
Development and validation of an interpretable delta radiomics-based model for predicting invasive ground-glass nodules in lung adenocarcinoma: a retrospective cohort study
Introduction
With the development of low-dose computed tomography (CT) screening, the widespread application of high-resolution chest CT, and expansion of artificial intelligence in the field of medical imaging in recent years, the detection rate and diagnostic accuracy for small nodules and ground-glass nodules (GGNs) have improved significantly, providing conditions for the early diagnosis and treatment of lung adenocarcinoma.
Patients with GGN-type lung adenocarcinoma have a good prognosis after surgical resection, with many studies reporting a postoperative 5-year survival rate of adenocarcinoma in situ (AIS) and minimally invasive adenocarcinoma (MIA) of up to 100% (1,2), which is significantly higher than that of invasive adenocarcinoma (IAC) (3). Therefore, accurately determining the evolution pattern of GGN invasiveness is critical in clinical decision-making. However, in clinical practice, radiologists typically determine the pathological subtypes of GGNs based solely on characteristics such as diameter, volume, density (heterogeneity), and overall morphology. Although many recent studies have used radiomics to evaluate the invasiveness of GGNs and achieved a degree of success, this method has not yet fully utilized the information on image changes over time during follow-up. This may prove critical, as, for example, patients with GGNs >6 mm in diameter are recommended to undergo routine follow-up according to the Fleischner Society pulmonary nodule recommendations (4). Thus, the value of combining radiomics with follow-up changes for GGN invasiveness prediction warrants further examination.
Delta radiomics (also known as delta texture analysis), in contrast to traditional radiomics methods, involves studying changes in texture features of patients after specific steps (i.e., specific treatments, time intervals, or biological events) (5). Previous studies (6,7) have shown that delta radiomics can improve lung cancer detection rate in screening and pulmonary nodule malignancy prediction. There are also researches suggesting that delta radiomics is valuable in predicting the invasiveness of GGNs, but the results supporting this may be unstable due to the relatively limited number of studies (8-10).
The unclear internal mechanism and interpretability of radiomics or delta radiomics models also hinder their general application. Interpretability typically serves as a means to engender trust, yet post hoc interpretability is sometimes used to obtain more useful information from a model (11). Shapley additive explanations (SHAP) is an approach based on game theory that can be used to explain the output of any machine learning model (12) and has been applied in various areas in radiomics research (13-16). By helping clinical doctors understand machine learning models, it may promote the use of this decision-making tool.
In this study, we incorporated information on clinical features and follow-up images to develop and validate a delta radiomics model that offers a more efficient, precise, and noninvasive method for GGN invasiveness prediction. Additionally, SHAP was used to improve model interpretability and visualization. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1711/rc).
Methods
Patients
This retrospective study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the institutional review board of Shanghai Chest Hospital, School of Medicine, Shanghai Jiao Tong University (No. KS1956). Informed consent requirements were waived because all patient data were used anonymously. CT images of 1,422 resected GGN-type lung adenocarcinomas were screened according to following inclusion criteria: (I) histologically confirmed as atypical adenomatous hyperplasia (AAH), AIS, MIA, or IAC; (II) a GGN size ≥5 mm and ≤30 mm; (III) completion of thin-section CT (section thickness ≤1.5 mm); and (IV) availability of at least one follow-up scan. Meanwhile, the exclusion criteria were as follows: (I) a severe artifact interfering with observation, (II) follow-up interval <1 month, (III) interval between the last CT scan and the surgical resection >1 month, and (IV) GGNs beyond scan range. If multiple GGNs were resected from one patient during this single surgery, the predominant GGN was included to this study. The predominant GGN was defined as follows: (I) the GGN with higher degree of invasiveness (IAC > MIA > AIS > AAH), (II) the GGN with bigger size when their pathology grade were the same, and (III) the GGN with higher mean CT value when they were about the same size. The process of patient selection is shown in Figure 1.
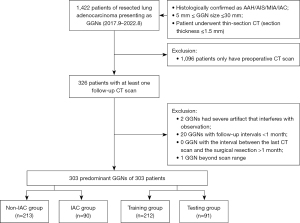
Finally, among the 303 predominant GGNs included from 303 patients that underwent surgical resection in Shanghai Chest Hospital between September 2017 and August 2022, 213 non-IAC GGNs (5 AAH, 98 AIS, and 110 MIA) and 90 IAC GGNs were divided into the training and test set randomly in a ratio of 7:3.
CT image acquisition
Thin-section CT scans performed at baseline and at the last follow-up before surgery were obtained with one the five following scanners: Revolution CT (GE HealthCare, Chicago, IL, USA), Discovery CT750 HD (GE HealthCare), Ingenuity CT (Philips, Amsterdam, the Netherlands), Brilliance iCT 256 (Philips), uCT 510 (United Imaging, Shanghai, China). All CT scans were performed with patients in the supine position at full inspiration under the following parameters: collimation, 0.625–1.25 mm; pitch, 0.64; section thickness, 0.625–1.25 mm without overlap; matrix, 512×512 or 1,024×1,024; field of view (FOV), 350–400 mm, tube voltage, 120 kVp; and tube current, 220–300 mA. Imaging data were reconstructed based on the standard algorithm.
GGN segmentation
CT Digital Imaging and Communications in Medicine (DICOM) images were exported from the picture archiving and communication system (PACS). A radiologist (T.X., with three years of experience in chest diagnosis) determined the region of interest (ROI) on every section of each GGN manually on the open-source software three-dimensional (3D) Slicer (RRID: SCR_005619) (version 5.2.2, Brigham and Women’s Hospital, Boston, MA, USA). One month later, 20 GGNs were randomly selected for segmentation by the same radiologist and another radiologist (X.Y., with 20 years of experience in chest diagnosis), and Bland-Altman plots were used to assess intraobserver and interobserver reproducibility. The two radiologists were blinded to the clinical data and outcomes.
Image preprocessing
The following measures were used to preprocess images before feature extraction: resampling images to isotropy with 1mm at X/Y/Z-spacing, discretizing voxel intensity using a bin width of 25, normalizing signal intensity to 1–500 Hounsfield units (HU); Z-score normalization, and denoising via Gaussian smoothing.
Radiomics feature extraction and selection
PyRadiomics, an open-source package (RRID: SCR_008394) in Python (Python Software Foundation, Wilmington, DE, USA), was used to extract the radiomics features of each patient’s baseline and follow-up CT images. Features were extracted from preprocessed images, exponential images, gradient images, local binary images, logarithmic images, and wavelet images and could be categorized into shape features, histogram features, and texture features including gray-level co-occurrence matrix (GLCM), gray-level run length matrix (GLRLM), gray-level size zone matrix (GLSZM), gray-level difference matrix (GLDM), and neighboring gray-tone difference matrix (NGTDM). Shape features were only extracted from ROI images and reflected the shape, size, volume, and other characteristics of the lesion area, while the remaining features were extracted from original images and derived images and reflected the overall intensity distribution and spatial distribution of pixels in the lesion area.
After Z-score normalization, the delta radiomics features were calculated as follows:
delta radiomics feature = (follow-up radiomics-baseline radiomics feature)/[time interval (days)/30]
(where is the feature after normalization, F is the original image, μ is the feature mean value, and σ is the feature standard deviation value).
Features of delta radiomics and general radiomics were both subjected to the following selection process. In the training set, t-test or the Mann-Whitney test was used to detect the correlation between each feature and the invasiveness of the lesion, with the radiomics feature being considered eligible for the following selection step if P<0.05. Subsequently, features screened via the t-test were input into a least absolute shrinkage and selection operator (LASSO) regression model and underwent penalty parameter adjustment via fivefold cross-validation to identify the optimal features of the nonzero coefficients. The selected optimal features from the training set were directly applied to the test set for evaluation.
Modeling and validation
We applied 10 different machine learning classification algorithms (support vector classifier, random forest classifier, K-nearest neighbor classifier, logistic regression classifier, decision tree classifier, Bernoulli Naïve Baye classifier, extreme gradient boosting classifier, linear discriminant analysis classifier, gradient boosting decision tree classifier, and AdaBoost classifier) and identified the optimal parameters of these models through fivefold cross-validation and grid search techniques. For each selected model, a receiver operating characteristic (ROC) curve was generated. The average area under the curve (AUC) was then calculated to evaluate predictive performance, while the learning curve was used to ascertain model overfitting. According to the classifier chosen, different measures were applied to prevent overfitting. In the training set, the best-performing classifier was selected among the established models based on the combination of the radiomics and delta radiomics features with the clinical and CT features of significance, which were then validated in the test set.
Model interpretation
SHAP is an open-source package (RRID: SCR_021362) in Python used to interpret models. It can quantify the contribution of each feature to the final prediction and determine whether the correlation is positive or negative. Summary plots and waterfall plots were generated in this study to provide global and local interpretations, respectively.
Statistical analysis
Statistical analysis was performed using SPSS v. 26.0) (RRID: SCR_016479) software (IBM Corp., Armonk, NY, USA). The Shapiro-Wilk test was applied for the normality test. Continuous variables with a normal distribution are expressed as the mean ± standard deviation, those abnormally distributed are expressed as the median and range, and categorical variables are expressed as counts and percentages. Differences in clinical and radiological features between two groups were analyzed with the independent samples t-test or the Mann-Whitney test for continuous variables and with the Pearson Chi-square test for categorical variables. P<0.05 was considered to indicate statistical significance. Those features demonstrating statistical significance were included in the model.
Results
Clinical characteristics and GGN CT features
The clinical characteristics and GGN CT features are shown in Table 1. Among 303 cases of GGN, 213 cases (70.3%) were non-IAC GGN (5 AAH, 98 AIS, 110 MIA), and 90 (29.7%) were IAC GGN. The average age of patients was 53.52±13.1 years old, and the median follow-up interval was 352 (range, 30–2,717) days. A comparison of the clinical characteristics and CT features of nodules between the non-IAC and IAC groups revealed significant differences in age (P<0.001), gender (P=0.008), mean preoperative diameter (P<0.001), mean CT value (P<0.001), volume (P<0.001), mass (P<0.001), 3D length (P<0.001), change in mean diameter (P=0.006), change in 3D length (P<0.001), change in volume (P<0.001), change in mass (P<0.001), morphological features such as lobulation (P=0.007), spiculation (P<0.001), air bronchogram (P<0.001), and pleural traction/indentation (P<0.001). However, no significant differences were found for location of GGN (P=0.22), vacuole/cavity (P>0.99), well-defined margin (P=0.53), and change in mean CT value (P=0.96). The clinical characteristics and GGN CT features after GGNs were divided randomly into a training set and test set are also provided in Table 1. The volume measurement was tested for intra- and interobserver consistency (Figure S1 and Table S1).
Table 1
| Clinical/CT features | Total (n=303) | Non-IAC group vs. IAC group | Training set vs. test set | |||||
|---|---|---|---|---|---|---|---|---|
| Non-IAC group (n=213) | IAC group (n=90) | P | Training set (n=212) | Test set (n=91) | P | |||
| Age (years) | 53.52±13.1 | 50.78±12.93 | 60.02±11.12 | <0.001† | 53 (14–77) | 54 (28–76) | 0.262§ | |
| Sex (%) | 0.008‡ | 0.532‡ | ||||||
| Male | 68 (22.4) | 39 (18.3) | 29 (32.2) | 45 (21.2) | 23 (25.3) | |||
| Female | 235 (77.6) | 174 (81.7) | 61 (67.8) | 167 (78.8) | 68 (74.7) | |||
| Location (%) | 0.22‡ | 0.0979‡ | ||||||
| Right upper lobe | 91 (30.0) | 58 (27.2) | 33 (36.7) | 60 (28.3) | 31 (34.1) | |||
| Left upper lobe | 96 (31.7) | 73 (34.3) | 23 (25.6) | 76 (35.8) | 20 (22.0) | |||
| Right middle lobe | 20 (6.6) | 16 (7.5) | 4 (4.4) | 11 (5.2) | 9 (9.9) | |||
| Right lower lobe | 53 (17.5) | 39 (18.3) | 14 (15.6) | 38 (17.9) | 15 (16.5) | |||
| Left lower lobe | 43 (14.2) | 27 (12.7) | 16 (17.8) | 27 (12.7) | 16 (17.6) | |||
| Morphological features (%) | ||||||||
| Lobulation | 232 (76.6) | 154 (72.3) | 78 (86.7) | 0.007‡ | 170 (80.2) | 62 (68.1) | 0.0337‡ | |
| Spiculation | 19 (6.3) | 3 (1.4) | 16 (17.8) | <0.001‡ | 10 (4.7) | 9 (9.9) | 0.149‡ | |
| Air bronchogram | 57 (18.8) | 27 (12.7) | 30 (33.3) | <0.001‡ | 39 (18.4) | 18 (19.8) | 0.903‡ | |
| Vacuole/cavity | 37 (12.2) | 26 (12.2) | 11 (12.2) | 0.997‡ | 29 (13.7) | 8 (8.8) | 0.317‡ | |
| Well-defined margin | 252 (83.2) | 179 (84.0) | 73 (81.1) | 0.534‡ | 177 (83.5) | 75 (82.4) | 0.951‡ | |
| Pleural retraction/ indentation | 100 (33.0) | 49 (23.0) | 51 (56.7) | 0.001‡ | 72 (34.0) | 28 (30.8) | 0.683‡ | |
| Follow-up interval (day) | 352 (30 to 2,717) | 372 (30 to 2,717) | 281 (33 to 2,546) | 0.51§ | 375 (30 to 2,659) | 284 (33 to 2,717) | 0.098 | |
| Preoperative diameter (mm) | 8.5 (4.0 to 30.0) | 8.0 (4.0 to 19.0) | 12.0 (4.5 to 30.0) | <0.001§ | 8.5 (4.5 to 30.0) | 8.5 (4.0 to 27.5) | 0.280§ | |
| Change in diameter (mm) | 0.0 (−6.0 to 21.5) | 0.0 (−3.0 to 7.5) | 0.5 (−6.0 to 21.5) | 0.006§ | 0.0 (−6.0 to 16.0) | 0.5 (−3.0 to 21.5) | 0.120§ | |
| Preoperative 3D length (mm) | 10.96 (6.0 to 42.06) | 10.1 (6.0 to 23.93) | 15.28 (6.07 to 42.06) | <0.001§ | 11.07 (6.07 to 42.06) | 10.82 (6.0 to 31.12) | 0.232§ | |
| Change in 3D length (mm) | 0.36 (−8.12 to 24.73) | 0.15 (−4.37 to 8.64) | 0.85 (−8.12 to 24.73) | <0.001§ | 0.41 (−8.12 to 20.89) | 0.31 (−4.37 to 24.73) | 0.404§ | |
| Preoperative CT value (HU) | −510.2 (−908.52 to 10.9) |
−549.1 (−845.8 to −155.87) |
−370.0 (−908.52 to 10.9) |
<0.001§ | −512.85 (−908.52 to 10.9) |
−496 (−795.1 to 9.6) |
0.335§ | |
| Change in CT value (HU) | 7.07 (−509.41 to 636.96) |
1.2 (−509.41 to 636.96) |
23.49 (−367 to 460) |
0.958§ | 6.55 (−509.41 to 449.88) |
12.12 (−422.68 to 636.96) |
0.488§ | |
| Preoperative volume (mm3) | 330.74 (20.64 to 10,833.19) |
258.25 (20.64 to 3,183.55) |
870.98 (103.87 to 10,833.19) |
<0.001§ | 362.6 (66.24 to 10,833.19) |
320.59 (20.64 to 10,106.12) |
0.225§ | |
| Change in volume (mm3) | 16.71 (−524.62 to 10,037.12) |
6.78 (−174.63 to 2,270.39) |
122.71 (−524.62 to 10,037.12) |
<0.001§ | 16.72 (−524.62 to 9,550.63) |
16.71 (−245.44 to 10,037.12) |
0.248§ | |
| Preoperative mass (mg) | 164.02 (11.68 to 10,695.61) |
122.79 (11.68 to 1,530.48) |
508.99 (47.10 to 10,695.61) |
<0.001§ | 172.86 (31.07 to 10,695.61) |
154.46 (11.68 to 8,223.35) |
0.369§ | |
| Change in mass (mg) | 10.73 (−465.47 to 10,006.34) |
2.19 (−119.88 to 616.33) |
92.98 (−465.47 to 10,006.34) |
<0.001§ | 9.99 (−465.47 to 2,052.72) |
12.48 (−148 to 10,006.34) |
0.341§ | |
Continuous variables are presented as the mean ± standard deviation or median (range). Categorical variables are presented as n (%). †, t-test; ‡, Chi-square test; §, Mann‑Whitney test. CT, computed tomography; IAC, invasive adenocarcinoma; 3D, three-dimensional; HU, Hounsfield unit.
Radiomics feature extraction and selection
A total of 1,409 radiomic features were extracted from the CT images of 303 GGNs, from which 34 general radiomics features and 10 delta radiomics features were selected via the t-test and LASSO regression (Figures 2,3), with clinical and radiological features being combined to construct a model. We selected the tuning parameter (λ) in LASSO regression using fivefold cross-validation via minimum criteria. The optimal λ value of 0.028 and 0.036 was selected, respectively, as shown in Figures S2,S3. The complete list of features after selection is shown in Table S2.
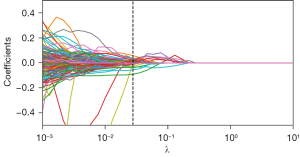
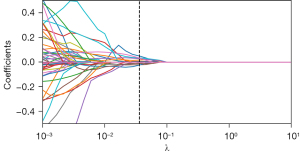
Modeling and validation
The 14 clinical and CT features of statistical significance, including age, gender, preoperative mean diameter, mean CT value, volume, mass, 3D length, change in mean diameter, change in 3D length, change in volume, change in mass, lobulation, spiculation, air bronchogram, and pleural traction/ indentation, were combined with the selected general and delta radiomics features, respectively, to predict the invasiveness of GGNs. Out of the 10 different machine learning classification algorithms tested, random forest classifier had the best performance, as shown in Figures S4,S5. Regularization was implemented to prevent overfitting by setting the following hyperparameters of the random forest model in the code: n_estimators, 100; max_depth, 5; min_sample_split, 10; and min_sample_leaf, 5.
Accuracy, recall, precision, and AUC were used for model evaluation, as shown in Table 2. The radiomics-clinical model and the delta radiomics-clinical models had AUCs of 0.986 [95% confidence interval (CI): 0.977–0.995] and 0.974 (95% CI: 0.959–0.987) in the training set, respectively, and 0.949 (95% CI: 0.908–0.978) and 0.927 (95% CI: 0.879–0.966) in the test set, respectively (Figure 4). The DeLong test of the two models showed statistical significance (P=0.03) in the training set but not in the test set (P=0.10). The calibration curve and the Brier score of the models are shown in Figure S6.
Table 2
| Model | Dataset | Accuracy | Recall | Precision | AUC (95% CI) |
|---|---|---|---|---|---|
| Radiomics-clinical | Train | 93.40% | 82.54% | 94.55% | 0.986 (0.977–0.995) |
| Test | 87.91% | 70.37% | 86.36% | 0.949 (0.908–0.978) | |
| Delta radiomics-clinical | Train | 90.57% | 76.19% | 90.57% | 0.974 (0.959–0.987) |
| Test | 81.32% | 62.96% | 70.83% | 0.927 (0.879–0.966) |
AUC, area under curve; CI, confidence interval.
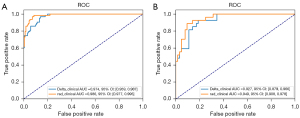
Interpreting the model
Global interpretation
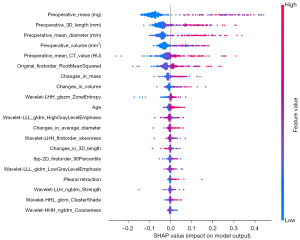
SHAP was used to output a summary plot for visualizing the global interpretation of the model, in which each feature’s contribution is indicated (Figure 5). In Figure 5, features are sorted from top to bottom by importance and characterized by a string of colored dots in the plot in which each dot represents a sample (GGN). For instance, blue dots—which represent samples of low feature value—of the most important feature “preoperative mass” have a negative effect on the model output, as the corresponding horizontal axis position has negative SHAP values, meaning these output a prediction of noninvasiveness. In the upper part of the plot, CT characteristics of the GGNs and their change, such as in preoperative mass, 3D length, mean diameter, mean CT value, mass, and volume, appear to play quite an important role in invasiveness prediction. Original_firstorder_RootMeanSquared is suggested to be the most vital delta radiomics feature, with the higher delta value (difference that occurs during follow-up) indicating a greater likelihood that the GGN is IAC.
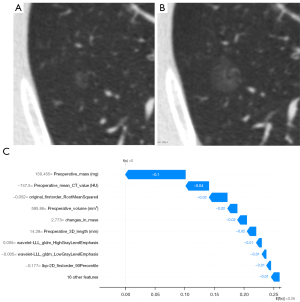
Local interpretation
A waterfall plot was employed to arrange all features in order according to the contribution of each to the final output of a particular GGN while showing the direction of their contribution by color. E[ƒ(z)], the base value, refers to the average SHAP values of model prediction. As shown in Figure 6, although growth, especially in diameter, is seen in this GGN during follow-up, the preoperative mass, mean CT value, and the lack of change in original_firstorder_RootMeanSquared do not support the prediction of invasiveness. The negative effects (blue) contribute to the current output ƒ(x) = 0, which is less than the base value E[ƒ(z)] = 0.26, indicating noninvasiveness.
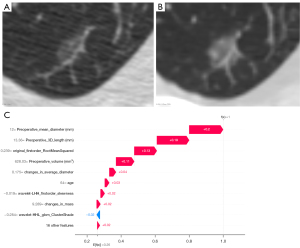
Similarly, preoperative mean diameter, 3D length, and original_firstorder_RootMeanSquared appear to be the more significant features, as indicated by the arrow’s length. Figure 7 includes a GGN which showed an increase in the overall density and a focal density at the right edge (9 o’clock direction of the nodule) during follow-up, which might have caused a significant change in original_firstorder_RootMeanSquared. The two features positively (red) contribute to output 1, which is greater than the base value 0.26, suggesting invasiveness.
Discussion
In this study, we developed and validated a delta radiomics-based model for predicting IAC and non-IAC GGNs, which in contrast to general radiomics models, incorporated the change in follow-up information. The delta radiomics-clinical model achieved good performance and was not inferior to the radiomics-clinical model in the test set. We used SHAP to enhance the interpretability and visibility of the model, which indicated that preoperative mass, 3D length, mean diameter, volume, mean CT value, and delta radiomics feature original_firstorder_RootMeanSquared were the relatively more important features in the model.
There has been limited research conducted on the application of delta radiomics in predicting invasive lung adenocarcinoma GGNs. Chen et al. (8) demonstrated that the use of nomogram coupled with radiographic features based on delta radiomics derived from non-contrast-enhanced CT (NECT) and contrast-enhanced CT (CECT) scans enhances the performance in differentiating IACs from AIS/MIAs in patients with part-solid nodules (PSNs). Ma et al. (9) reported a radiomics signature that could aid in distinguishing between preinvasive GGNs (AAH/AIS) and invasive GGNs (MIA/IAC), with the delta radiomics signature demonstrating a higher AUC than the radiomics signature in identifying invasive GGNs. Lv et al. (10) observed that their delta radiomics model showed satisfactory diagnostic efficiency and superiority compared to the clinical model in distinguishing between invasive adenocarcinoma (IA) and preinvasive lesion (PIL)/MIA in GGN-like lung adenocarcinoma. However, its diagnostic efficiency was slightly lower than that of the radiomics or combined models, which seems to contradict the findings of Ma et al. In our study, although the delt radiomics-clinical model and the general radiomics-clinical model both demonstrated excellent performance, the former included fewer features yet did not exhibit a performance advantage, which, to some extent, aligns with the findings of Lv et al.’s study. However, Ma et al.’s study reported a relatively different result, which we attribute to the different groupings of the two studies. It is possible that significant radiomics changes occur during the transition from an AIS to an MIA, coinciding with the emergence of invasiveness. This discrepancy warrants further investigation and exploration.
The model-interpretation tool, SHAP, quantifies the importance of features and presents them visually, thereby enhancing the interpretability of the model. Interpretability can be broadly classified into transparency interpretability and post hoc interpretability (11). Although SHAP does little to solve the black box issue associated with transparency interpretability, it does offer a more extensive framework for post hoc interpretability. This aspect may hold greater clinical value, as it allows for the exploration of the conversion from nonsemantic features to clinical explanations, which may be highly valuable and warrants further investigation. Wang et al. (14) used the SHAP method to interpret their radiomics model for assessing the treatment response of whole-brain radiation therapy (WBRT). The authors found that SHAP identified 3D contrast-enhanced T1-weighted [CET1-w (3D)]_firstorderM as the most influential nonsemantic feature, which represents the median percentile of gray values within the volume. They hypothesized that the lower values of CET1-w (3D)_firstorderM in the nonresponding group compared to the responding group may indicate the absence of a gadolinium-based contrast agent in tumors due to inadequate vascular supply. Additionally, de Moura (17) evaluated the use of SHAP in radiomics-based machine learning classification models for coronavirus disease 2019 (COVID-19) pneumonia and identified middle left–first order–kurtosis as the most crucial feature. As kurtosis describes the peakedness of the distribution of the values (18), the low first-order kurtosis in COVID-19 makes sense, as the consolidation and ground-glass opacification observed in COVID-19 often induce a distribution with lighter tails and a flatter peak. By establishing a connection between nonsemantic features and the microscopic pathological changes and mechanisms involved in the growth of GGN and elucidating this relationship from a clinical standpoint, this approach becomes more akin to the process employed by clinicians in assessing the invasiveness of GGNs based on morphological characteristics. This not only enhances clinicians’ comprehension and utilization of radiomics models in clinical practice but also facilitates a deeper understanding of the pathological pathogenesis and evolutionary process of GGNs. In our study, original_firstorder_RootMeanSquared was identified as the most significant delta radiomics feature. Root mean square (RMS) is a statistical measure that represents the square root of the average of all squared intensity values, serving as an indicator of the magnitude of image values (19). In comparison to the arithmetic mean, RMS provides a more accurate reflection of data dispersion and is particularly useful when dealing with datasets containing both positive and negative values. This may be particularly relevant for examining PSNs characterized by a ground-glass area of low density ranging from –800 to –600 HU and a solid area of density ranging from 0 to 50 HU. The presence of significant RMS change, as evidenced by the red dots in the summary plot (Figure 5), suggested an increase in the focal or overall density of GGNs or even the emergence of a solid component during follow-up, positively influencing the model to generate an “IAC” output. Moreover, the use of SHAP analysis allowed us to ascertain that the model places considerable importance on CT characteristics and some of their changes, such as in mass, 3D length, mean diameter, mean CT value, and volume. This finding, to some extent, is consistent with our clinical judgment based on experience. The local interpretation provided by SHAP analysis is particularly valuable in comprehending the rationale behind a clinical decision made for a specific patient, especially when radiomic changes during the follow-up and clinical and CT characteristics are taken into consideration.
This study involved certain limitations that should be addressed. First, the inclusion of solely surgical cases might have introduced selection bias, and the surgical inclination toward high-risk nodules could have also contributed to bias in the selection process. Additionally, the median follow-up time interval of around one year might not have allowed for sufficient changes in the radiomic features of some GGNs. Furthermore, the use of different CT machines (between different patients and between the two scans of the same patient) could have potentially influence the results, even with the image data being normalized. Finally, our findings need to be verified by multicenter data.
Conclusions
The use of delta radiomics features represents a viable approach for predicting both IAC and non-IAC GGNs and is an effective, precise, and noninvasive prediction method that includes the follow-up context. The incorporation of SHAP in the model’s interpretation and visualization enhances the comprehensibility of the model from a clinical standpoint and aids in facilitating its practical implementation. With further research, the integration of these two methodologies may contribute to a more profound comprehension of the pathological pathogenesis and evolutionary progression of ground-glass lung adenocarcinoma.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-1711/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1711/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This retrospective study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the institutional review board of Shanghai Chest Hospital, School of Medicine, Shanghai Jiao Tong University (No. KS1956). Informed consent requirements were waived because all patient data were used anonymously.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yotsukura M, Asamura H, Motoi N, Kashima J, Yoshida Y, Nakagawa K, Shiraishi K, Kohno T, Yatabe Y, Watanabe SI. Long-Term Prognosis of Patients With Resected Adenocarcinoma In Situ and Minimally Invasive Adenocarcinoma of the Lung. J Thorac Oncol 2021;16:1312-20. [Crossref] [PubMed]
- Travis WD, Asamura H, Bankier AA, Beasley MB, Detterbeck F, Flieder DB, Goo JM, MacMahon H, Naidich D, Nicholson AG, Powell CA, Prokop M, Rami-Porta R, Rusch V, van Schil P, Yatabe Y; International Association for the Study of Lung Cancer Staging and Prognostic Factors Committee and Advisory Board Members. The IASLC Lung Cancer Staging Project: Proposals for Coding T Categories for Subsolid Nodules and Assessment of Tumor Size in Part-Solid Tumors in the Forthcoming Eighth Edition of the TNM Classification of Lung Cancer. J Thorac Oncol 2016;11:1204-23.
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB, Geisinger K, Hirsch FR, Ishikawa Y, Kerr KM, Noguchi M, Pelosi G, Powell CA, Tsao MS, Wistuba I, Panel WHO. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J Thorac Oncol 2015;10:1243-60. [Crossref] [PubMed]
- MacMahon H, Naidich DP, Goo JM, Lee KS, Leung ANC, Mayo JR, Mehta AC, Ohno Y, Powell CA, Prokop M, Rubin GD, Schaefer-Prokop CM, Travis WD, Van Schil PE, Bankier AA. Guidelines for Management of Incidental Pulmonary Nodules Detected on CT Images: From the Fleischner Society 2017. Radiology 2017;284:228-43. [Crossref] [PubMed]
- Nardone V, Reginelli A, Grassi R, Boldrini L, Vacca G, D'Ippolito E, Annunziata S, Farchione A, Belfiore MP, Desideri I, Cappabianca S. Delta radiomics: a systematic review. Radiol Med 2021;126:1571-83. [Crossref] [PubMed]
- Cherezov D, Hawkins SH, Goldgof DB, Hall LO, Liu Y, Li Q, Balagurunathan Y, Gillies RJ, Schabath MB. Delta radiomic features improve prediction for lung cancer incidence: A nested case-control analysis of the National Lung Screening Trial. Cancer Med 2018;7:6340-56. [Crossref] [PubMed]
- Alahmari SS, Cherezov D, Goldgof D, Hall L, Gillies RJ, Schabath MB. Delta Radiomics Improves Pulmonary Nodule Malignancy Prediction in Lung Cancer Screening. IEEE Access 2018;6:77796-806.
- Chen W, Wang R, Ma Z, Hua Y, Mao D, Wu H, Yang Y, Li C, Li M. A delta-radiomics model for preoperative prediction of invasive lung adenocarcinomas manifesting as radiological part-solid nodules. Front Oncol 2022;12:927974. [Crossref] [PubMed]
- Ma Y, Ma W, Xu X, Cao F. How Does the Delta-Radiomics Better Differentiate Pre-Invasive GGNs From Invasive GGNs? Front Oncol 2020;10:1017. [Crossref] [PubMed]
- Lv Y, Ye J, Yin YL, Ling J, Pan XP. A comparative study for the evaluation of CT-based conventional, radiomic, combined conventional and radiomic, and delta-radiomic features, and the prediction of the invasiveness of lung adenocarcinoma manifesting as ground-glass nodules. Clin Radiol 2022;77:e741-8. [Crossref] [PubMed]
- Lipton ZC. The Mythos of Model Interpretability: In machine learning, the concept of interpretability is both important and slippery. Queue 2018;16:31-57.
- Lundberg S, Lee SI. A Unified Approach to Interpreting Model Predictions. NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems 2017:4768-77.
- Jiang Z, Yin J, Han P, Chen N, Kang Q, Qiu Y, Li Y, Lao Q, Sun M, Yang D, Huang S, Qiu J, Li K. Wavelet transformation can enhance computed tomography texture features: a multicenter radiomics study for grade assessment of COVID-19 pulmonary lesions. Quant Imaging Med Surg 2022;12:4758-70. [Crossref] [PubMed]
- Wang Y, Lang J, Zuo JZ, Dong Y, Hu Z, Xu X, Zhang Y, Wang Q, Yang L, Wong STC, Wang H, Li H. The radiomic-clinical model using the SHAP method for assessing the treatment response of whole-brain radiotherapy: a multicentric study. Eur Radiol 2022;32:8737-47. [Crossref] [PubMed]
- Yang H, Liu H, Lin J, Xiao H, Guo Y, Mei H, Ding Q, Yuan Y, Lai X, Wu K, Wu S. An automatic texture feature analysis framework of renal tumor: surgical, pathological, and molecular evaluation based on multi-phase abdominal CT. Eur Radiol 2024;34:355-66. [Crossref] [PubMed]
- Zhao Y, Wei J, Xiao B, Wang L, Jiang X, Zhu Y, He W. Early prediction of acute pancreatitis severity based on changes in pancreatic and peripancreatic computed tomography radiomics nomogram. Quant Imaging Med Surg 2023;13:1927-36. [Crossref] [PubMed]
- de Moura LV, Mattjie C, Dartora CM, Barros RC, Marques da Silva AM. Explainable Machine Learning for COVID-19 Pneumonia Classification With Texture-Based Features Extraction in Chest Radiography. Front Digit Health 2021;3:662343. [Crossref] [PubMed]
- DeCarlo LT. On the meaning and use of kurtosis. Psychological Methods 1997;2:292-307.
- Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. [Crossref] [PubMed]


