving the Efficiency of Deep Metric Learning Loss Functions for the Optimal Embedding of histopathological images. J Pathol Inform 2020;11:38. [Crossref] [PubMed]
- Javed S, Mahmood A, Werghi N, Benes K, Rajpoot N. Multiplex Cellular Communities in Multi-Gigapixel Colorectal Cancer Histology Images for Tissue Phenotyping. IEEE Trans Image Process 2020; Epub ahead of print. [Crossref] [PubMed]
- Jiang Z, Dong Z, Fan J, Yu Y, Xian Y, Wang Z. Breast TransFG Plus: Transformer-based fine-grained classification model for breast cancer grading in Hematoxylin-Eosin stained pathological images. Biomed Signal Process Control 2023;86:105284. [Crossref]
- Liu W, Juhas M, Zhang Y. Fine-Grained Breast Cancer Classification With Bilinear Convolutional Neural Networks (BCNNs). Front Genet 2020;11:547327. [Crossref] [PubMed]
- Li L, Pan X, Yang H, Liu Z, He Y, Li Z, Fan Y, Cao Z, Zhang L. Multi-task deep learning for fine-grained classification and grading in breast cancer histopathological images. Multimed Tools Appl 2020;79:14509-28. [Crossref]
- Wang Chaofeng, Shi Jun, Zhang Qi, Ying Shihui. Histopathological image classification with bilinear convolutional neural networks. Annu Int Conf IEEE Eng Med Biol Soc 2017;2017:4050-3. [Crossref] [PubMed]
- Fan M, Chakraborti T, Chang EI-C, Xu Y, Rittscher J. Microscopic Fine-Grained Instance Classification Through Deep Attention. In: Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga MA, Zhou SK, Racoceanu D, Joskowicz L. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. Lecture Notes in Computer Science, Springer, 2020;12265:490-9.
- Mahapatra D. Registration of histopathogy images using structural information from fine grained feature maps. arXiv preprint arXiv: 200702078, 2020.
- Chattopadhyay S, Dey A, Singh PK, Oliva D, Cuevas E, Sarkar R. MTRRE-Net: A deep learning model for detection of breast cancer from histopathological images. Comput Biol Med 2022;150:106155. [Crossref] [PubMed]
- Zhang X, Su H, Yang L, Zhang S. Fine-grained histopathological image analysis via robust segmentation and large-scale retrieval. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 2015:5361-8.
- Yan R, Ren F, Wang Z, Wang L, Zhang T, Liu Y, Rao X, Zheng C, Zhang F. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 2020;173:52-60. [Crossref] [PubMed]
- Wu J, Chang D, Sain A, Li X, Ma Z, Cao J, Guo J, Song YZ. Bi-directional feature reconstruction network for fine-grained few-shot image classification. Proceedings of the AAAI Conference on Artificial Intelligence 2023;37:2821-9. [Crossref]
- Lee S, Moon W, Heo JP. Task discrepancy maximization for fine-grained few-shot classification. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2022:5331-40.
- Doersch C, Gupta A, Zisserman A. CrossTransformers: spatially-aware few-shot transfer. Part of Advances in Neural Information Processing Systems 33 (NeurIPS 2020) 2020;33:21981-93.
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. Part of Advances in Neural Information Processing Systems 30 (NIPS 2017), 2017;30.
- Chen Y, Liu Z, Xu H, Darrell T, Wang X. Meta-baseline: Exploring simple meta-learning for few-shot learning. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV), 2021:9062-71.
- Gidaris S, Komodakis N. Dynamic few-shot visual learning without forgetting. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2018:4367-75.
- Borkowski AA, Bui MM, Thomas LB, Wilson CP, DeLand LA, Mastorides SM. Lung and colon cancer histopathological image dataset (lc25000). arXiv preprint arXiv: 191212142, 2019.
- Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A Dataset for Breast Cancer Histopathological Image Classification. IEEE Trans Biomed Eng 2016;63:1455-62. [Crossref] [PubMed]
- Kather JN, Halama N, Marx A. 100,000 histological images of human colorectal cancer and healthy tissue. Zenodo10 2018;5281.
- Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, Marx A, Boor P, Tacke F, Neumann UP, Grabsch HI, Yoshikawa T, Brenner H, Chang-Claude J, Hoffmeister M, Trautwein C, Luedde T. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054-6. [Crossref] [PubMed]
- Orlov NV, Chen WW, Eckley DM, Macura TJ, Shamir L, Jaffe ES, Goldberg IG. Automatic classification of lymphoma images with transform-based global features. IEEE Trans Inf Technol Biomed 2010;14:1003-13. [Crossref] [PubMed]
- Chen WY, Liu YC, Kira Z, Wang YCF, Huang JB. A closer look at few-shot classification. arXiv preprint arXiv: 190404232, 2019.
- Wang Y, Chao WL, Weinberger KQ, Van Der Maaten L. Simpleshot: Revisiting nearest-neighbor classification for few-shot learning. arXiv preprint arXiv: 191104623, 2019.
- Ye HJ, Hu H, Zhan DC, Sha F. Few-shot learning via embedding adaptation with set-to-set functions. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2020:8808-17.
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L. Pytorch: An imperative style, high-performance deep learning library. Part of Advances in Neural Information Processing Systems 32 (NeurIPS 2019), 2019;32.




