
Deep learning-based target tracking with X-ray images for radiotherapy: a narrative review
Introduction
Cancer is becoming the dominant cause of human death and the most prominent barrier to a longer life worldwide (1). According to GLOBOCAN, an online database that provides global cancer statistics and estimates of morbidity and mortality for 36 types of cancers in 185 countries, approximately 19.3 million new cancer cases and 9.96 million cancer deaths occurred in 2020 (1).
To control the progression of malignant tumors and increase the survival rate and life quality of cancer patients, in addition to surgical resection (2-4), radiotherapy (RT) (also known as radiation therapy) has become a routine treatment method (5-7). In the course of treatment, it is crucial to ensure that the prescribed dose is deposited in the gross tumor volume (GTV). Meanwhile, the fewer organs at risk (OARs) irradiated, the better. Thus, it is of vital importance that the GTV and OARs are localized in a precise and timely manner.
To ensure that the patient treatment set up follows the planning computed tomography (CT) scan, modern linear accelerators (LINACs) are typically equipped with gantry-mounted kV imaging systems (e.g., the On-Board Imager system) that provide submillimeter resolution X-ray images of the patient’s anatomy. This enables the continuous capture of two-dimensional (2D) kV X-ray images for cone beam CT reconstruction, as well as imaging during irradiation (8). Unlike most modern LINACs, the CyberKnife® (9) employs room-mounted dual X-ray imagers and in-floor built detectors (10). Before irradiating the GTV, the CyberKnife® system captures kV images to verify the real-time tumor spatial location and then adjusts the robot to precisely irradiate the region of the tumor. These imaging systems open up possibilities for using commercially available equipment to realize the marker-less tracking of targets with 2D kV X-ray images.
X-ray target tracking refers to the process of continuously monitoring and accurately tracking the position and motion of the target in an X-ray image or sequence of X-ray images (11). X-ray target tracking typically involves the following steps: image acquisition, pre-processing, target localization, motion estimation, tracking, and updating. First, the target object is exposed to X-ray radiation, and the resulting image is captured in the subsequent steps. The acquired X-ray image may need to be pre-processed later to enhance the image quality and reduce noise. Next, the target is located in the X-ray image. This can involve techniques such as image automatic segmentation (12), edge detection (13), and template matching (14). This step aims to identify the target’s boundaries and estimate the target’s position accurately. Once the target has been localized in the X-ray image, a sequence of X-ray images can be analyzed to estimate the target’s motion. Finally, the target’s position is continuously updated with new X-ray images acquired, providing real-time information on target’s displacement. X-ray target tracking is commonly used in various applications, including medical imaging, industrial inspection, security systems, etc. (11,15,16).
However, it is still challenging to perform tumor tracking with 2D kV X-ray images due to poor soft tissue contrast, organ overlapping, and organ motion. In such cases, metal fiducials are implanted into or near the tumor to provide a more precise spatial location of the GTV (14,17,18). The use of metal fiducials has been proven to be effective; however, some potential issues have also been noted. First, the implementation of metal fiducials is invasive and carries risks for the patient (e.g., bleeding and inflammation). Second, metal fiducials may lead to metal artifacts in the planning CT scan, resulting in a decrease in image quality. Third, it can still be difficult to track the OARs when mental fiducials have been implanted, as the OARs may have different shift compared with the GTV. Therefore, marker-less target tracking is urgently needed.
Some optical surface imaging systems have been developed and applied in the clinic, including Align RT® (19) and C-Rad® Catalyst (19). These monitoring systems provide real-time monitoring of the patient’s surface posture and enable non-invasive positioning; however, these systems cannot directly track soft tissues and tumors. Once the geometric relationship between the tumor and the skin surface changes, such methods may face the issue of inaccurate positioning. Thus, the question of how to track the soft tissues and tumors directly during treatment requires investigation.
Further, various motions and uncertainties may occur during irradiation, such as respiration-induced motion, cardiac-induced motion, and residual set-up errors. Several methods have been proposed to control respiratory movement (20), of which, the commonly employed approaches are abdominal compression (21), respiratory-gating (22), breath-holding (23), etc. However, even with the employment of respiratory-gating, the re-setup accuracy sometimes still exceeds 5 mm (with an average of 1.5 mm) (24). The use of respiratory-gating technology also increases the time of the intervention and the discomfort of the patient. Additionally, the heartbeat-induced motion amplitude ranges from 0.2 to 2.6 mm (25). Due to frequency and phase discrepancies, cardiac motion cannot be captured even with respiration-correlated four-dimensional (4D)-CT. Thus, cardiac motion cannot be addressed in the design of treatment plans. If both tumors and OARs could be tracked directly and in a timely manner in the daily verification images or during treatment, then adaptive RT could be applied to provide more precise dose delivery.
Deep learning is an essential branch of machine learning that focuses on training neural networks with multiple layers to extract complex features from the data and perform tasks automatically. It is inspired by the structure and function of the human brain. The frameworks of deep-learning networks differ depending on the application scenario. Convolutional neural networks (CNNs) (26) are one of the most commonly used architectures. CNNs consist of the following key components: an initial input layer, hidden layers, and a final output layer. Hidden layers usually include batch normalization, convolutional, pooling, and activation layers. CNNs significantly reduce the parameters of the hidden layer by sharing kernels. The encoder-decoder architecture is widely used in CNNs for medical imaging applications, such as image registration (27-29), image segmentation (30-33), and image synthesis (34-36).
U-Net (37) (see Figure 1A) is a typical example of an encoder-decoder architecture. The encoder gradually down-samples the input image, while extracting high-level context and abstract features. The decoder, which is symmetric to the encoder, gradually up-samples the feature maps to make accurate predictions. Primarily, skip connections are employed to establish direct links between corresponding encoder and decoder layers, mitigating information loss during propagation and aiding in precise predictions. However, U-Net also has some limitations. Notably, U-Net primarily focuses on local features and may have limited capability in capturing global context or long-range dependencies. This limitation can affect model performance in cases in which a broader understanding of the entire image is necessary. In addition, U-Net may have relatively high memory requirements, especially for deeper and wider architectures. This can pose challenges when deploying U-Net on resource-constrained devices or working with large-scale datasets.
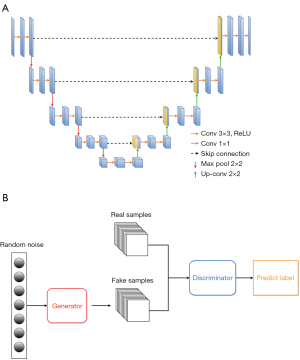
The generative adversarial network (GAN) is another popular model in deep-learning methods. The GAN (see Figure 1B) (38) consists of two main components: a generator and a discriminator. The generator takes random noise as input to generate fake samples, and the discriminator aims to distinguish between real and fake samples synthesized by the generator. The generator and discriminator are trained concurrently in a competitive manner, whereby the generator strives to synthesize more realistic samples, while the discriminator endeavors to improve its capability to differentiate between real and fake samples. The GAN excels at generating new data samples that resemble the training data distribution that could be applied to data augmentation. GANs can generate high-quality synthetic images under unsupervised learning, which makes them particularly useful when labeled data is scarce or expensive. However, GANs also have some limitations. GAN training can be challenging and unstable. The generator and discriminator are trained iteratively and balancing these two components may not be easy. The network architecture and training strategies must be carefully designed. Additionally, the GAN training process typically requires substantial computational resources, including powerful graphics processing units (GPUs) with large amounts of memory. It can also be time consuming, especially for complex datasets. In addition to U-Net and the GAN, more advanced models, such as the recurrent neural network (RNN) (39), region-based convolutional neural network (R-CNN) (40), graph neural network (GCN) (41), and you only look once (YOLO) (42), are also being successfully applied to address diverse medical imaging tasks.
In recent years, a few novel methods based on deep learning have been proposed for real-time target tracking with 2D kV X-ray images (43-48) to help perform online adaptive RT. To the best of our knowledge, to date, no review has sought to summarize the latest developments and the overall situation of deep learning-based real-time target tracking with 2D kV X-ray images. Mylonas et al. (49), Zhao et al. (50), and Salari et al. (51) reviewed the topic of artificial intelligence (AI)-based motion tracking. However, their reviews focused on AI-based methods for target tracking, including machine-learning and deep-learning methods, and considered diverse image modalities, such as magnetic resonance imaging, CT, ultrasound, and X-ray. Conversely, the present article sought to briefly review the progress made in deep learning-based target tracking with 2D kV X-ray images. Unlike previous reviews on similar topics, we focused on applying deep-learning methods to perform real-time marker-less target tracking with 2D X-ray images only. The existing limitations, the application challenges, possible solutions, and future developments of real-time marker-less target tracking were also considered. We present this article in accordance with the Narrative Review reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1489/rc).
Methods
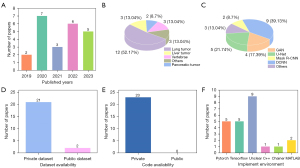
Studies on deep learning-based target tracking have been published constantly in recent years. To ensure as many relevant studies were included in this review as possible, a range of keywords were employed in our searches. The keywords used to search for English-language studies included the following terms: “radiotherapy, radiation therapy, motion tracking, target tracking, motion estimation, motion monitoring, X-ray images, digitally reconstructed radiographs, deep learning, convolutional neural network, and deep neural network”. First, manual searches of the Web of Science, and PubMed, and Google Scholar were carried out. Next, the abstract of each article was reviewed, and any irrelevant articles were excluded. Ultimately, 23 articles that were closely relevant to deep learning-based target tracking with 2D kV X-ray images were included in this review. Table 1 provides details of our predetermined search strategy. Figure 2 shows the results of the statistical analysis of the included articles.
Table 1
| Items | Specification |
|---|---|
| Dates of searches | June 29th, 2023; December 4th, 2023 |
| Databases and other sources searched | Web of Science, PubMed, and Google Scholar |
| Search terms used | “Radiotherapy” OR “radiation therapy” AND “motion tracking” OR “target tracking” OR “motion estimation” OR “motion monitoring” AND “X-ray images” OR “digitally reconstructed radiograph” AND “deep learning” OR “convolutional neural network” OR “deep neural network” |
| Timeframe | March 2019 to December 2023 |
| Inclusion and exclusion criteria | Inclusion: articles closely related to deep learning-based target tracking with two-dimensional kV X-ray images (or other similar descriptive words, such as motion tracking, and motion management) |
| Exclusion: unpublished articles, non-English articles, and/or articles using other modality images (e.g., cone beam computed tomography and magnetic resonance imaging) | |
| Selection process | The literature search and selection were conducted by X.L., and the selection process was discussed by all authors. Differences were resolved by consensus |
Deep learning-based target tracking
At present, target tracking is indirect in clinical practice, as it is based on the assumption that the geometric correlation between the target and bony structures or metal fiducials during the course of irradiation is consistent with the planning CT scan (51). Unfortunately, it is difficult to maintain the invariance of geometric correlation due to respiratory movement, etc. The question of how to instantaneously and precisely localize the tumor and OARs on poor-quality images is a challenging problem that continues to be of concern in RT. In this part of the article, we focus on the development of deep learning-based target tracking with 2D kV X-ray images for RT.
To better elucidate and compare the relevant studies, we categorized the retrieved studies into the following four subgroups: U-Net, GANs, deep CNNs, and other neural networks. Give that U-Net and GANs were the most commonly used models in previous studies, we separated them from the subgroup of deep CNNs to facilitate the performance of the comparison. Each study was allocated to one subgroup only. If a study used a deep CNN as its basic model, it was categorized under the deep CNN subgroup if it was unclear whether it was a U-Net or GAN. The “other neural network” subgroup mainly included studies that used GCNs or RNNs as their basic models.
U-Net-based target tracking
Liang et al. (52) designed a U-Net-based scheme to automatically localize the fiducial marker and evaluate intra-fraction motion with orthogonal kV X-ray images acquired by the CyberKnife® system. First, U-net was trained to automatically detect the bounding boxes of the fiducial marker, and the central coordinates of the bounding boxes from two orthogonal projections were then obtained and used to derive the three-dimensional (3D) coordinates of the fiducial marker. Finally, the intra-fraction motion was evaluated by rigidly registering the floating fiducial cohort with the reference fiducial cohort.
Shao et al. (53) also tried to improve the accuracy of automatic liver tumor tracking using U-Net. Their method can be divided into three steps: initial 2D-3D deformable registration, liver boundary deformation vector field (DVF) optimization, and intra-liver tumor motion tracking. In their method, U-net was used as a tool to learn the motion correlation between cranial and caudal liver boundaries and then to optimize the liver boundary DVFs. Subsequently, after optimization, the liver boundary DVFs were fed into a biomechanical model to obtain the intra-liver DVFs for liver tumor tracking using the finite element method.
Kim et al. (54) employed U-Net to automatically detect and segment the lumbar vertebrae on X-ray images. Their method comprised three steps: spine localization, segmentation of lumbar vertebra, and fine-tuning segmentation. First, a CNN-based Pose-net was employed to automatically localize the center of five lumbar vertebrae, and the bounding boxes of the five lumbar vertebrae were then extracted as the input for the next step according to their center positions predicted by the Pose-net. Next, a M-net designed on the basis of U-net was trained to segment the five lumbar vertebrae. Finally, a level-set method was used to fine-tune the segmentation.
Terunuma et al. (43) applied U-Net to transform kV X-ray fluoroscopic images into projected clinical target volume (CTV) images, and then identify the position of the tumor for real-time tracking. They attempted to focus the model’s attention on soft tissues instead of bones through the artificial difference of co-occurrence probability, so that the model could accurately identify the projected CTV on X-ray images.
Recently, Grama et al. (44) used Siamese networks, comprising twin subnetworks, to track lung tumors. Each subnetwork shared the same weights and processed images with different time frames. In their methods, U-Net was used as the backbone of the subnetworks to extract tumor features separately from two input images; that is, the target image and the search image. By comparing the tumor features and measuring the similarities between the target image and the search image consecutively, the model could estimate the tumor’s position in the search image and track its motion over time.
U-Net is a deep-learning network frequently used for image segmentation tasks, but it also has the potential to be applied to target tracking tasks after modification or combination with other methods. U-Net can capture contextual information in an image through the use of skip connections. This architecture can help to accurately locate and track the targets, especially when the distinction between the target and the background is not clear. However, the pooling operations used in the U-Net may potentially blur the boundaries of the tracked target, resulting in less clear contours. This issue poses challenges for target tracking tasks that require precise boundaries. Therefore, it is crucial to thoroughly consider the advantages and disadvantages and make appropriate modifications to enhance the suitability of U-Net for target tracking tasks.
GAN-based target tracking
Lei et al. (45) translated the target tracking task into a 2D-to-3D image synthesis task. If high-quality volumetric images can be generated from 2D images, it is possible that the task of target tracking would become simpler, as the overlapping or occlusion of organs or structures would be alleviated in the volumetric images. Similar to most image generation tasks, they employed a generative adversarial learning strategy to enable more realistic 2D-to-3D transformation, showing the potential for real-time tumor tracking during treatment. The 2D-to-3D transformation network (named TransNet) comprised three parts: the encoding module, transformation module, and decoding module. To improve the performance of the model, they devised the conventional loss function of the GAN by introducing the perceptual loss.
He et al. (55) tried to accomplish the target tracking task by extracting single spine images from 2D kV X-ray images. They introduced residual network (ResNet) GAN (ResNetGAN) to automatically decompose the spine images from the 2D kV X-ray images. As the name suggests, ResNetGAN included seven ResNet blocks in its generator to learn the discrepancy between the input and the actual distribution. As for the loss function, it consisted of four terms calculated in two domains; that is, the image domain and the feature domain. In the image domain, the following three terms were considered: the mean squared error and the Pearson correlation between the decomposed spine images generated by the network and the reference spine digitally reconstructed radiographs (DRRs), and the Pearson correlation of the soft tissue. The fourth term of the loss function was the difference between the decomposed spine image and the ground truth in the feature domain.
Peng et al. (56) considered the tumor tracking task in the 2D fluoroscopic projection images as a video object segmentation task. In their scheme, a GAN was also adopted. In the generator, two U-nets were cascaded to predict the tumor location from coarse to fine. Specifically, convolutional long short-term memory modules were introduced in the skip connection of the first U-Net and in the bottom layer of two U-nets to assist the model to capture temporal information in the 2D fluoroscopic image sequence. Additionally, a convolutional long short-term memory module was introduced before the fully connected layer in the discriminator. In terms of the loss function, they adopted a multiscale hybrid loss function that combined the generative adversarial loss, L1 loss, structural similarity index loss, and intersection over union loss.
Recently, Fu et al. (46) tried to perform tumor tracking by enhancing the visibility of the target on the X-ray images. They employed a conditional GAN to learn the mapping between the onboard X-ray images to target specific DRRs generated from the planning CT scan. U-Net was used as the generator to synthesize target specific DRRs with enhanced visibility. Comparing the target specific DRRs and real-time kV X-ray images, the intra-fractional tumor motion could be estimated and managed.
GAN is a deep-learning network commonly used for image synthesis tasks, but its performance in target tracking tasks has also been explored. When applied to target tracking tasks, it can be broadly divided into two categories. The first category involves the direct processing of 2D images, using a GAN to generate visually enhanced 2D images, thereby enabling target segmentation and then target tracking on the 2D images. The second category involves using a GAN to generate 3D images from 2D images, enabling image registration and object tracking in the 3D domain. GANs are capable of synthesizing pseudo images that closely resemble real images, but the training can be challenging and unstable. Further, mode collapse may occur when a generator fails to capture the diversity of the target data distribution, limiting the overall quality of the generated images. It is important to address the challenges and limitations associated with GANs when using them to solve ill-posed problems. One common method is modifying the loss function to further improve their effectiveness and reliability.
Deep CNN-based target tracking
In addition to using U-Net or GANs as basic models, some studies have also attempted to use other deep CNNs to address target tracking. Hirai et al. (57) developed a tracking algorithm based on an encoder-decoder structure to perform marker-less tumor tracking on fluoroscopic images. Sub-images of fluoroscopic images were fed into the model and a target probability map was generated to calculate the target position. Finally, the linear regression model was used to correct the tumor position in the lateral direction.
Wei et al. (58) attempted to predict the DVF relative to the planning CT scan from a single X-ray projection. In their method, a principal component analysis (PCA) was first conducted on 4D-CT, after which, any DVF could be linearly represented as the average motion vector field plus the combination of three PCA eigenvectors and their corresponding PCA coefficients. A CNN was used to predict the corresponding PCA coefficients from the input X-ray projection. Thereafter, the DVF could be calculated and the tumor location could then be estimated. Subsequently, they modified their model to address the issue of tumor localization at arbitrary gantry angles (59). The first modification was that an angle-dependent binary region of interest mask was applied on each extracted feature map, which addressed the issue of modeling the intricate mapping between the X-ray projection and tumor motion. The second modification was the use of a gantry angle-dependent fully connection layer, which was applied to recover the specific mapping from the extracted features maps to the tumor motion for each projection angle.
Takahashi et al. (60) tried to perform real-time tumor tracking using a variant of a fully convolutional network. They replaced the original deconvolution layers with a pixel shuffle layer to speed up the processing and perform real-time tracking. Motley et al. (61) first applied a YOLO framework to automatically detect fiducial markers in pelvis kV projection images. In their framework, the input image was first divided into grid regions. Each grid region was then input into the CNN to generate several bounding boxes with a confidence score. Finally, the bounding boxes with a confidence score above the set threshold was selected, and the central coordinates of these bounding boxes were considered as the marker position coordinates.
Lei et al. (62) designed a center-ness matching network to localize tumor position with two orthogonal 2D projections. They constructed feature extractors with CNNs and used them to extract features representing the probability map of the tumor’s center-of-mass. Next, the extracted feature maps were fused through 3D rotating back to 3D space, and the center-ness map was then generated. Thereafter, the rotated feature maps were re-sampled to match the size of 3D volume via a tensor re-sizing operator. Finally, the CNN-based detection module was used to predict the 3D location of the tumor’s center-of-mass with the center-ness map as the network input.
Zhou et al. (47) also investigated the feasibility of deep learning-based real-time tumor tracking. Their method used a ResNet and a feature pyramid network to extract features to predict the contour of the GTV on X-ray images. Next, the 3D position of the GTV was calculated according to the centroids of the predicted contours in two orthogonal images.
Ahmed et al. (63) used a CNN for automatic fiducial marker tracking. In their study, a sliding window technique was employed to determine the search area from the input kV images, and the sub-images cropped from the search area were then input into the CNN to classify them as either fiducial markers or background. Finally, the central position of the markers was estimated by aggregating the positions of all the sub-images classified as fiducial markers. They also investigated the performance of a pre-trained YOLO framework and a hybrid CNN-YOLO. In the hybrid framework, the CNN was first used to detect the markers. In cases in which the CNN failed to detect the markers, the YOLO would take over the detection process.
Recently, Dai et al. (64) attempted to address the issue of tumor tracking via 2D to 3D elastic registration. The 2D projection image was first fed into the ResNet to form a 3D feature map for subsequent image registration. Next, the 3D feature map was cropped into patches that served as inputs for the registration network. The registration network integrated Swin transformer blocks into its encoder path to effectively capture features with an attention mechanism, significantly reducing the computational complexity. Skip connections were also introduced between the corresponding encoder and decoder layers. Finally, a 3D DVF was predicted by the registration network, and it was then used to generate a 3D warped image for tumor tracking.
Researchers have also introduced the region proposal strategy in models to assist in target tracking. Zhao et al. (65) used a pre-trained CNN (VGG16) combined with a region proposal network (RPN) to perform marker-less tumor tracking in pancreatic cancer. The VGG16 was used to extract high-dimensional features, and the features were then selected by the RPN to generate proposals for later region-based target detection. Subsequently, they investigated its performance in prostate cancer with the same deep-learning framework (66). Roggen et al. (67) used the ResNet as the backbone to construct the mask R-CNN model to track the vertebrae. They optimized their network weighting parameters by pre-training the network using the Common Object in Context (COCO) dataset (68). Zhou et al. (48) also constructed a mask R-CNN model to perform real-time pancreatic tumor tracking. In their method, the ResNet and a feature pyramid network were used as the backbone to extract features, and the model was also pre-trained using the COCO dataset.
The use of CNNs has been widely explored in target tracking tasks. CNNs can automatically learn hierarchical feature representations of a target through successive convolutional layers, which enables CNNs to improve the accuracy of target recognition and tracking. Additionally, CNNs can effectively leverage contextual information around a target by expanding the receptive field and extracting features at multiple levels. However, deep CNNs may require high computational resources, which limits the use of CNNs in real-time target tracking. In complex target tracking scenarios, it may be necessary to combine a CNN with other strategies and methods to improve tracking efficiency and accuracy.
Other neural network-based target tracking
The use of RNN-based models has also been explored in target tracking tasks. Wang et al. (69) designed an RNN-based framework to localize lung tumors. Their model comprised three main parts: a CNN, an RNN, and a flexible calibration mechanism. According to their design, a series of delta images that represented the difference between a current projection and a previous projection were calculated to act as the input of the CNN. Extracted features related to tumor motion by the CNN were combined with gantry and time-stamp information to generate motion feature vectors that served as input for the following RNN. The RNN predicted the 3D tumor locations by parsing the feature vectors and calculating the motion amplitude along the anteroposterior, lateral, and superior-inferior directions. Finally, to improve the performance, the output of the RNN in superior-inferior direction was frequently corrected using the cross-correlation registration technique.
Finally, GCNs have also been used to perform target tracking tasks. Shao et al. (70) used a GCN to directly predict liver boundary DVFs. The ResNet-50 combined with a perceptual feature pooling layer was employed as a feature extraction subnetwork to extract hidden image features from onboard X-ray projections. Pooled image features were subsequently fed into the GCN to predict the liver boundary DVFs. Just like in their previous work (53), the predicted liver boundary DVFs were finally fed into a biomechanical model to obtain the intra-liver DVFs for liver tumor localization.
RNNs are suitable for undertaking target tracking tasks with temporal structures, as they can leverage the temporal evolution and dynamic changes of the tracking target. However, RNNs may suffer from long-term dependency. Further, the computation of RNNs proceeds in a step-by-step manner, which may limit their ability to handle real-time target tracking, especially in longer sequences. GCNs can capture inter-target relationships by performing information propagation and aggregation among nodes in the graph structure, thus improving the accuracy of target tracking. However, it is more complex to construct and define graph structures with GCNs than CNNs. In applying GCNs to target tracking tasks, it is necessary to accurately model and define the relationships between targets. Similarly, GCNs usually entail high computational complexity, especially for large-scale images, which limits their application in real-time target tracking tasks. Compared to CNN-based models, fewer studies have used RNNs or GCNs to perform target tracking tasks. We expect that more studies using various state-of-the-art models will be conducted in the future.
Table 2 provides a detailed summary of the aforementioned works related to deep learning-based target tracking with 2D kV X-ray images, and includes details related to the dataset, network, input, tracking targets, equipment, results, and research highlights.
Table 2
| References | Datasets | Network | Input | Tracking targets | Equipment | Results | Research highlights |
|---|---|---|---|---|---|---|---|
| Liang et al. (52) | 5,927 real images and 13 patients | U-Net | Real X-ray images | Fiducial marker | CyberKnife | The mean centroid error between the predictions and the ground truth was 0.25±0.47 pixels for the test data. A precision rate of 98.6%, and a recall rate of 95.6% was achieved by the fiducial marker detection model | Using a fully convolutional network to predict the fiducial marker bounding boxes and reconstructing the 3D positions of the fiducial marker with the prediction |
| Shao et al. (53) | 34 patients | U-Net | 3D DVFs | Liver tumor | Unknown | The tumor center-of-mass-error was 1.7±0.4 mm for the model, and the mean HD and DSC were 4.5±1.3 mm and 0.78±0.03, respectively | Developing U-Net-based network to optimize the liver boundary DVFs to improve the accuracy of subsequent biomechanical modeling and automatic liver tumor localization |
| Kim et al. (54) | 797 real images | U-Net | DR or CR X-ray images | Lumbar vertebrae | FCR5000 (Fujifilm) and Discovery XR656 (GE Healthcare) | The model achieved a DSC of 91.60%±2.22%, and a mean center position error of 5.07±2.17 mm for lumbar vertebra identification, when compared the predictions with the labels created by radiologists | Using a CNN-based Pose-net to localize the center of five lumbar vertebrae, and a U-Net-based M-net to segment the five lumbar vertebrae |
| Terunuma et al. (43) | 10 patients | U-Net | DRRs or X-ray fluoroscopic images | CTV of lung cancer | Optima 580W (GE Healthcare) | The model had a 3D 95 percentile tracking error of 1.3–3.9 mm, a Jaccard index of 0.85–0.94, and a HD of 0.6–4.9 mm | Using the artificial difference of co-occurrence probability to assist U-Net to focus on soft tissues, and accurately synthesizing projected CTV images from original X-ray fluoroscopic image for tumor tracking |
| Grama et al. (44) | 6 patients and a thorax phantom | U-Net | 2D DRRs or 2D kV images | Lung tumor | Discovery CT590 RT Scanner (GE Healthcare) | The MAE was 0.57–0.79 mm compared to the ground truth when tested with the phantom data. As for the patient data, a correlation coefficient of 0.71–0.98 was achieved when compared the tumor location predicted by the model with the records of Respiratory Motion System | Using Siamese networks to capture tumor features, and estimating the position of the lung tumor by comparing, and measuring the similarity between consecutive volumes |
| Lei et al. (45) | 24 patients | GAN | 2D projections from 3D CT | Lung tumor | SOMATOM Definition AS CT scanner (Siemens) | The MAE, PSNR, and SSIM between the 3D images generated by the model and the ground truth were 99.3±14.1 HU, 15.4±2.5 dB, and 0.839±0.090, respectively, and the mean center of the mass distance of the tumor was 1.26 mm | Developing a novel GAN, named TransNet, to transform the 2D projections into 3D images, providing the potential for real-time tumor tracking during treatment |
| He et al. (55) | 24 patients | GAN | 2D kV images from raw CBCT data | Spine structure | TrueBeam LINAC (Varian Medical Systems) | The decomposed spine images generated by the model obtained a mean PSNR of 60.08 dB, an SSIM of 0.99, and a mean error of 0.13 and 0.12 mm in the X- and Y-directions, respectively, when matched with the reference spine DRRs | Proposing a novel GAN, introducing ResNet blocks in the generator and constraining the loss function in two domains (i.e., the image domain and the feature domain) |
| Peng et al. (56) | X-CAT phantom | GAN | 2D fluoroscopic images | Lung tumor | N/A | For the group-based model, an average IOU of 0.93 and an average DSC of 0.96 were achieved by the model when evaluating the overlapping between the tracked region of the tumor by the model and the ground truth. The tumor’s average center-of-mass difference was 1.6 and 0.7 mm for the SI and LR directions, respectively | Using a GAN combined with convolutional long short-term memory modules, a cascaded U-Net structure, and hybrid loss to capture temporal and spatial information to predict tumor location |
| The patient-specific model achieved an average IOU of 0.98, an average DSC of 0.99, and an average center-of-mass difference of 3 and 1 mm for the SI and LR directions, respectively | |||||||
| Fu et al. (46) | LUNGMAN phantom and 2 patients | GAN | 2D DRRs | Lung tumor or spine tumor | TrueBeam LINAC (Varian Medical Systems) | The MAE of tumor tracking in X direction was 0.11±0.05 and 0.1±0.3 mm for spine phantom and lung phantom, respectively, and in Y direction, the MAE was 0.25±0.08 and 0.1±0.3 mm for spine phantom and lung phantom, respectively | Using a conditional GAN to synthesize target specific DRRs from kV X-ray images to enhance target visibility and then track tumors |
| Hirai et al. (57) | 10 patients | CNN | 2D sub-images of fluoroscopic images | Lung tumor or liver tumor | PaxScan 3030 (Varian Medical Systems) | The average tracking accuracy was 1.90±0.65 and 1.37±0.81 mm for lung cases and liver cases, respectively | Employing a CNN to generate target probability map and then calculating the target position according to the target probability map |
| Wei et al. (58) | 3 patients and X-CAT phantom | CNN | 2D DRRs | Lung tumor | VersaHD LINAC (Elekta) | The model could locate the tumor with a 3D mean error of less than 0.13 mm at three different projection angles (0°, 45°, and 90°) | Combining a principal component analysis and CNN to predict the DVF from input 2D projection to estimate tumor location |
| Wei et al. (59) | 15 patients | CNN | 2D DRRs | Lung tumor | TrueBeam LINAC (Varian Medical Systems) | The mean error of tumor localization was under 1.8 and 1.0 mm in the SI and LR directions, respectively | Applying an angle-dependent binary region of interest mask on every extracted feature map, and introducing a gantry angle-dependent fully connection layer to address the issue of tumor localization at arbitrary angles |
| Takahashi et al. (60) | X-CAT phantom | CNN | 2D DRRs | Lung tumor | N/A | The mean tracking error was less than 0.2 mm for X-CAT digital phantom and less than 1 mm for epoxy phantom | Using a pixel shuffle layer to replace deconvolution layers to reduce calculation time, and introducing random translation and noise to DRRs to simulate X-ray images |
| Motley et al. (61) | 14 patients | CNN | 2D projections from 3D CBCT | Fiducial marker | XVI system (Elekta) | The detection model achieved a mean accuracy of 97.8% when applied to compute displacements, and an average deviation of 2.0±0.9 mm was found for inter-fraction marker migration | Applying a YOLO framework to predict the position of the marker for image-guiding radiotherapy and inter-fraction motion tracking |
| Lei et al. (62) | 10 patients | CNN | 2D projections from 3D CT | Lung tumor | SOMATOM Definition AS CT scanner (Siemens) | A mean 3D position error of 2.6±0.7 mm was obtained when compared the center of mass of the tumor predicted by the model with the ground truth | Proposing a center-ness matching network to predict the 3D location of the tumor’s center-of-mass using the input of one kV 2D projection and its orthogonal MV 2D projections |
| Zhou et al. (47) | 10 patients | CNN | 2D DRR or X-ray images | Lung tumor | Vero4DRT system (Hitachi Ltd. and Brainlab AG) | The median 3D position deviation between the model prediction and the ground truth was 2.27 mm | Predicting the contour of the GTV dynamically on real clinical X-ray images for the first time |
| Ahmed et al. (63) | 13 patients | CNN | Sub-images of kV X-ray images | Fiducial marker | TrueBeam LINAC (Varian Medical Systems) | The MAE of marker tracking was less than 0.88±0.11 mm in the AP, LR, and SI directions | Evaluating three deep-learning methods (CNN, YOLO, and hybrid CNN-YOLO) for the detection and tracking of fiducial markers in pancreatic cancer |
| Dai et al. (64) | Patient data from TCIA and CIRS phantom data | CNN | 2D DRRs or real projections | Lung tumor | Unknown | For real CBCT X-ray projections, the RMSE of the tumor centroid was less than 1.5 mm | Introducing Swin transformer blocks into the encoder path to effectively capture the features and accurately localize the tumor |
| Zhao et al. (65) | 2 patients | RPN | 2D DRRs | Pancreatic tumor | Unknown | The mean absolute difference between the model predictions and actual positions was less than 2.60 mm in the AP, LR and SI directions | Using VGG16 to extract high-dimensional features and a RPN to automatically generate proposals for following region-based target detection |
| Roggen et al. (67) | 13 patients | Mask R-CNN | 2D kV projections from 3D CBCT | Vertebrae | TrueBeam LINAC (Varian Medical Systems) | The model was able to detect the positional shifts within a range of 1.5 mm and to identify the rotations above 1 degree | Proposing a fast deep learning-based vertebra detection model, and evaluating the performance on a patient-like full-body phantom with vertebrae |
| Zhou et al. (48) | 14 patients | Mask R-CNN | 2D DRRs | CTV of the pancreatic tumor | Vero4DRT system (Hitachi Ltd. and Brainlab AG) | A mean DSC of 0.98 was achieved by the model when evaluating the overlapping between the predicted contour and the ground truth. The mean 3D error was 0.29 mm between the position predicted by the model and the actual situation, and the mean CTV contouring calculation time was 55 ms per image | Proposing a mask R-CNN using ResNnet and the feature pyramid network as the backbone to extract features, and conducting pre-training using the COCO dataset |
| Wang et al. (69) | 13 patients | RNN | 2D kV projection sequences | Lung tumor | TrueBeam LINAC (Varian Medical Systems) | The 3D position error between model prediction and ground truth was 1.3±1.4 mm, and the calculation time was 20 ms for one projection | Designing an RNN-based algorithm to calculate 3D tumor motion from extracted feature vectors, and integrating cross-correlation-based registrations at every 10° gantry rotation into the algorithm to enhance calculation performance |
| Shao et al. (70) | 10 patients | GCN | 2D projections from 3D CT | Liver tumor | Unknown | The center-of-mass error between the model calculation and ground truth was less than 1.2 mm, and the HD was less than 2.9 mm, and the DSC was approximately 0.9 when assessing the relative overlap of the tumor contour between the model-generated and actual situations | Using a GCN to predict liver boundary DVFs and then employing a biomechanical model to solve the intra-liver DVFs to realize liver tumor precise localization |
3D, three-dimensional; DVF, deformation vector field; HD, Hausdorff distance; DSC, dice similarity coefficient; DR, digital radiography; CR, computed radiography; CNN, convolutional neural network; DRR, digitally reconstructed radiography; CTV, clinical target volume; MAE, mean absolute error; GAN, generative adversarial network; 2D, two-dimensional; CT, computed tomography; PSNR, peak signal-to-noise ratio; SSIM, structural similarity index metric; TransNet, transformation network; ResNet, residual network; CBCT, cone beam computed tomography; LINAC, linear accelerator; N/A, not applicable; IOU, intersection over union; SI, superior-inferior; LR, left-right; YOLO, you only look once; GTV, gross tumor volume; AP, anterior-posterior; TCIA, The Cancer Imaging Archive; RMSE, root mean square error; RPN, region proposal network; R-CNN, region convolutional neural network; COCO, Common Object in Context; RNN, recurrent neural network; GCN, graph neural network.
Discussion
In current RT, intra-fraction motion management still lacks a satisfactory commercial solution for LINACs because of the demand for instantaneity and precision. Deep learning-based methods excel at extracting high-dimensional features from training datasets and learning the hidden correlation between abstract features and targets. With the continuous application of deep learning in medical image processing, it is possible to fill in the gaps for intra- and inter-fraction motion management using deep learning-based methods. In the past 5 years, there has been a surge of studies focusing on marker-less deep learning-based target tracking with 2D kV X-ray images. The employment of marker-less target tracking is beneficial for patients due to its non-invasiveness. According to the statistics of this review, marker-less target tracking using deep learning-based methods with 2D kV X-ray images has been investigated for RT of the pancreas (48,65), prostate (66,71), liver (53,57,70), lungs (43-47,56,58-60,62,64,69), and vertebrae (54,67). There is no doubt that challenges and opportunities coexist. Some progress has been made in marker-less target tracking with 2D X-ray images, but many limitations and challenges remain.
Current limitations
Limitations related to datasets
First, because of the lack of any available projection images or kV X-ray images and corresponding ground truth before the actual RT, each patient’s planning CT images have to be used to generate simulated images to train the patient-specific models. Currently, almost all researchers have trained their models using simulated images rather than real 2D kV X-ray images acquired from the clinic. Given that actual projection images will be employed to predict the instantaneous target position, these simulated projection images for training ought to be similar to the acquired in-treatment 2D kV X-ray images in terms of image quality. However, the actual kV X-ray images obtained by flat panel detectors are contaminated by scattering and noise, and unlike simulated images acquired by the ray-tracing method, they are not ideal. When using the actual in-treatment images with degraded image quality to predict the target spatial position, it is uncertain whether such models can maintain their accuracy and precision.
Some studies (52,54,57,63) have used real 2D kV X-ray images to train their models, thus mitigating the potential effects resulting from discrepancies between the simulated images and real images; however, some limitations persisted. It should be noted that the tracking target of reference (52) is the fiducial marker, and the question of whether it is efficient for marker-less target tracking requires further investigation. And Kim et al. (54) localized the lumbar vertebrae only and reported a mean center position error of 5.07±2.17 mm; however, this slightly large error could limit the application of the model in the clinic. Hirai et al. (57) and Ahmed et al. (63) also used real 2D kV images, but the input of their models was the sub-images cropped from the real images. The use of sub-images as input may limit the ability of the deep-learning model to extract global features and possibly decrease the tracking accuracy, especially when tumor and organ positions inter-fractionally change.
These issues could be addressed by using Monte-Carlo simulations that generate more realistic simulated projection images for training models. However, it should be noted that the time required to generate projection data increases substantially using the Monte-Carlo method. Intensity correction between DRRs and real images could also assist to reduce this discrepancy. The scattering and noise could affect the intensity distribution of the acquired images. If the intensity correction relationship could be established, simulated images could be generated that are closer to the real images.
Another possible approach is to leverage deep learning-based methods. In recent years, extensive research on medical image synthesis tasks using deep learning-based methods has achieved promising results in transforming images between different image modalities (72-74). Attempts could be made to establish a mapping relationship between DRRs and actual in-treatment images using deep learning-based methods to enable the deep-learning models to generate a large dataset that could then be used to develop the target tracking algorithms. By applying post-processing techniques, the DRRs could be corrected and synthetic images that closely resemble real images could be generated, thereby addressing the issues that arise from differences between the images. Exploring unsupervised learning or contrastive learning combined with popular image generation models, such as GANs, might be a promising research direction that could solve the dataset issue. Dai et al. (64) showed the effectiveness of this method.
Limitations related to ground truth
The question of how to impersonally determine the clinically optimal ground truth on X-ray images to train deep-learning networks is also challenging. As is well known, the image quality of the 2D kV X-ray images is poor due to noise and scattering, and the contrast of the soft tissue boundary is worse compared to that of CT, which is not conducive to determining the optimal segmentation result in the clinic. Further, given the intrinsic nature of X-ray images (e.g., the large overlapping of anatomical structures, complicated texture patterns, and fuzzy boundaries), even expert radiologists may make mistakes sometimes. Further, just like contouring structures on planning CT images, the quality of the manual segmentation is dependent on the prior knowledge and experience of the expert, which means that the greatest inconsistencies could arise in the course of manual contouring, and such inconsistencies could affect the training of a network.
Currently, except for a few studies, the ground truth for target tracking has been transformed from contours on planning CT images. When transforming the contour from the planning CT images to the current 2D kV image, 2D-3D registration is commonly used. As is commonly known, the 2D-3D registration is an ill-posed problem that can affect the quality of the ground truth, which in turn affects the learning process of the model. Wang et al. (69) trained their model using data recorded by the CalypsoTM system during treatment, which provided a more accurate position for tracking the target as the ground truth, resulting in a more reliable model. When using the CalypsoTM system, an electromagnetic transponder is implanted in or near the target area. Despite the fact that the implanted transponders are significantly smaller than the size of the tumor, the presence of the transponders can still have some influence on the training and learning process of the model. Thus, further studies need to be conducted to determine whether a model developed in this case can perform well in marker-less scenarios.
Improving the quality of X-ray images could be conducive to generating more precise ground truth. Future research should seek to use a deep-learning algorithm to improve the 2D kV X-ray image quality using the same approach as that adopted for cone beam CT (75). Undoubtedly, enhanced image quality has the potential to facilitate the more accurate and precise contouring of structures on 2D kV X-ray images directly. Training the model with labels directly generated on the 2D kV X-ray images would enhance the credibility of the model.
Another possible solution is to develop advanced image deformable registration algorithms. If satisfactory deformable registration could be achieved between the X-ray image and the planning CT scan (76), the contour generated by projection on the planning CT scan could be migrated to the X-ray image, which could significantly mitigate the manual labeling effort. Even if the contour obtained through deformable registration could not be used directly, it could be implemented in the clinic after further modification, which could also reduce the labeling effort. Certainly, it is essential to develop international clinical guidelines to eliminate the effects of inconsistencies.
Limitations related to robustness
The third limitation is related to the robustness of the model. To train a robust model, substantial datasets are required. However, collecting large amounts of high-quality data, especially for medical images, is challenging and time consuming. Terunuma et al. (43), Grama et al. (44), Fu et al. (46), and Takahashi et al. (60) all attempted to migrate this limitation by training patient-specific models. With this strategy, the trained models can be further personalized. However, training a patient-specific model may take a few hours, and thus delays in RT treatment may occur in the clinic with inadequate computer resources. Lei et al. (45), Zhou et al. (47), and Zhao et al. (65) even trained an angle-specific model for each patient. Angle-specific models, which are limited to tracking targets at specific angles for specific patients, may reduce the clinical practicality of such models and impose additional demands on computing resources. By leveraging more powerful GPU cards, the training process could be accelerated, addressing this issue to some extent.
However, another point worth noting is that the patient-specific models were all trained by simulated DRR datasets. Given that the performance of deep learning-based models may be related to the unique dataset employed for training, the robustness of models is a significant concern. For example, due to the upgrading of the hardware or software in an onboard imaging device, the acquired imaging data could change in some unknown way. This dataset change could affect the performance of the deep-learning models that were trained with the dataset prior to the change. Further, if the test dataset is outside the distribution of the training dataset, the performance of the model may be inferior. These potential issues limit the use of such models in the clinic due to the data heterogeneity of different patients.
The development of large publicly annotated image datasets through multi-center cooperation will address this limitation and improve the robustness of models. If robust models (rather than patient-specific models) could be developed, it is likely that such models would have widespread application in the clinic. However, it is difficult to establish a large public dataset with high annotation quality in a short time. In principle, the larger the dataset, the greater the robustness of the trained network. However, it has not yet been established how large a dataset would need to be to adequately train a robust model. Given this, the strategy of continual learning may address this problem to some extent. A model could be fine-tuned with new incoming data, such that the robustness of that model would become stronger as the dataset becomes larger.
Common challenges
Challenges related to real-time tracking
The first challenge relates to the high demand for computing efficiency to achieve marker-less real-time target tracking. The AAPM Task Group 264 (77) defines “real time” as a system latency below 500 ms. Thus, deep learning-based models need to complete target tracking within an extremely short time. The greatest effort should be made to minimize the latency of target tracking to enable real-time motion management. Using more powerful GPU cards and multi-GPU systems could accelerate the image analysis to obtain the real-time target spatial position. Additionally, the network architecture also affects the computation time. Currently, several studies (48,65,67) have employed region proposal strategies in their models to track the target in real time and have achieved promising results. The use of a region proposal strategy may reduce the inference time, as the model only needs to search for the target in specific regions rather than the entire image. This could have benefits for practical applications because it is necessary to have minimal system latency.
Research should also seek to improve the efficiency of deep-learning models in feature extraction, which when applied to target tracking, might also contribute to decreasing a model’s inference time. Recently, transformer models have been attracting increasing attention due to their powerful feature extraction capabilities. Some studies (78-80) have explored the effectiveness of transformer models in medical imaging. A common strategy is to combine transformers with CNNs to leverage both strengths. CNNs could extract the local features from images, after which, transformer modules could model the global context and fuse features. Such hybrid networks could preserve local information while capturing the long-range dependencies within the images. Dai et al. (64) investigated the performance of the combination of a Swin transformer and CNN in real-time target tracking. A Swin transformer is a transformer variant with a hierarchical structure that divides the input image into smaller non-overlapping patches. These patches are processed hierarchically, such that the information is gradually aggregated across different levels of the network. This hierarchical processing allows the model to effectively capture local and global information, enabling it to understand spatial relationships in the image. According to their comparative results, introducing the Swin transformer improved the tracking accuracy. Additionally, the calculation time was only 51 ms per image; thus, this model holds great promise in realizing real-time demands. Thus, consideration should be given to conducting further research in this direction.
Moreover, serious consideration should be given to accelerating X-ray image data acquisition and image reconstruction. In general, the time it takes to acquire an X-ray image is much longer than the time it takes to process it. If research is conducted to enable the target to be tracked instantaneously, efforts should be made to minimize the time spent in each step as much as possible. Studies could also seek to develop a more efficient detector and propose a more advanced reconstruction algorithm based on AI to alleviate the extent of patient repositioning during image processing.
Challenges related to interpretability
Due to the lack of good interpretability, deep learning-based methods are considered black-box algorithms. Thus, it is challenging to fully determine how and which factors result in inferior performance. In other words, a deep-learning model may localize the target in an unpredictable way during the actual application, which is dangerous in the clinic. Common methods can be used to interpret deep-learning models, such as gradient-weighted class activation mapping, and feature importance analyses. Visualizing the activations and feature maps of the intermediate layers can help us to understand how a model processes input data and visualize a model’s attention on specific regions. Feature importance analyses, such as gradient-based methods or feature importance ranking, can be employed to determine which features have a greater effect on a model’s predictions and help us to better understand the process.
Challenges related to transportability
The transportability of deep-learning models across different institutions is also worth considering. Different institutions have different training practices and image acquisition protocols, which could affect the image quality, variability of the contours etc., which could ultimately affect the performance of deep-learning models. This issue could be addressed with federated learning. It is well known that federated learning can realize joint modeling and improve the efficacy of AI models while ensuring data privacy and security. Additionally, a comprehensive set of guidelines could also address these issues and promote the development of deep learning-based algorithms in this field. Notably, Mongan et al. (81) proposed comprehensive guidelines; that is, the Checklist for Artificial Intelligence in Medical Imaging. If researchers working on the same topics followed this checklist, the generalizability and reproducibility of models could be improved.
Once the target tracking has finished, consideration should be given as to how this result can be used to accommodate any motion. Thus, steps need to be taken to ensure that the results produced by deep-learning models can be imported consistently and accurately into image-guided RT systems. Additionally, the X-ray images captured in real time during treatment must be exported quickly into the model for further monitoring. Thus, it is necessary to establish an exhaustive quality assurance system to constantly manage patients’ risks throughout treatment and ensure patient safety.
Challenges related to ethics
Last but not least, ethical and legal concerns need to be considered. Patients may have concerns about data privacy, and thus it is of vital importance that strict consensus regulations and guidelines be established for the clinical implementation of deep-learning models. Prior to implementation in clinical practice, deep learning-based methods should be subjected to pre-market review and post-market surveillance. All of the above-mentioned studies are initial proof of concept studies that generally applied off-the-shelf algorithms, adopted from other fields, such as computer vision. It is necessary to conduct performance analyses and prospective studies. Additionally, it is essential to clearly state the technical limitations of automatic target tracking algorithms to ensure user awareness and enable vendors to address these limitations effectively.
There is no doubt that the use of deep learning in real-time target tracking is still in its early stages. At the time of this review, the number of articles published on real-time marker-less target tracking with 2D kV X-ray images was modest compared to the number of articles published on other RT topics, such as automatic segmentation and image registration. As far as we know, all the studies mentioned in this review were retrospective, and to date, no prospective studies have been conducted. There is a significant scarcity of prospective studies based on deep-learning methods in medical imaging. The difficulties in conducting prospective research include the following: (I) data collection and processing challenges. Prospective studies require real-time data collection and processing, and are thus more challenging than retrospective studies; (II) ethical approval and privacy protection. Prospective studies have more strict ethical approval and privacy protection measures; (III) validation issues. The results of prospective studies need to be validated in future practice, which poses a challenge for deep-learning methods. The performance of deep-learning models usually varies with changes in data, and prospective research data will only be available in the future; and (IV) period. Prospective studies typically require that study subjects be tracked for longer periods to observe and record changes. Thus, extended periods may be required to train and validate deep-learning models.
Overall, there is still a long way to go before deep learning-based real-time target tracking can be implemented in clinical practice. Comprehensive consideration needs to be given to the limitations and challenges of deep learning-based methods and efforts need to be made to solve these limitations and challenges to enable applications of such methods in the clinic under the premise of ensuring safety.
Conclusions
Deep learning-based real-time target tracking has become a promising method in real-time organ motion management during RT. In this review, recent progress relevant to deep-learning based target tracking with 2D kV X-ray images was summarized. Our statistical analysis demonstrated that the models proposed in the identified studies primarily adopted the structures of U-Net, GANs, or deep CNNs. The majority of the identified studies employed simulated projection images as datasets to train and test their models due to a lack of labeled 2D kV images. All in all, the use of deep-learning based methods has been shown to be feasible in markers-less real-time target tracking. However, it is still quite challenging to achieve real-time target tracking in view of the latency in the X-ray imaging system, calculation algorithm, and system response time. The continued improvement of deep learning-based methods is crucial to improve the calculation efficiency, accuracy, and robustness of such models. We anticipate an increase in the number of studies in this area, along with the further refinement of methods. Finally, all of the limitations should be considered before any of these models are implemented in clinical practice to ensure the maximum safety of cancer patients.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-1489/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1489/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Schneider M, Hackert T, Strobel O, Büchler MW. Technical advances in surgery for pancreatic cancer. Br J Surg 2021;108:777-85. [Crossref] [PubMed]
- Chen C, Kolbe J, Christmas T. Surgical treatment of non-small-cell lung cancer in octogenarians: a single-centre retrospective study. Intern Med J 2021;51:596-9. [Crossref] [PubMed]
- Suzuki K, Watanabe SI, Wakabayashi M, Saji H, Aokage K, Moriya Y, Yoshino I, Tsuboi M, Nakamura S, Nakamura K, Mitsudomi T, Asamura HWest Japan Oncology Group and Japan Clinical Oncology Group. A single-arm study of sublobar resection for ground-glass opacity dominant peripheral lung cancer. J Thorac Cardiovasc Surg 2022;163:289-301.e2. [Crossref] [PubMed]
- Gao L, Zheng H, Cai Q, Wei L. Autophagy and Tumour Radiotherapy. Adv Exp Med Biol 2020;1207:375-87. [Crossref] [PubMed]
- Steinmeier T, Schulze Schleithoff S, Timmermann B. Evolving Radiotherapy Techniques in Paediatric Oncology. Clin Oncol (R Coll Radiol) 2019;31:142-50. [Crossref] [PubMed]
- Chandra RA, Keane FK, Voncken FEM, Thomas CR Jr. Contemporary radiotherapy: present and future. Lancet 2021;398:171-84. [Crossref] [PubMed]
- Bertholet J, Knopf A, Eiben B, McClelland J, Grimwood A, Harris E, Menten M, Poulsen P, Nguyen DT, Keall P, Oelfke U. Real-time intrafraction motion monitoring in external beam radiotherapy. Phys Med Biol 2019;64:15TR01. [Crossref] [PubMed]
- Shirato H, Shimizu S, Shimizu T, Nishioka T, Miyasaka K. Real-time tumour-tracking radiotherapy. Lancet 1999;353:1331-2. [Crossref] [PubMed]
- Kuo JS, Yu C, Petrovich Z, Apuzzo ML. The CyberKnife stereotactic radiosurgery system: description, installation, and an initial evaluation of use and functionality. Neurosurgery 2003;53:1235-9; discussion 1239. [Crossref] [PubMed]
- Qin G, Qin J, Xia Q, Zou J, Lin P, Ren C, Wang R. Dynamic Target Tracking Method Based on Medical Imaging. Front Physiol 2022;13:894282. [Crossref] [PubMed]
- Li L, Hu Z, Huang Y, Zhu W, Wang Y, Chen M, Yu J. Automatic multi-plaque tracking and segmentation in ultrasonic videos. Med Image Anal 2021;74:102201. [Crossref] [PubMed]
- Bergholm F. Edge focusing. IEEE Trans Pattern Anal Mach Intell 1987;9:726-41. [Crossref] [PubMed]
- Campbell WG, Miften M, Jones BL. Automated target tracking in kilovoltage images using dynamic templates of fiducial marker clusters. Med Phys 2017;44:364-74.
- Jiao L, Wang D, Bai Y, Chen P, Liu F. Deep Learning in Visual Tracking: A Review. IEEE Trans Neural Netw Learn Syst 2023;34:5497-516. [Crossref] [PubMed]
- Marvasti-Zadeh SM, Cheng L, Ghanei-Yakhdan H, Kasaei S. Deep Learning for Visual Tracking: A Comprehensive Survey. IEEE Transactions on Intelligent Transportation Systems 2022;23:3943-68.
- Li R, Mok E, Chang DT, Daly M, Loo BW Jr, Diehn M, Le QT, Koong A, Xing L. Intrafraction verification of gated RapidArc by using beam-level kilovoltage X-ray images. Int J Radiat Oncol Biol Phys 2012;83:e709-15. [Crossref] [PubMed]
- van der Horst A, Wognum S, Dávila Fajardo R, de Jong R, van Hooft JE, Fockens P, van Tienhoven G, Bel A. Interfractional position variation of pancreatic tumors quantified using intratumoral fiducial markers and daily cone beam computed tomography. Int J Radiat Oncol Biol Phys 2013;87:202-8. [Crossref] [PubMed]
- Li G. Advances and potential of optical surface imaging in radiotherapy. Phys Med Biol 2022;67: [Crossref] [PubMed]
- Keall PJ, Mageras GS, Balter JM, Emery RS, Forster KM, Jiang SB, Kapatoes JM, Low DA, Murphy MJ, Murray BR, Ramsey CR, Van Herk MB, Vedam SS, Wong JW, Yorke E. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med Phys 2006;33:3874-900. [Crossref] [PubMed]
- Heinzerling JH, Anderson JF, Papiez L, Boike T, Chien S, Zhang G, Abdulrahman R, Timmerman R. Four-dimensional computed tomography scan analysis of tumor and organ motion at varying levels of abdominal compression during stereotactic treatment of lung and liver. Int J Radiat Oncol Biol Phys 2008;70:1571-8. [Crossref] [PubMed]
- Wong JW, Sharpe MB, Jaffray DA, Kini VR, Robertson JM, Stromberg JS, Martinez AA. The use of active breathing control (ABC) to reduce margin for breathing motion. Int J Radiat Oncol Biol Phys 1999;44:911-9. [Crossref] [PubMed]
- Remouchamps VM, Vicini FA, Sharpe MB, Kestin LL, Martinez AA, Wong JW. Significant reductions in heart and lung doses using deep inspiration breath hold with active breathing control and intensity-modulated radiation therapy for patients treated with locoregional breast irradiation. Int J Radiat Oncol Biol Phys 2003;55:392-406. [Crossref] [PubMed]
- Hostettler A, Nicolau SA, Forest C, Soler L, Remond Y. Real Time Simulation of Organ Motions Induced by Breathing: First Evaluation on Patient Data. In: Harders M, Székely G. editors. Biomedical Simulation. Berlin, Heidelberg: Springer Berlin Heidelberg, 2006.
- Chen T, Qin S, Xu X, Jabbour SK, Haffty BG, Yue NJ. Frequency filtering based analysis on the cardiac induced lung tumor motion and its impact on the radiotherapy management. Radiother Oncol 2014;112:365-70. [Crossref] [PubMed]
- LeCun Y, Kavukcuoglu K, Farabet C. Convolutional networks and applications in vision. Proceedings of 2010 IEEE International Symposium on Circuits and Systems. Paris: IEEE, 2010.
- Lu J, Jin R, Song E, Ma G, Wang M. Lung-CRNet: A convolutional recurrent neural network for lung 4DCT image registration. Med Phys 2021;48:7900-12. [Crossref] [PubMed]
- Xiao H, Teng X, Liu C, Li T, Ren G, Yang R, Shen D, Cai J. A review of deep learning-based three-dimensional medical image registration methods. Quant Imaging Med Surg 2021;11:4895-916. [Crossref] [PubMed]
- Fu Y, Lei Y, Wang T, Higgins K, Bradley JD, Curran WJ, Liu T, Yang X. LungRegNet: An unsupervised deformable image registration method for 4D-CT lung. Med Phys 2020;47:1763-74. [Crossref] [PubMed]
- Liu X, Li KW, Yang R, Geng LS. Review of Deep Learning Based Automatic Segmentation for Lung Cancer Radiotherapy. Front Oncol 2021;11:717039. [Crossref] [PubMed]
- Liu Y, Qin C, Yu Z, Yang R, Suqing T, Liu X, Ma X. Double-branch U-Net for multi-scale organ segmentation. Methods 2022;205:220-5. [Crossref] [PubMed]
- Wu W, Lei R, Niu K, Yang R, He Z. Automatic segmentation of colon, small intestine, and duodenum based on scale attention network. Med Phys 2022;49:7316-26. [Crossref] [PubMed]
- Zhang S, Wang H, Tian S, Zhang X, Li J, Lei R, Gao M, Liu C, Yang L, Bi X, Zhu L, Zhu S, Xu T, Yang R. A slice classification model-facilitated 3D encoder-decoder network for segmenting organs at risk in head and neck cancer. J Radiat Res 2021;62:94-103. [Crossref] [PubMed]
- Liu X, Yang R, Xiong T, Yang X, Li W, Song L, Zhu J, Wang M, Cai J, Geng L. CBCT-to-CT Synthesis for Cervical Cancer Adaptive Radiotherapy via U-Net-Based Model Hierarchically Trained with Hybrid Dataset. Cancers (Basel) 2023;15:5479. [Crossref] [PubMed]
- Yu B, Wang Y, Wang L, Shen D, Zhou L. Medical Image Synthesis via Deep Learning. Adv Exp Med Biol 2020;1213:23-44. [Crossref] [PubMed]
- Spadea MF, Maspero M, Zaffino P, Seco J. Deep learning based synthetic-CT generation in radiotherapy and PET: A review. Med Phys 2021;48:6537-66. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing, 2015.
- Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y. Generative adversarial nets. Proceedings of the 27th International Conference on Neural Information Processing Systems - Volume 2. Cambridge, MA, USA: MIT Press, 2014:2672-80.
- Mikolov T, Joulin A, Chopra S, Mathieu M, Ranzato MA. Learning Longer Memory in Recurrent Neural Networks. arXiv 2014. arXiv:1412.7753.
- Girshick R, Donahue J, Darrell T, Malik J. Rich Feature Hierarchies for Accurate Object Detection and Semantic Segmentation. 2014 IEEE Conference on Computer Vision and Pattern Recognition. Columbus, OH, USA: IEEE, 2014:580-7.
- Scarselli F, Gori M, Tsoi AC, Hagenbuchner M, Monfardini G. The graph neural network model. IEEE Trans Neural Netw 2009;20:61-80. [Crossref] [PubMed]
- Redmon J, Divvala S, Girshick R, Farhadi A. You Only Look Once: Unified, Real-Time Object Detection. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). Las Vegas, NV, USA: IEEE, 2016:779-88.
- Terunuma T, Sakae T, Hu Y, Takei H, Moriya S, Okumura T, Sakurai H. Explainability and controllability of patient-specific deep learning with attention-based augmentation for markerless image-guided radiotherapy. Med Phys 2023;50:480-94. [Crossref] [PubMed]
- Grama D, Dahele M, van Rooij W, Slotman B, Gupta DK, Verbakel WFAR. Deep learning-based markerless lung tumor tracking in stereotactic radiotherapy using Siamese networks. Med Phys 2023;50:6881-93. [Crossref] [PubMed]
- Lei Y, Tian Z, Wang T, Higgins K, Bradley JD, Curran WJ, Liu T, Yang X. Deep learning-based real-time volumetric imaging for lung stereotactic body radiation therapy: a proof of concept study. Phys Med Biol 2020;65:235003. [Crossref] [PubMed]
- Fu Y, Fan Q, Cai W, Li F, He X, Cuaron J, Cervino L, Moran JM, Li T, Li X. Enhancing the target visibility with synthetic target specific digitally reconstructed radiograph for intrafraction motion monitoring: A proof-of-concept study. Med Phys 2023;50:7791-805. [Crossref] [PubMed]
- Zhou D, Nakamura M, Mukumoto N, Matsuo Y, Mizowaki T. Feasibility study of deep learning-based markerless real-time lung tumor tracking with orthogonal X-ray projection images. J Appl Clin Med Phys 2023;24:e13894. [Crossref] [PubMed]
- Zhou D, Nakamura M, Mukumoto N, Yoshimura M, Mizowaki T. Development of a deep learning-based patient-specific target contour prediction model for markerless tumor positioning. Med Phys 2022;49:1382-90. [Crossref] [PubMed]
- Mylonas A, Booth J, Nguyen DT. A review of artificial intelligence applications for motion tracking in radiotherapy. J Med Imaging Radiat Oncol 2021;65:596-611. [Crossref] [PubMed]
- Zhao W, Shen L, Islam MT, Qin W, Zhang Z, Liang X, Zhang G, Xu S, Li X. Artificial intelligence in image-guided radiotherapy: a review of treatment target localization. Quant Imaging Med Surg 2021;11:4881-94. [Crossref] [PubMed]
- Salari E, Wang J, Wynne J, Chang CW, Yang X. Artificial Intelligence-based Motion Tracking in Cancer Radiotherapy: A Review. arXiv 2023. arXiv:2309.09333.
- Liang Z, Zhou Q, Yang J, Zhang L, Liu D, Tu B, Zhang S. Artificial intelligence-based framework in evaluating intrafraction motion for liver cancer robotic stereotactic body radiation therapy with fiducial tracking. Med Phys 2020;47:5482-9. [Crossref] [PubMed]
- Shao HC, Huang X, Folkert MR, Wang J, Zhang Y. Automatic liver tumor localization using deep learning-based liver boundary motion estimation and biomechanical modeling (DL-Bio). Med Phys 2021;48:7790-805. [Crossref] [PubMed]
- Kim KC, Cho HC, Jang TJ, Choi JM, Seo JK. Automatic detection and segmentation of lumbar vertebrae from X-ray images for compression fracture evaluation. Comput Methods Programs Biomed 2021;200:105833. [Crossref] [PubMed]
- He X, Cai W, Li F, Fan Q, Zhang P, Cuaron JJ, Cerviño LI, Li X, Li T. Decompose kV projection using neural network for improved motion tracking in paraspinal SBRT. Med Phys 2021;48:7590-601. [Crossref] [PubMed]
- Peng T, Jiang Z, Chang Y, Ren L. Real-time Markerless Tracking of Lung Tumors based on 2-D Fluoroscopy Imaging using Convolutional LSTM. IEEE Trans Radiat Plasma Med Sci 2022;6:189-99. [Crossref] [PubMed]
- Hirai R, Sakata Y, Tanizawa A, Mori S. Real-time tumor tracking using fluoroscopic imaging with deep neural network analysis. Phys Med 2019;59:22-9. [Crossref] [PubMed]
- Wei R, Zhou F, Liu B, Bai X, Fu D, Li Y, Liang B, Wu Q. Convolutional Neural Network (CNN) Based Three Dimensional Tumor Localization Using Single X-Ray Projection. IEEE Access 2019;7:37026-38.
- Wei R, Zhou F, Liu B, Bai X, Fu D, Liang B, Wu Q. Real-time tumor localization with single x-ray projection at arbitrary gantry angles using a convolutional neural network (CNN). Phys Med Biol 2020;65:065012. [Crossref] [PubMed]
- Takahashi W, Oshikawa S, Mori S. Real-time markerless tumour tracking with patient-specific deep learning using a personalised data generation strategy: proof of concept by phantom study. Br J Radiol 2020;93:20190420. [Crossref] [PubMed]
- Motley R, Ramachandran P, Fielding A. A feasibility study on the development and use of a deep learning model to automate real-time monitoring of tumor position and assessment of interfraction fiducial marker migration in prostate radiotherapy patients. Biomed Phys Eng Express 2022;8: [Crossref] [PubMed]
- Lei Y, Tian Z, Qiu R, Wang T, Roper J, Higgins K, Bradley JD, Liu T, Yang X. Deep-learning-based markerless tumor localization using 2D KV/MV image. SPIE Medical Imaging. 2022. doi: https://doi.org/
10.1117/12.2611823 . - Ahmed AM, Gargett M, Madden L, Mylonas A, Chrystall D, Brown R, Briggs A, Nguyen T, Keall P, Kneebone A, Hruby G, Booth J. Evaluation of deep learning based implanted fiducial markers tracking in pancreatic cancer patients. Biomed Phys Eng Express 2023;9: [Crossref] [PubMed]
- Dai J, Dong G, Zhang C, He W, Liu L, Wang T, Jiang Y, Zhao W, Zhao X, Xie Y, Liang X. Volumetric tumor tracking from a single cone-beam X-ray projection image enabled by deep learning. Med Image Anal 2024;91:102998. [Crossref] [PubMed]
- Zhao W, Shen L, Han B, Yang Y, Cheng K, Toesca DAS, Koong AC, Chang DT, Xing L. Markerless Pancreatic Tumor Target Localization Enabled By Deep Learning. Int J Radiat Oncol Biol Phys 2019;105:432-9. [Crossref] [PubMed]
- Zhao W, Shen L, Wu Y, Han B, Yang Y, Xing L. Automatic marker-free target positioning and tracking for image-guided radiotherapy and interventions. SPIE Medical Imaging. 2019. doi: https://doi.org/
10.1117/12.2512166 . - Roggen T, Bobic M, Givehchi N, Scheib SG. Deep Learning model for markerless tracking in spinal SBRT. Phys Med 2020;74:66-73. [Crossref] [PubMed]
- Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, Zitnick CL. Microsoft COCO: Common Objects in Context. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T. editors. Computer Vision – ECCV 2014. Cham: Springer International Publishing, 2014.
- Wang C, Hunt M, Zhang L, Rimner A, Yorke E, Lovelock M, Li X, Li T, Mageras G, Zhang P. Technical Note: 3D localization of lung tumors on cone beam CT projections via a convolutional recurrent neural network. Med Phys 2020;47:1161-6. [Crossref] [PubMed]
- Shao HC, Wang J, Bai T, Chun J, Park JC, Jiang S, Zhang Y. Real-time liver tumor localization via a single x-ray projection using deep graph neural network-assisted biomechanical modeling. Phys Med Biol 2022;67: [Crossref] [PubMed]
- Zhao W, Han B, Yang Y, Buyyounouski M, Hancock SL, Bagshaw H, Xing L. Incorporating imaging information from deep neural network layers into image guided radiation therapy (IGRT). Radiother Oncol 2019;140:167-74. [Crossref] [PubMed]
- Ren G, Zhang J, Li T, Xiao H, Cheung LY, Ho WY, Qin J, Cai J. Deep Learning-Based Computed Tomography Perfusion Mapping (DL-CTPM) for Pulmonary CT-to-Perfusion Translation. Int J Radiat Oncol Biol Phys 2021;110:1508-18. [Crossref] [PubMed]
- Li W, Xiao H, Li T, Ren G, Lam S, Teng X, Liu C, Zhang J, Kar-Ho Lee F, Au KH, Ho-Fun Lee V, Chang ATY, Cai J. Virtual Contrast-Enhanced Magnetic Resonance Images Synthesis for Patients With Nasopharyngeal Carcinoma Using Multimodality-Guided Synergistic Neural Network. Int J Radiat Oncol Biol Phys 2022;112:1033-44. [Crossref] [PubMed]
- Li W, Lam S, Wang Y, Liu C, Li T, Kleesiek J, Cheung AL, Sun Y, Lee FK, Au KH, Lee VH, Cai J. Model Generalizability Investigation for GFCE-MRI Synthesis in NPC Radiotherapy Using Multi-Institutional Patient-Based Data Normalization. IEEE J Biomed Health Inform 2024;28:100-9. [Crossref] [PubMed]
- Wu W, Qu J, Cai J, Yang R. Multiresolution residual deep neural network for improving pelvic CBCT image quality. Med Phys 2022;49:1522-34. [Crossref] [PubMed]
- Zheng S, Yang X, Wang Y, Ding M, Hou W. Unsupervised Cross-Modality Domain Adaptation Network for X-Ray to CT Registration. IEEE J Biomed Health Inform 2022;26:2637-47.
- Keall PJ, Sawant A, Berbeco RI, Booth JT, Cho B, Cerviño LI, Cirino E, Dieterich S, Fast MF, Greer PB, Munck Af Rosenschöld P, Parikh PJ, Poulsen PR, Santanam L, Sherouse GW, Shi J, Stathakis S. AAPM Task Group 264. The safe clinical implementation of MLC tracking in radiotherapy. Med Phys 2021;48:e44-64. [Crossref] [PubMed]
- Yang B, Liu Y, Zhu J, Dai J, Men K. Deep learning framework to improve the quality of cone-beam computed tomography for radiotherapy scenarios. Med Phys 2023;50:7641-53. [Crossref] [PubMed]
- Chen J, Frey EC, He Y, Segars WP, Li Y, Du Y. TransMorph: Transformer for unsupervised medical image registration. Med Image Anal 2022;82:102615. [Crossref] [PubMed]
- Zhao X, Yang T, Li B, Zhang X. SwinGAN: A dual-domain Swin Transformer-based generative adversarial network for MRI reconstruction. Comput Biol Med 2023;153:106513. [Crossref] [PubMed]
- Mongan J, Moy L, Kahn CE Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol Artif Intell 2020;2:e200029. [Crossref] [PubMed]


