
Automated detection and classification of coronary atherosclerotic plaques on coronary CT angiography using deep learning algorithm
Introduction
Coronary artery disease (CAD) is the leading cause of mortality worldwide (1,2). Accurate diagnosis of CAD presence and severity is pivotal in determining appropriate clinical management for symptomatic patients (3). Traditionally, digital subtraction angiography has served as the gold standard for diagnosing CAD (4). Other coronary plaque imaging methods, such as intravascular ultrasound and optical coherence tomography, have also been employed to identify culprit coronary atherosclerosis (5,6). However, these invasive imaging techniques may entail complications such as arrhythmia, myocardial infarction, or strokes (7,8).
In contrast, coronary computed tomographic angiography (CCTA) is a non-invasive technique capable of accurately detecting obstructive CAD and evaluating atherosclerotic plaque burden in the coronary artery (7,9-13). Despite its reliability as a diagnostic tool, utilizing CCTA for coronary artery assessment remains time-consuming and requires extensive expertise (14,15).
Recent advancements in deep learning technology have shown promising results in enhancing the efficiency of CAD plaque assessment (14,16-24). For the optical coherence tomography images, Liu et al. proposed a framework which automatically segmented lumen and calcified plaque, by leveraging deep learning techniques (25). For CCTA images, a significant contribution to this field is the development of a deep learning-based technology that leverages an enhanced three-dimensional (3D) U-Net architecture. This advanced version of the 3D U-Net incorporates a bottle-neck design for fully automated segmentation, alongside a connected growth prediction model (CGPM) for the repair of segmentation fractures (3,26). Crucial to its design, this architecture is augmented with attention mechanisms and residual modules, which are not explicitly mentioned but integral to its operation. The attention mechanisms enable focused analysis of salient image features critical for the accurate identification of obstructive CAD, while the residual modules facilitate the training of deeper networks by alleviating the vanishing gradient problem, thus enhancing the model’s ability to learn complex patterns in the data. This nuanced architecture has demonstrated the capability to accurately and effectively identify obstructive CAD (21), showcasing the potential of deep learning in revolutionizing CAD plaque assessment. However, the widespread application of this technology necessitates rigorous validation of its diagnostic efficiency. Moreover, it is imperative to benchmark the accuracy and reliability of this deep learning approach against the diagnostic performance of physicians at various levels of expertise.
This study has a dual purpose: firstly, to determine the capability of deep learning technology in detecting coronary plaques by comparing it with physicians at varying diagnostic levels; secondly, to assess its ability to distinguish between different types of plaques. We present this article in accordance with the STARD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1513/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Nanjing Drum Tower Hospital Clinical College of Nanjing University of Chinese Medicine (No. 2022-547-01), and individual consent for this retrospective analysis was waived. Between January 2019 and September 2020, CCTA images of 773 consecutive patients with suspected CAD from Nanjing Drum Tower Hospital Clinical College of Nanjing University of Chinese Medicine were collected. First, of 773 patients, 38 patients with poor quality CCTA images were excluded. Second, 59 patients with a history of coronary stenting or coronary artery bypass grafting were excluded. Thirdly, 7 patients with severe cardiac or coronary anatomical variants were excluded. Finally, CCTA images from a total of 669 patients were included in this study (Figure 1). These patients were divided into two groups to evaluate the performance of the deep learning algorithm: Group 1, consisting of 106 patients, for plaque detection, and Group 2, comprising 563 patients, for plaque type characterization.
In Group 1, the algorithm’s results were compared with assessments made by physicians with varying levels of expertise: a cardiovascular research radiologist (K.Y., <2 years of clinical experience), a junior cardiovascular radiologist (W.C., 3 years of clinical experience), an intervention cardiologist (R.G., 5 years of clinical experience), and a senior cardiovascular radiologist (H.Y., 5 years of clinical experience). Diagnostic outcomes from two expert cardiovascular radiologists (H.L. and D.M., >10 years of clinical experience) were considered the gold standard for comparison. Any disparities were resolved through a consensus process among them. Moreover, these two expert radiologists interpreted CCTA images from the remaining 563 patients (Group 2), categorizing plaques as absent, calcified, non-calcified, or mixed calcified per segment, following Society of Cardiovascular Computed Tomography (SCCT) guidelines (27,28) (see Figure 2). The algorithm’s outputs were then compared to these expert interpretations. All physicians were blind to the patients’ clinical information.
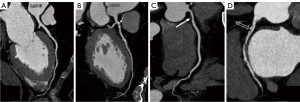
Imaging acquisition and analysis
All CCTA scans were conducted using a 256-slice computed tomography (CT) scanner (Brilliance iCT, Philips Healthcare, Cleveland, OH, USA) using electrocardiography (ECG)-gated prospective-triggered technology. The following acquisition parameters were applied: a tube voltage of 100 kVp, detector collimation of 128×0.625 mm, a rotation time of 0.27 seconds and automatic tube current modulation enabled (DoseRight with index setting of 16, Philips Healthcare). A total of 65 mL of iopromide with an iodine concentration of 370 mg/mL (Ultravist, Bayer, Guangzhou, China) was intravenously administered at 4–5 mL/s, followed by 50 mL of normal saline at same injection speed. The bolus tracking technique was employed, with a delay of 7 seconds after reaching a trigger threshold of 130 Hounsfield unit (HU) in the proximal descending aorta.
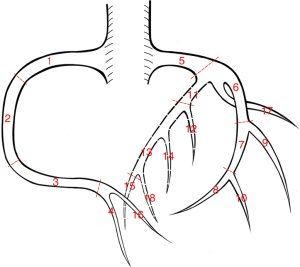
As per SCCT guidelines (27,28), atherosclerotic plaque analysis in the coronary artery segments was performed using a dedicated workstation (IntelliSpace Portal, Philips Healthcare). The evaluation covered 18 coronary artery segments: proximal right coronary artery (pRCA, 1), middle right coronary artery (mRCA, 2), distal right coronary artery (dRCA, 3), right posterior descending artery (R-PDA, 4), left main (LM, 5), proximal left anterior descending (pLAD, 6), middle left anterior descending (mLAD, 7), distal left anterior descending (dLAD, 8), diagonal 1 (D1, 9), diagonal 2 (D2, 10), proximal circumflex (pCx, 11), obtuse marginal 1 (OM1, 12), mid and distal left circumflex (LCx, 13), obtuse marginal 2 (OM2, 14), posterior descending artery from LCx (L-PDA, 15), right posterior lateral branch (R-PLB, 16), ramus intermedius (RI, 17), and posterior lateral branch from LCx (L-PLB, 18) (Figure 3).
The interpretation of CCTA images involved the following steps:
- Image loading and review: loading the CCTA image series onto the workstation for reviewing the original axial images to assess quality and identify anatomical landmarks.
- Heart segmentation stage: manual correction or re-segmentation, if required.
- Coronary extraction: verification, correction, or manual definition of coronary artery segmentation, vessel centerlines, and their labels. A straightened multi-planar reconstruction (MPR) image of the selected artery was provided.
- Coronary analysis: including plaque detection, diameter and area measurements of stenosis, and selection of reference locations. Volume-rendered images, curved MPR images, straightened MPR images, and cross-section images of vessels were provided for analysis.
- Plaque analysis: calculation and display of plaque content as the percent (%) of calcified plaque, mixed plaque, and non-calcified plaque based on pre-defined HU thresholds. The interpretation time was recorded from image loading to plaque detection or plaque analysis.
Deep learning algorithm
A deep learning-based system (Coronary Doc, ShuKun Technology, Beijing, China) was employed for coronary artery segmentation and plaque detection. This system primarily utilized 3D U-Net1 architecture with a bottle-neck design for fully automated segmentation and a CGPM for segmentation fracture repair (26,29). After transferring the original axial images to the system, various image reconstructions, such as multiple planar reformat, straightened rendering, curve planar reformat, maximum intensity projection, and volume rendering images, were created based on the segmentation.
Further enhancing the 3D U-Net1 architecture, a dedicated 3D U-Net model (U-Net2) was trained specifically for recognizing plaques within the coronary arteries. Data cubes sized 60×60 (within the cross-sectional plane) ×128 (along the centerline) were extracted from the straightened arteries and input into the model. This model consisted of a five-layer encoding structure, a five-layer decoding structure, and a max-pooling output layer. It underwent 100 epochs of training using stochastic gradient descent as the optimizer, starting with a learning rate of 0.01 and halving it every 20 epochs. The output layer of U-Net2 generated two probability curves, each 128 pixels in length, corresponding to calcified and non-calcified (soft) plaques (16,18,21,22,26,30,31) (Figure 4). In this context, a calcified plaque referred to a plaque with a CT value of ≥130 HU, commonly used to quantify calcification burden in clinical trials and identify calcified plaques in CCTA images. Conversely, a non-calcified plaque was defined by a CT value of <130 HU, while a mixed plaque denoted the overlap between calcified and non-calcified plaque positions. Plaque locations were determined by areas in the probability curve exceeding the cut-off value.
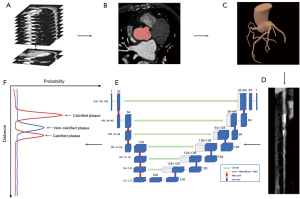
This deep learning-based system was integrated into the picture archiving and communication system (PACS), automatically analyzing coronary axial computed tomographic angiography (CTA) images transferred to PACS. It provided preliminary results, including comprehensive 3D reconstructions for curved plane reconstruction (CPR), lumen, and multi-cross sections, along with location identification, qualitative and quantitative plaque analyses. The interpretation time using this system involved opening the initial report, reviewing reconstructed images, and confirming the final report.
Statistical analysis
Statistical analysis was conducted to evaluate the deep learning model’s performance using R (version 4.1.1, R Foundation for Statistical Computing, Vienna). Sensitivity, specificity, positive predictive value (PPV), negative predictive values (NPV), accuracy, and Cohen’s kappa were calculated. Cohen’s kappa>0.61 was considered to have strong consistency. Categorical variables were presented as counts or percentages, while continuous variables were described as means with standard deviations (SD). McNemar’s test was utilized for continuous, two-sided, paired variables, with a two-tailed P value <0.05 considered significant.
Results
The 669 patients were divided into two groups to evaluate the performance of the deep learning algorithm: Group 1, consisting of 106 patients, for plaque detection, and Group 2, comprising 563 patients, for plaque type characterization. Patient demographics are summarized in Table 1.
Table 1
| Demographics | Group 1 | Group 2 | |||
|---|---|---|---|---|---|
| All | Values† | All | Values† | ||
| No. of patients | 106 | 563 | |||
| Female | 39 (36.8) | 225 (40.0) | |||
| Age, years | 63±10 | 61±12 | |||
| Indication | |||||
| Chest pain | 19 (17.9) | 139 (24.7) | |||
| Chest tightness | 31 (29.2) | 163 (29.0) | |||
| CAD | 28 (26.4) | 155 (27.5) | |||
| Lower extremity edema | 6 (5.7) | 18 (3.2) | |||
| Shortness of breath | 10 (9.4) | 14 (2.5) | |||
| Others | 12 (11.3) | 74 (13.1) | |||
| History of CAD | 70 | 52 (74.3) | 400 | 330 (82.5) | |
| BMI, kg/m2 | 72 | 374 | |||
| <23 (normal) | 25 (34.7) | 85 (22.7) | |||
| 23.00–27.49 (overweight) | 21 (29.2) | 122 (32.6) | |||
| ≥27.5 (obesity) | 26 (36.1) | 167 (44.7) | |||
| Ever smoke (former and current) | 53 | 14 (26.4) | 271 | 94 (34.7) | |
| Diabetes | 93 | 33 (35.5) | 292 | 106 (36.3) | |
| Hypertension | 96 | 67 (69.8) | 364 | 271 (74.5) | |
| Hyperlipidemia | 83 | 23 (27.7) | 270 | 90 (33.3) | |
| Cardiovascular events | 72 | 56 (77.8) | 249 | 190 (76.3) | |
†, data are presented as mean ± SD or n (%). The indication “CAD” represents the suspicious diagnosis made by the doctor during the outpatient visit, based on the patient’s symptoms, laboratory tests, and other relevant factors. A history of CAD refers to a patient who has previously been diagnosed with CAD. Group 1: for plaque detection; Group 2: for plaque type characterization. CAD, coronary artery disease; SD, standard deviation; BMI, body mass index.
Coronary plaque detection efficiency of deep learning compared to physicians
The average sensitivity, specificity, PPV, NPV, and accuracy of the deep learning system were 92% [95% confidence interval (CI): 89.5–94.1%], 87% (95% CI: 84.2–88.5%), 79% (95% CI: 76.1–82.4%), 95% (95% CI: 93.4–96.3%), and 89% (95% CI: 86.9–90.0%), respectively. Comparative data for cardiovascular radiologists with varying experience levels were as follows: those with 5 years of clinical experience achieved an average sensitivity of 82% (95% CI: 78.3–84.7%), specificity of 86% (95% CI: 84.0–88.3%), PPV of 77% (95% CI: 73.6–80.4%), NPV of 89% (95% CI: 87.2–89.3%), and accuracy of 85% (95% CI: 82.8–86.4%); those with 3 years of clinical experience had an average sensitivity of 80%(95% CI: 76.4–83.2%), specificity of 82% (95% CI: 79.7–84.6%), PPV of 72% (68.1–75.2%), NPV of 88% (95% CI: 85.6–89.9%), and accuracy of 81% (95% CI: 79.5–83.3%); while the research cardiovascular radiologist with less than 2 years of clinical experience achieved an average sensitivity of 72% (95% CI: 68.4–75.9%), specificity of 79% (95% CI: 76.9%-81.9%), PPV of 67% (95% CI: 62.7–70.2%), NPV of 84% (95% CI: 81.0–85.8%), and accuracy of 77% (95% CI: 74.8–78.9%). The intervention cardiologist with five years of clinical experience reached an average sensitivity of 78% (95% CI: 73.1–80.2%), specificity of 86% (95% CI: 83.8–88.2%), PPV of 76% (95% CI: 72.0–79.2%), NPV of 87% (95% CI: 84.5–88.8%), and accuracy of 83% (95% CI: 80.8–84.6%). For detailed results, refer to Table 2.
Table 2
| Segments | Total | Ref. standard (2 radiologists with more than 10 years of clinical experience) |
Deep learning | Clinical experience | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiologist with 5 years |
Radiologist with 3 years |
Radiologist with ≤2 years |
Intervention cardiologist with 5 years |
|||||||||||||||||||||||||
| Positive | Negative | TP | TN | FP | FN | TP | TN | FP | FN | TP | TN | FP | FN | TP | TN | FP | FN | TP | TN | FP | FN | |||||||
| pRCA | 106 | 56 | 50 | 54 | 47 | 3 | 2 | 49 | 46 | 4 | 7 | 49 | 43 | 7 | 7 | 50 | 42 | 8 | 6 | 44 | 44 | 6 | 12 | |||||
| mRCA | 106 | 45 | 61 | 40 | 46 | 15 | 5 | 35 | 53 | 8 | 10 | 35 | 48 | 13 | 10 | 42 | 54 | 7 | 3 | 37 | 51 | 10 | 8 | |||||
| dRCA | 97 | 19 | 78 | 16 | 59 | 19 | 3 | 13 | 61 | 17 | 6 | 15 | 62 | 16 | 4 | 16 | 74 | 4 | 3 | 16 | 77 | 1 | 3 | |||||
| R-PDA | 95 | 32 | 63 | 29 | 62 | 1 | 3 | 27 | 61 | 2 | 5 | 24 | 56 | 7 | 8 | 22 | 50 | 13 | 10 | 25 | 59 | 4 | 7 | |||||
| LM | 106 | 41 | 65 | 40 | 49 | 16 | 1 | 28 | 56 | 9 | 13 | 31 | 55 | 10 | 10 | 20 | 43 | 22 | 21 | 32 | 59 | 6 | 9 | |||||
| pLAD | 106 | 67 | 39 | 62 | 38 | 1 | 5 | 64 | 35 | 4 | 3 | 58 | 34 | 5 | 9 | 63 | 37 | 2 | 4 | 58 | 28 | 11 | 9 | |||||
| mLAD | 106 | 55 | 51 | 52 | 36 | 15 | 3 | 47 | 38 | 13 | 8 | 46 | 44 | 7 | 9 | 51 | 44 | 7 | 4 | 49 | 47 | 4 | 6 | |||||
| dLAD | 106 | 12 | 94 | 10 | 81 | 13 | 2 | 7 | 80 | 14 | 5 | 7 | 59 | 35 | 5 | 2 | 82 | 12 | 10 | 5 | 84 | 10 | 7 | |||||
| D1 | 106 | 35 | 71 | 32 | 58 | 13 | 3 | 23 | 60 | 11 | 12 | 27 | 62 | 9 | 8 | 14 | 42 | 29 | 21 | 27 | 55 | 16 | 8 | |||||
| D2 | 93 | 32 | 61 | 29 | 55 | 6 | 3 | 23 | 47 | 14 | 9 | 22 | 47 | 14 | 10 | 15 | 41 | 20 | 17 | 21 | 50 | 11 | 11 | |||||
| pCx | 106 | 56 | 50 | 53 | 48 | 2 | 3 | 56 | 48 | 2 | 0 | 50 | 42 | 8 | 6 | 39 | 35 | 15 | 17 | 48 | 39 | 11 | 8 | |||||
| OM1 | 101 | 27 | 74 | 26 | 72 | 2 | 1 | 25 | 64 | 10 | 2 | 23 | 60 | 14 | 4 | 15 | 54 | 20 | 12 | 21 | 67 | 7 | 6 | |||||
| LCx | 106 | 27 | 79 | 25 | 59 | 20 | 2 | 11 | 68 | 11 | 16 | 17 | 66 | 13 | 10 | 20 | 66 | 13 | 7 | 19 | 73 | 6 | 8 | |||||
| OM2 | 78 | 12 | 66 | 12 | 63 | 3 | 0 | 10 | 60 | 6 | 2 | 7 | 59 | 7 | 5 | 6 | 56 | 10 | 6 | 5 | 56 | 10 | 7 | |||||
| L-PDA | 11 | 5 | 6 | 4 | 4 | 2 | 1 | 4 | 3 | 3 | 1 | 3 | 3 | 3 | 2 | 2 | 4 | 2 | 3 | 1 | 3 | 3 | 4 | |||||
| R-PLB | 81 | 26 | 55 | 23 | 55 | 0 | 3 | 23 | 50 | 5 | 3 | 22 | 51 | 4 | 4 | 19 | 44 | 11 | 7 | 18 | 45 | 10 | 8 | |||||
| RI | 45 | 11 | 34 | 9 | 32 | 2 | 2 | 10 | 29 | 5 | 1 | 10 | 28 | 6 | 1 | 6 | 24 | 10 | 5 | 5 | 24 | 10 | 6 | |||||
| L-PLB | 25 | 12 | 13 | 9 | 10 | 3 | 3 | 11 | 13 | 0 | 1 | 10 | 12 | 1 | 2 | 10 | 11 | 2 | 2 | 7 | 9 | 4 | 5 | |||||
| Sensitivity, % | 92 (89.5–94.1) | 82 (78.3–84.7) | 80 (76.4–83.2) | 72 (68.4–75.9) | 78 (73.1–80.2) | |||||||||||||||||||||||
| Specificity, % | 87 (84.2–88.5) | 86 (84.0–88.3) | 82 (79.7–84.6) | 79 (76.9–81.9) | 86 (83.8–88.2) | |||||||||||||||||||||||
| PPV, % | 79 (76.1–82.4) | 77 (73.6–80.4) | 72 (68.1–75.2) | 67 (62.7–70.2) | 76 (72.0–79.2) | |||||||||||||||||||||||
| NPV, % | 95 (93.4–96.3) | 89 (89.3–87.2) | 88 (85.6–89.9) | 84 (81.0–85.8) | 87 (84.5–88.8) | |||||||||||||||||||||||
| Accuracy, % | 89 (86.9–90.0) | 85 (82.8–86.4) | 81 (79.5 –83.3) | 77 (74.8–78.9) | 83 (80.8–84.6) | |||||||||||||||||||||||
Data in parentheses are 95% confidence interval. CCTA, coronary computed tomographic angiography; TP, true positive; TN, true negative; FP, false positive; FN, false negative; PPV, positive predictive value; NPV, negative predictive value; pRCA, proximal right coronary artery; mRCA, middle right coronary artery; dRCA, distal right coronary artery; R-PDA, right posterior descending artery; LM, left main; pLAD, proximal left anterior descending; mLAD, middle left anterior descending; dLAD, distal left anterior descending; D1, diagonal 1; D2, diagonal 2; pCx, proximal circumflex; OM1, obtuse marginal 1; LCx, mid and distal left circumflex; OM2, obtuse marginal 2; L-PDA, posterior descending artery from LCx; R-PLB, right posterior lateral branch; RI, ramus intermedius; L-PLB, posterior lateral branch from LCx.
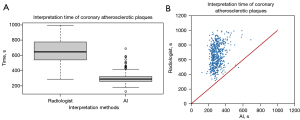
Deep learning time efficiency on plaque classification
When radiologists classified plaques per patient, interpretation time ranged from 280 to 995 s, with a mean of 658±150 s. In contrast, using the deep learning model, interpretation time ranged from 125 to 689 s, with a mean of 298±60 s. Significantly shorter interpretation time was observed with deep learning compared to radiologists (McNemar’s test, P<0.001) (Figure 5).
Deep learning performance on plaque classification
Among 563 patients, there were 1,233 calcified plaques, 416 non-calcified plaques, and 130 mixed plaques. The overall sensitivity, specificity, PPV, NPV, and accuracy were 94% (95% CI: 92.3–94.7%), 90% (95% CI: 88.8–90.3%), 70% (95% CI: 68.3–72.1%), 98% (95% CI: 97.8–98.5%), and 90% (95% CI: 89.8–91.1%), respectively, demonstrating substantial agreement (κ=0.74, 95% CI: 0.70–0.78) between the deep learning model and two expert cardiovascular radiologists in coronary atherosclerosis classification (Table 3).
Table 3
| Segments | Total | Ref. standard (2 radiologists with more than 10 years of clinical experience) | Deep learning | Sensitivity (95% CI), % |
Specificity (95% CI), % |
PPV (95% CI), % |
NPV (95% CI),% |
Accuracy (95% CI), % |
κ (95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-plaque | Calcified plaques | Non-calcified plaques | Mixed plaques | TP | TN | FP | FN | |||||||||
| pRCA | 563 | 364 | 113 | 66 | 20 | 179 | 299 | 75 | 10 | 95 (90.2–97.3) | 80 (75.5–83.8) | 70 (64.4–75.9) | 97 (93.9–98.3) | 85 (81.7– 87.6) | 0.74 (0.69–0.79) | |
| mRCA | 563 | 377 | 129 | 49 | 8 | 168 | 291 | 97 | 7 | 96 (91.6–98.2) | 75 (70.3–79.2) | 63 (57.2–69.1) | 97 (95.0–98.9) | 82 (78.1– 84.5) | 0.68 (0.62–0.73) | |
| dRCA | 526 | 412 | 75 | 35 | 4 | 108 | 356 | 60 | 2 | 98 (92.9–99.7) | 86 (81.7–88.7) | 64 (56.5–71.4) | 99 (97.8–99.9) | 88 (85.2–90.7) | 0.73 (0.66–0.79) | |
| R-PDA | 485 | 445 | 32 | 6 | 2 | 40 | 407 | 38 | 0 | 100 (89.1–100) | 91 (88.4–93.8) | 51 (39.8–62.7) | 100 (98.9–100) | 92 (89.4–94.3) | 0.65 (0.55–0.75) | |
| LM | 563 | 430 | 108 | 19 | 6 | 122 | 412 | 22 | 7 | 94 (88.7–97.6) | 95 (92.3–96.7) | 85 (77.6–89.9) | 98 (96.4–99.3) | 95 (92.7–96.4) | 0.87 (0.82–0.91) | |
| pLAD | 563 | 291 | 157 | 73 | 42 | 220 | 258 | 54 | 31 | 88 (82.8–91.3) | 83 (77.9–86.6) | 80 (75.0–84.7) | 89 (85.0–92.5) | 85 (81.7–87.6) | 0.76 (0.71–0.81) | |
| mLAD | 563 | 311 | 176 | 62 | 14 | 222 | 250 | 72 | 19 | 92 (87.8–95.1) | 78 (72.6–81.9) | 75 (70.1–80.2) | 93 (89.0–95.5) | 84 (80.6–86.7) | 0.74 (0.69–0.79) | |
| dLAD | 563 | 540 | 18 | 5 | 0 | 21 | 507 | 33 | 2 | 91 (70.5–98.5) | 94 (91.4–95.7) | 39 (26.2–53.1) | 99 (98.4–99.9) | 94 (91.5–95.5) | 0.52 (0.39–0.66) | |
| D1 | 554 | 475 | 65 | 13 | 1 | 76 | 442 | 33 | 3 | 96 (88.5–99) | 93 (90.3–95.1) | 70 (60.1–77.9) | 99 (97.9–99.9) | 93 (91.1–95.3) | 0.79 (0.71–0.85) | |
| D2 | 399 | 354 | 35 | 10 | 0 | 44 | 342 | 12 | 1 | 98 (86.8–99.9) | 97 (94.0–98.2) | 79 (65.2–88.0) | 99 (98.1–99.9) | 97 (94.5–98.1) | 0.86 (0.78–0.93) | |
| pCx | 563 | 425 | 113 | 17 | 8 | 124 | 384 | 44 | 11 | 92 (85.6–95.7) | 90 (86.4–92.3) | 74 (66.4–80.1) | 97 (94.9–98.5) | 90 (87.5–92.4) | 0.77 (0.71–0.83) | |
| OM1 | 493 | 449 | 37 | 7 | 0 | 44 | 413 | 36 | 0 | 100 (90.0–100) | 92 (90.0–94.2) | 55 (43.5–66.0) | 100 (98.9–100) | 93 (90.0–94.7) | 0.69 (0.58–0.78) | |
| LCx | 563 | 438 | 100 | 21 | 4 | 118 | 397 | 44 | 4 | 97 (91.3–98.9) | 90 (86.7–92.6) | 73 (65.2–79.3) | 99 (97.3–99.7) | 91 (88.9–93.5) | 0.79 (0.73–0.85) | |
| OM2 | 303 | 280 | 19 | 4 | 0 | 23 | 264 | 16 | 0 | 100 (82.2–100) | 94 (90.7–96.6) | 59 (42.2–74.2) | 100 (98.2–100) | 95 (91.5–96.8) | 0.72 (0.59–0.85) | |
| L-PDA | 78 | 64 | 6 | 3 | 5 | 12 | 59 | 5 | 2 | 86 (56.2–97.5) | 92 (82.0–97.1) | 70 (44.0–88.6) | 97 (87.6–99.4) | 91 (82.3–95.9) | 0.74 (0.56–0.92) | |
| R-PLB | 462 | 422 | 18 | 15 | 7 | 30 | 417 | 10 | 5 | 86 (69.0–94.6) | 98 (95.6–98.8) | 75 (58.5–86.8) | 99 (97–99.6) | 97 (94.7–98.1) | 0.79 (0.61–0.97) | |
| RI | 258 | 229 | 25 | 4 | 0 | 28 | 221 | 8 | 1 | 96 (80.4–99.8) | 96 (93.0–98.4) | 78 (60.4–89.3) | 99 (97.1–99.9) | 97 (93.4–98.3) | 0.85 (0.75–0.94) | |
| L-PLB | 101 | 78 | 7 | 7 | 9 | 14 | 72 | 12 | 3 | 82 (55.8–95.3) | 86 (76–92.1) | 54 (33.7–72.9) | 96 (87.9–99.0) | 85 (76.8–90.9) | 0.63 (0.48–0.79) | |
The mean sensitivity, specificity, PPV, NPV, and accuracy are 94% (95% CI: 92.3–94.7%) 90% (95% CI: 88.8–90.3%), 70% (95% CI: 68.3–72.1%), 98% (95% CI: 97.8–98.5%), and 90% (95% CI: 89.8–91.1%). Overall, there is substantial agreement between deep learning and the radiologists in the identification of the coronary atherosclerotic plaques (κ=0.74, 95% CI: 0.70–0.78). CCTA, coronary computed tomographic angiography; TP, true positive; TN, true negative; FP, false positive; FN, false negative; PPV, positive predictive value; NPV, negative predictive value; CI, confidence interval; pRCA, proximal right coronary artery; mRCA, middle right coronary artery; dRCA, distal right coronary artery; R-PDA, right posterior descending artery; LM, left main; pLAD, proximal left anterior descending; mLAD, middle left anterior descending; dLAD, distal left anterior descending; D1, diagonal 1;D2, diagonal 2; pCx, proximal circumflex; OM1, obtuse marginal 1; LCx, mid and distal left circumflex; OM2, obtuse marginal 2; L-PDA, posterior descending artery from LCx; R-PLB, right posterior lateral branch; RI, ramus intermedius; L-PLB, posterior lateral branch from LCx.
For calcified plaques, sensitivity, specificity, PPV, NPV, and accuracy were 93% (95% CI: 91.9–94.7%), 96% (95% CI: 95.6–96.5%), 81% (95% CI: 78.6–82.8%), 99% (95% CI: 98.5–99.0%) and 96% (95% CI: 95.2–96.1%), respectively (Table 4). Lowest PPV, particularly in the dLAD and R-PLB areas, was the primary factor contributing to the overall lower PPV in characterizing calcified plaques with deep learning. In characterizing non-calcified plaques, mean sensitivity, specificity, PPV, NPV, and accuracy were 84% (95% CI: 79.7–87.0%), 95% (95% CI: 94.7–95.7%), 48% (95% CI: 44.6–52.1%), 99% (95% CI: 98.8–99.3%) and 95% (95% CI: 94.1–95.1%), respectively (Table 4). Similarly, the lowest PPV in the dLAD and R-PDA areas contributed to the lower average PPV in characterizing non-calcified plaques. Concerning mixed plaques, mean sensitivity, specificity, PPV, NPV, and accuracy were 72% (95% CI: 62.8–78.9%), 99% (95% CI: 99.4–99.7%), 79% (95% CI: 70.1–85.6%), 99% (95% CI: 99.2–99.6%), 81% (95% CI: 79.7–81.4%), respectively (Table 4).
Table 4
| Segments | Total | Coronary calcified plaques | Coronary non-calcified plaques | Coronary mixed plaques | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | TP | TN | FP | FN | TP | TN | FP | FN | ||||
| pRCA | 563 | 105 | 435 | 15 | 8 | 57 | 438 | 59 | 9 | 17 | 542 | 1 | 3 | ||
| mRCA | 563 | 122 | 414 | 20 | 7 | 40 | 442 | 72 | 9 | 6 | 550 | 5 | 2 | ||
| dRCA | 526 | 74 | 421 | 30 | 1 | 32 | 462 | 29 | 3 | 2 | 521 | 1 | 2 | ||
| R-PDA | 485 | 32 | 436 | 17 | 0 | 6 | 459 | 20 | 0 | 2 | 482 | 1 | 0 | ||
| LM | 563 | 101 | 440 | 15 | 7 | 17 | 537 | 7 | 2 | 4 | 557 | 0 | 2 | ||
| pLAD | 563 | 141 | 383 | 23 | 16 | 53 | 463 | 27 | 20 | 26 | 517 | 4 | 16 | ||
| mLAD | 563 | 162 | 365 | 22 | 14 | 49 | 453 | 48 | 13 | 11 | 547 | 2 | 3 | ||
| dLAD | 563 | 17 | 523 | 22 | 1 | 4 | 547 | 11 | 1 | – | – | – | – | ||
| D1 | 554 | 62 | 471 | 18 | 3 | 13 | 527 | 14 | 0 | 1 | 552 | 1 | 0 | ||
| D2 | 399 | 34 | 356 | 8 | 1 | 10 | 385 | 4 | 0 | – | – | – | – | ||
| pCx | 563 | 104 | 435 | 15 | 9 | 14 | 519 | 27 | 3 | 6 | 553 | 2 | 2 | ||
| OM1 | 493 | 37 | 435 | 21 | 0 | 7 | 472 | 14 | 0 | 0 | 492 | 1 | 0 | ||
| LCx | 563 | 95 | 441 | 22 | 5 | 19 | 522 | 20 | 2 | 4 | 557 | 2 | 0 | ||
| OM2 | 303 | 19 | 273 | 11 | 0 | 4 | 294 | 5 | 0 | – | – | – | – | ||
| L-PDA | 78 | 5 | 70 | 2 | 1 | 3 | 73 | 2 | 0 | 4 | 72 | 1 | 1 | ||
| R-PLB | 462 | 14 | 436 | 8 | 4 | 10 | 445 | 2 | 5 | 6 | 455 | 0 | 1 | ||
| RI | 258 | 24 | 231 | 2 | 1 | 4 | 248 | 6 | 0 | – | – | – | – | ||
| L-PLB | 101 | 4 | 91 | 3 | 3 | 6 | 89 | 5 | 1 | 4 | 88 | 4 | 5 | ||
| Sensitivity (95% CI), % | 93 (91.9–94.7) | 84 (79.7–87.0) | 72 (62.8–78.9) | ||||||||||||
| Specificity (95% CI), % | 96 (95.6–96.5) | 95 (94.7–95.7) | 99 (99.4–99.7) | ||||||||||||
| PPV (95% CI), % | 81 (78.6–82.8) | 48 (44.6–52.1) | 79 (70.1–85.6) | ||||||||||||
| NPV (95% CI), % | 99 (98.5–99.0) | 99 (98.8–99.3) | 99 (99.2–99.6) | ||||||||||||
| Accuracy (95% CI), % | 96 (95.2–96.1) | 95 (94.1–95.1) | 81 (79.7–81.4) | ||||||||||||
CCTA, coronary computed tomographic angiography; TP, true positive; TN, true negative; FP, false positive; FN, false negative; CI, confidence interval; pRCA, proximal right coronary artery; mRCA, middle right coronary artery; dRCA, distal right coronary artery; R-PDA, right posterior descending artery; LM, left main; pLAD, proximal left anterior descending; mLAD, middle left anterior descending; dLAD, distal left anterior descending; D1, diagonal 1;D2, diagonal 2; pCx, proximal circumflex; OM1, obtuse marginal 1; LCx, mid and distal left circumflex; OM2, obtuse marginal 2; L-PDA, posterior descending artery from LCx; R-PLB, right posterior lateral branch; RI, ramus intermedius; L-PLB, posterior lateral branch from LCx; PPV, positive predictive value; NPV, negative predictive value.
Discussion
In our study, the employed deep learning-based system (Coronary Doc, ShuKun Technology, Beijing, China) offers multiple advantages over conventional algorithms. It utilizes a 3D U-Net1 architecture, providing a more comprehensive spatial context for accurate plaque detection compared to existing 2D models (21,29). Additionally, it significantly reduces subjectivity and time consumption associated with manual or semi-automatic plaque detection methods, thus enhancing efficiency and reproducibility. It has been tested on an NVIDIA GeForce GTX 1080 Ti graphics card with which it demonstrates the ability to complete the diagnosis process for CAD within 120 seconds. This timeframe includes the full range of computations, from initial image processing to the final output. Using more advanced graphics cards can further reduce this computation time, indicating that the model’s efficiency improves with better hardware. This scalability suggests the approach is not only effective with current technology but will also benefit from future advancements in graphics processing unit (GPU) capabilities. Furthermore, the integration of a CGPM for segmentation fracture repair notably improved plaque detection and segmentation (21,22). The dedicated 3D U-Net model (U-Net2) specifically trained for coronary arteries offers enhanced precision in plaque detection, providing a more detailed view of plaque morphology, composition, and distribution.
For deep learning-based coronary plaque detection models, the most basic function is to identify plaques as well as stenosis in the vessel lumen. Only when this basic function achieves a high degree of stability, the accurate classification of plaques may be further realized. In the first part of this study, the deep learning-based model demonstrated the significantly enhanced efficiency in coronary artery plaque detection when compared to radiologists or an intervention cardiologist with up to 5 years of clinical experience. In the second part of this study, the model exhibited high consistency in classifying coronary atherosclerotic plaques compared to expert radiologists while achieving this in a notably shorter interpretation time. Furthermore, our analysis revealed not only good sensitivity and accuracy but also excellent NPV in diagnosing coronary atherosclerotic plaques. This is particularly valuable in excluding culprit atherosclerotic plaques.
The implications of our study are significant. Firstly, in daily practice, prompt diagnosis and appropriate treatment of coronary atherosclerosis are crucial to prevent myocardial infarction and sudden death. In densely populated countries like China, efficiently identifying individuals at risk of CAD and providing effective treatment to those with the disease are imperative. CCTA, more accurately detecting CAD (32), holds clinical significance in identifying high-risk CAD patients using efficient deep learning techniques. Moreover, for CAD patients, quickly determining the location and hemodynamic status of culprit blood vessels may allow cardiovascular physicians sufficient time for precise treatment. Secondly, variations in CAD therapy among hospitals or physicians may lead to different outcomes for CAD patients. Therefore, accurate and precise CAD identification is crucial. The deep learning technology in our study holds the potential to minimize disparities and enhance the precision of CAD treatment among different medical institutions. Thirdly, leveraging the capabilities of deep learning to distinguish plaque types and its excellent negative prediction ability may help avoid unnecessary standardized treatments for some CAD patients. Additionally, the extensive use of deep learning could become an indispensable tool for training physicians in CAD diagnosis and treatment. The efficacy of our study's deep learning for diagnostics suggests that it can provide relatively reliable results and potentially help physicians of different expertise levels shorten the time required for CAD diagnosis training.
There are limitations in this study. Firstly, as a retrospective study, there might be data insufficiencies, leading to the exclusion of some clinically significant indicators from the analysis. Secondly, we did not evaluate whether deep learning can effectively assess the degree of coronary artery stenosis or analyze plaque burden and stability, both crucial CAD predictors. Additionally, the single-hospital dataset suggests a need for a larger, multi-center study to validate and expand our findings. Moreover, while our model has shown promising results, we acknowledge using a dataset with a high plaque prevalence might influence the model’s performance, potentially creating bias towards identifying plaques. Validation of our algorithm on datasets with varied plaque prevalence will ensure its robustness and generalizability for broader clinical use. Finally, the study also highlighted a lower PPV in the identification of coronary artery plaques, primarily attributed to the lower PPV in identifying calcified plaques in the dLAD and R-PLB, and non-calcified plaques in the dLAD and R-PDA. To improve the detection of lipid-rich and calcified plaques in these smaller branches, further training of this automated system with a larger number of plaques is warranted. Such plaque characterization may assist in identifying vulnerable plaques (33) and categorizing associated risks (34,35).
Conclusions
In conclusion, the deep learning algorithm demonstrates high efficacy in detecting coronary plaques, comparable to expert radiologists and superior to junior radiologists or interventional cardiologists. Moreover, the automated classification of coronary plaques using deep learning is time-saving and reliable, potentially aiding in triaging patients with acute coronary symptoms. This study lays the foundation for further development and utilization of deep learning in cardiac diagnostics and treatment, offering promising avenues for improving patient care.
Acknowledgments
The authors thank all the study investigators, clinicians, nurses, and technicians for dedicating their time and skills to the completion of this study.
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-1513/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1513/coif). Z.S. and X.C. are employees of Philips Healthcare. C.Z. is an employee of Shukun (Beijing) Network Technology. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Nanjing Drum Tower Hospital Clinical College of Nanjing University of Chinese Medicine (No. 2022-547-01), and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zhou M, Wang H, Zeng X, Yin P, Zhu J, Chen W, et al. Mortality, morbidity, and risk factors in China and its provinces, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 2019;394:1145-58. [Crossref] [PubMed]
- Ralapanawa U, Sivakanesan R. Epidemiology and the Magnitude of Coronary Artery Disease and Acute Coronary Syndrome: A Narrative Review. J Epidemiol Glob Health 2021;11:169-77. [Crossref] [PubMed]
- Cappelletti A, Latib A, Mazzavillani M, Magni V, Calori G, Colombo A, Margonato A. Coronary Artery risk factors Profile and Prognostic localization (CAPP) Study. Severity and prognostic localization of critical coronary artery stenoses: correlation with clinical control of major traditional risk factors. Coron Artery Dis 2012;23:455-9. [Crossref] [PubMed]
- Motoyama S, Sarai M, Harigaya H, Anno H, Inoue K, Hara T, Naruse H, Ishii J, Hishida H, Wong ND, Virmani R, Kondo T, Ozaki Y, Narula J. Computed tomographic angiography characteristics of atherosclerotic plaques subsequently resulting in acute coronary syndrome. J Am Coll Cardiol 2009;54:49-57. [Crossref] [PubMed]
- Nakanishi R, Motoyama S, Leipsic J, Budoff MJ. How accurate is atherosclerosis imaging by coronary computed tomography angiography? J Cardiovasc Comput Tomogr 2019;13:254-60. [Crossref] [PubMed]
- Nogic J, Prosser H, O'Brien J, Thakur U, Soon K, Proimos G, Brown AJ. The assessment of intermediate coronary lesions using intracoronary imaging. Cardiovasc Diagn Ther 2020;10:1445-60. [Crossref] [PubMed]
- Achenbach S, Daniel WG. Current role of cardiac computed tomography. Herz 2007;32:97-107. [Crossref] [PubMed]
- Sajjadieh A, Hekmatnia A, Keivani M, Asoodeh A, Pourmoghaddas M, Sanei H. Diagnostic performance of 64-row coronary CT angiography in detecting significant stenosis as compared with conventional invasive coronary angiography. ARYA Atheroscler 2013;9:157-63.
- Abazid RM, Romsa JG, Akincioglu C, Warrington JC, Bureau Y, Kiaii B, Vezina WC. Coronary artery calcium progression after coronary artery bypass grafting surgery. Open Heart 2021;8:e001684. [Crossref] [PubMed]
- Aldrovandi A, Cademartiri F, Menozzi A, Ugo F, Lina D, Maffei E, Palumbo A, Fusaro M, Crisi G, Ardissino D. Evaluation of coronary atherosclerosis by multislice computed tomography in patients with acute myocardial infarction and without significant coronary artery stenosis: a comparative study with quantitative coronary angiography. Circ Cardiovasc Imaging 2008;1:205-11. [Crossref] [PubMed]
- Andreini D, Conte E, Serruys PW. Coronary plaque features on CTA can identify patients at increased risk of cardiovascular events. Curr Opin Cardiol 2021;36:784-92. [Crossref] [PubMed]
- Aziz MU, Singh S. Computed tomography of coronary artery atherosclerosis: A review. J Med Imaging Radiat Sci 2021;52:S19-39. [Crossref] [PubMed]
- Chow CK, Sheth T. What is the role of invasive versus non-invasive coronary angiography in the investigation of patients suspected to have coronary heart disease? Intern Med J 2011;41:5-13. [Crossref] [PubMed]
- Choi AD, Marques H, Kumar V, Griffin WF, Rahban H, Karlsberg RP, Zeman RK, Katz RJ, Earls JP. CT Evaluation by Artificial Intelligence for Atherosclerosis, Stenosis and Vascular Morphology (CLARIFY): A Multi-center, international study. J Cardiovasc Comput Tomogr 2021;15:470-6. [Crossref] [PubMed]
- Liu K, Hsieh C, Zhuang N, Gao Y, Li Z, Ren X, Yang L, Zhang J, Budoff MJ, Lu B. Current utilization of cardiac computed tomography in mainland China: A national survey. J Cardiovasc Comput Tomogr 2016;10:76-81. [Crossref] [PubMed]
- Candemir S, White RD, Demirer M, Gupta V, Bigelow MT, Prevedello LM, Erdal BS. Automated coronary artery atherosclerosis detection and weakly supervised localization on coronary CT angiography with a deep 3-dimensional convolutional neural network. Comput Med Imaging Graph 2020;83:101721. [Crossref] [PubMed]
- Ji F, Zhou S, Bi Z. Computed Tomography Angiography under Deep Learning in the Treatment of Atherosclerosis with Rapamycin. J Healthc Eng 2021;2021:4543702. [Crossref] [PubMed]
- Kolossváry M, De Cecco CN, Feuchtner G, Maurovich-Horvat P. Advanced atherosclerosis imaging by CT: Radiomics, machine learning and deep learning. J Cardiovasc Comput Tomogr 2019;13:274-80. [Crossref] [PubMed]
- Zhao H, Yuan L, Chen Z, Liao Y, Lin J. Exploring the diagnostic effectiveness for myocardial ischaemia based on CCTA myocardial texture features. BMC Cardiovasc Disord 2021;21:416. [Crossref] [PubMed]
- Fu F, Wei J, Zhang M, Yu F, Xiao Y, Rong D, Shan Y, Li Y, Zhao C, Liao F, Yang Z, Li Y, Chen Y, Wang X, Lu J. Rapid vessel segmentation and reconstruction of head and neck angiograms using 3D convolutional neural network. Nat Commun 2020;11:4829. [Crossref] [PubMed]
- Chen M, Wang X, Hao G, Cheng X, Ma C, Guo N, Hu S, Tao Q, Yao F, Hu C. Diagnostic performance of deep learning-based vascular extraction and stenosis detection technique for coronary artery disease. Br J Radiol 2020;93:20191028. [Crossref] [PubMed]
- Han D, Liu J, Sun Z, Cui Y, He Y, Yang Z. Deep learning analysis in coronary computed tomographic angiography imaging for the assessment of patients with coronary artery stenosis. Comput Methods Programs Biomed 2020;196:105651. [Crossref] [PubMed]
- Liu Y, Cui C, Li Y, Wang Y, Hu Y, Bai M, Huang D, Zheng Q, Liu L. Predictive value of the echocardiographic noninvasive myocardial work index for left ventricular reverse remodeling in patients with multivessel coronary artery disease after percutaneous coronary intervention. Quant Imaging Med Surg 2022;12:3725-37. [Crossref] [PubMed]
- Xu C, Guo H, Xu M, Duan M, Wang M, Liu P, Luo X, Jin Z, Liu H, Wang Y. Automatic coronary artery calcium scoring on routine chest computed tomography (CT): comparison of a deep learning algorithm and a dedicated calcium scoring CT. Quant Imaging Med Surg 2022;12:2684-95. [Crossref] [PubMed]
- Liu Y, Nezami FR, Edelman ER. A Framework for Automated Quantification of Calcified Coronary Artery from Intravascular Optical Coherence Tomography Images. Pittsburgh, PA, USA: 2023 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI); 2023:1-4. doi:
10.1109/BHI58575.2023.10313463 . - Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, et al. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods 2019;16:67-70. [Crossref] [PubMed]
- Raff GL, Abidov A, Achenbach S, Berman DS, Boxt LM, Budoff MJ, Cheng V, DeFrance T, Hellinger JC, Karlsberg RP. SCCT guidelines for the interpretation and reporting of coronary computed tomographic angiography. J Cardiovasc Comput Tomogr 2009;3:122-36. [Crossref] [PubMed]
- Raff GL, Chinnaiyan KM, Cury RC, Garcia MT, Hecht HS, Hollander JE, O'Neil B, Taylor AJ, Hoffmann USociety of Cardiovascular Computed Tomography Guidelines Committee. SCCT guidelines on the use of coronary computed tomographic angiography for patients presenting with acute chest pain to the emergency department: a report of the Society of Cardiovascular Computed Tomography Guidelines Committee. J Cardiovasc Comput Tomogr 2014;8:254-71. [Crossref] [PubMed]
- Jiang B, Guo N, Ge Y, Zhang L, Oudkerk M, Xie X. Development and application of artificial intelligence in cardiac imaging. Br J Radiol 2020;93:20190812. [Crossref] [PubMed]
- van Velzen SGM, Lessmann N, Velthuis BK, Bank IEM, van den Bongard DHJG, Leiner T, de Jong PA, Veldhuis WB, Correa A, Terry JG, Carr JJ, Viergever MA, Verkooijen HM, Išgum I. Deep Learning for Automatic Calcium Scoring in CT: Validation Using Multiple Cardiac CT and Chest CT Protocols. Radiology 2020;295:66-79. [Crossref] [PubMed]
- Livne M, Rieger J, Aydin OU, Taha AA, Akay EM, Kossen T, Sobesky J, Kelleher JD, Hildebrand K, Frey D, Madai VI. A U-Net Deep Learning Framework for High Performance Vessel Segmentation in Patients With Cerebrovascular Disease. Front Neurosci 2019;13:97. [Crossref] [PubMed]
- CT coronary angiography in patients with suspected angina due to coronary heart disease (SCOT-HEART): an open-label, parallel-group, multicentre trial. Lancet 2015;385:2383-91. [Crossref] [PubMed]
- Kitahara S, Kataoka Y, Miura H, Nishii T, Nishimura K, Murai K, et al. The feasibility and limitation of coronary computed tomographic angiography imaging to identify coronary lipid-rich atheroma in vivo: Findings from near-infrared spectroscopy analysis. Atherosclerosis 2021;322:1-7. [Crossref] [PubMed]
- Mu D, Bai J, Chen W, Yu H, Liang J, Yin K, Li H, Qing Z, He K, Yang HY, Zhang J, Yin Y, McLellan HW, Schoepf UJ, Zhang B. Calcium Scoring at Coronary CT Angiography Using Deep Learning. Radiology 2022;302:309-16. [Crossref] [PubMed]
- Greenland P, Blaha MJ, Budoff MJ, Erbel R, Watson KE. Coronary Calcium Score and Cardiovascular Risk. J Am Coll Cardiol 2018;72:434-47. [Crossref] [PubMed]



