
Synthesis of virtual monoenergetic images from kilovoltage peak images using wavelet loss enhanced CycleGAN for improving radiomics features reproducibility
Introduction
Radiomics has been applied to extract information from image data as biomarkers for outcome prediction and treatment assessment (1-3). Morphological, intensity-based, textural features, and wavelets features can be extracted and assessed to uncover imaging patterns exceeding the ordinary visual image interpretation by the naked eye (1,4).
In recent years, due to the widespread applications of deep learning in the field of medical imaging, the latest achievements have been incorporated into medical image synthesis and translation involving computed tomography (CT). These works were categorized into 2 main groups: synthesis of CT from other modalities (5-8) and intra-modality synthesis (9-11). Many works have indicated that image quality or construction configuration have a significant influence on radiomic feature values (12,13). The reproducibility of radiomics is sensitive to scanning parameters such as kilovoltage (kV), tube current (mAs), slice thickness, spatial resolution, and image reconstruction algorithm. The standardization of imaging protocols is a solution to overcome the challenge of reproducibility in radiomics (13). However, this idea requires more manual intervention and has not yet provided widely-used guidelines for the clinical applications of scanning protocols. Recently, several works have suggested the promising effectiveness of image synthesis or conversion through deep learning methods to improve the reproducibility and accuracy of radiomic features. A convolutional neural network (CNN) was employed to convert CT images with different reconstruction kernels for improving radiomics reproducibility (14). Generative adversarial networks (GAN) have been applied to standardize radiomics features extracted from images obtained from imaging devices produced by different manufacturers (15) or devices of different hospitals (16). The influence of 4-dimensional cone beam CT (4D-CBCT) image quality on radiomics has been investigated and the temporally coherent GANs for video super-resolution (TecoGAN) has been applied to enhance 4D-CBCT for improving radiomics accuracy (17). In addition, the impact of CT image slice thickness on the radiomic features’ reproducibility has also been studied (18). A super-resolution approach based on a CNN model was proposed to synthesize the CT images with slice thickness of 1 mm from that of 3 and 5 mm.
Dual-energy CT (DECT), also known as spectral, multi-energy, or polychromatic CT, employs dual image acquisition at different kV levels of the same scan volume to enable material separation (19). Virtual monoenergetic images (VMIs) from DECT are incrementally used in routine clinical practice (20). Recently, DECT images have been used to produce imaging features for radiomics analysis in several publications (21,22). Due to the difference of reconstruction approaches and parameters, conventional CT images and VMI may have distinct pixel distributions. Distributions of pixels in various VMI may also differ from one another. The VMI produced by the DECT can provide different image contrasts: low energy (40–60 keV) for high soft tissue contrast and iodine attenuation; high energy (120–200 keV) to depress beam hardening and metal artifacts (23-25). Therefore, different levels of VMI obtained from 1 scan may provide different values of the same radiomics feature by DECT device. This facilitates the extraction of different dimensions of information from the same image, thus contributing to radiomics analysis. Existing DECT acquisition techniques include dual-source, fast voltage switching, dual-layer, and dual-spiral (25).
Several deep learning approaches have been employed to address VMI of DECT. ResNet was used to produce virtual monoenergetic CT from polyenergetic (single-spectrum) CT (26). Conditional generative adversarial networks (cGAN) can synthesize pseudo low monoenergetic CT images from a single-tube voltage CT scanner (27). Material decomposition in DECT from a kilovoltage CT could be performed through deep convolutional generative adversarial networks (DCGAN) (28). CNN has been used to estimate DECT images from single-energy CT (SECT) (29).
In this study, we compared radiomics feature reproducibility between ground-truth VMI, conventional SECT, and our generated synthetic VMI (sVMI). sVMIs were generated from SECT using our proposed wavelet loss-enhanced CycleGAN approach and evaluated against ground-truth VMI for improvement in precision of radiomics analysis. Our main contributions of this work are the following:
- The impact of VMI on radiomics features was investigated. Radiomics feature values were extracted from the region of interest (ROI) of conventional CT images and VMIs. The concordance correlation coefficient was used to evaluate the difference.
- The CycleGAN model was employed to improve radiomics reproducibility against virtual mono energy. The conventional CT images were fed to the network and then monoenergetic images were synthesized.
- We defined the wavelet loss of 2 images to evaluate the high-frequency detailed differences. The proposed wavelet loss was incorporated into the CycleGAN model to enhance the synthesis performance.
Methods
Datasets
We retrospectively collected image data from 45 patients obtained using detector-based dual-layer DECT scanner of the IQon Spectral CT System (Philips Healthcare, Cleveland, OH, USA). For each patient, we obtained pretreatment VMIs at various energy levels and conventional CT images under the standard tube-voltage setting. The following CT protocols were used for all image acquisitions: tube voltage, 120 kilovoltage peak (kVp); slice thickness, 3 mm; pixel spacing, 0.8 mm; protocol name, Chest + ABD + C/Thorax. The collected data is described in Table 1.
Table 1
| Variables | Values (n=45) |
|---|---|
| Sex | |
| Men | 37 (82.2) |
| Women | 8 (17.8) |
| Age (years) | 67.4±9.2 |
| ROI volume (mm3) | 22.9±18.1 |
| ROI surface area (mm2) | 53.5±28.0 |
| Primary site | |
| Upper thoracic | 16 |
| Middle thoracic | 23 |
| Lower thoracic | 11 |
| Clinical stage | |
| II | 4 |
| III | 38 |
| IV | 3 |
| T stage | |
| T1 | 1 |
| T2 | 4 |
| T3 | 12 |
| T4 | 28 |
| N stage | |
| N0 | 15 |
| N1 | 13 |
| N2 | 16 |
| N3 | 1 |
| M stage | |
| M0 | 43 |
| M1 | 2 |
Unless otherwise stated, data are the number of patients; data in parentheses are percentages. ROI, region of interest.
The ROIs were segmented and delineated manually by the physician using the open-source medical image software 3D Slicer (version 4.10.2) (30).
VMIs are produced from the original projection images using the material decomposition algorithm. Therefore, VMIs at various keV levels and conventional CT images are pixel-wise corresponding for each patient and image registration is not required as a preprocessing operation.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shandong Cancer Hospital and Institute, and the requirement for individual consent for this retrospective analysis was waived.
Deep learning system/pipeline
The basic purpose was to evaluate the effectiveness of deep learning model for improving feature reproducibility between VMIs (kVp images) and conventional CT images. We trained the CycleGAN-based deep learning model to convert conventional CT images to VMIs, and vice versa.
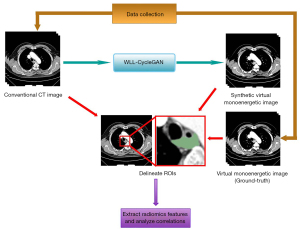
The workflow of the study consists of 5 steps (see Figure 1):
- Data collection: acquire detector-based dual-layer spectral CT images including conventional kVp images and VMIs.
- The correlation coefficients of radiomics features between conventional CT images and VMIs at various energy levels were computed to investigate the impact of material decomposition in detector-based dual-energy spectral CT on radiomic features.
- Image conversion employing CycleGAN: conventional CT images were served as input, and the corresponding VMIs were served as output.
- Radiomics extraction: radiomic features were extracted from the ROI of each patient.
- Performance evaluation: intensity histograms, difference image, and sVMIs were compared to radiomic features extracted from VMIs.
CycleGAN-based deep learning network
The network architecture
A classical GAN model uses the generator and discriminator networks competing with each other. In a study (31), researchers developed the CycleGAN model to perform translation in 2 directions between 2 image domains. In this work, we leveraged the CycleGAN with paired loss proposed in previous research (32,33) as our benchmark model to learn the mapping between images with different energetic configurations. Furthermore, we defined the wavelet loss of 2 images and the proposed wavelet loss item was incorporated into the benchmark model to enhance the effect.
Energy-spectrum CT images, including both conventional CT images and VMIs, are reconstructed from the same projection data acquired from a single scan (34). Unlike conventional CT images, the reconstruction process of VMIs requires material decomposition algorithms (35,36). There is a pixel-wise correspondence between a series of VMIs and conventional (kVp) images. Therefore, paired data training is available in our work.
The cycle and synthesis loss
The total cycle and synthetic consistency loss function L(cyc+syn) used in the proposed model is represented as:
where Gcon-VM is the generator converting conventional CT images to VMIs, GVM−con denotes the generator converting VMIs to conventional CT images, Icon denotes conventional CT images, IVM denotes VMIs, λMPL is the weighting coefficient of mean pixel loss (MPL), and λGML is the coefficient of gradient magnitude loss (GML) (32) item.
The losses in equation (1) are defined as follows:
where Icon is conventional CT images, IVM is VMIs; gradient magnitude distance (GMD) (32) denotes the Euclidean distance of image gradients.
The wavelet loss
The wavelet domain images produced by wavelet transform can depict the details multi-level frequency components of the original images. We proposed the wavelet loss (WLL) to improve the performance of the model.
First of all, the sub bands are computed by the discrete wavelet transformation (DWT).
where I denote the original image, A, V, H and D denote the sub-bands containing wavelet coefficients for average, vertical, horizontal, and diagonal details, respectively.
The wavelet loss between 2 images is defined as:
where Irdenotes the real image, Is denotes the synthetic image, SBi(Ir) denotes the sub-brand of the real image, SBi(Is) denotes the sub-brand of the synthetic image, MSE denotes the mean square error operator of 2 images, and BatNor is the batch normalization operator using the min-max method, defined as follows:
where A and B are the images to be normalized, A’ and B’ are the normalized images. The normalization process including the following steps:
where pmax denotes the maximal pixel value of the 2 input images, pmin denotes the minimal pixel value of the 2 input images, and i and j denote the coordinates of the pixels in the image.
The computation of wavelet loss includes 5 steps:
- Discrete wavelet transform is applied on the real images and synthetic images to generate 4 wavelet sub-bands images.
- The images of the same sub-band are grouped in 1 group.
- Normalization is performed on intragroup images in each group. Pixel values in different sub-bands groups have large difference. For optimizing loss from each sub-band uniformly, intragroup normalization is performed. Meanwhile, intragroup normalization can reserve the difference between the sub-bands image deduced from real and synthetic images.
- The mean square error is computed for normalized intragroup images.
- The sum of all 4 sub-bands groups is obtained as the wavelet loss item. The overall process of obtaining WLL is revealed in Figure 2.
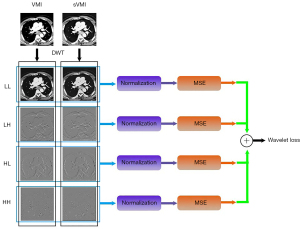
The wavelet loss is incorporated into the framework of synthetic consistency and cycle consistency.
where WLLs denotes the synthetic consistency of WLL, WLLcdenotes the cycle consistency of WLL, and WLLs+c denotes the sum of WLLs and WLLc.
The total cycle and synthetic consistency loss function in equation 1 will be extended to the new form as:
The adversarial loss
The adversarial loss function, depending on the output of the discriminators, applies to both the Con-to-VMI generator and the VMI-to-Con generator. The adversarial loss for both directions is represented as follows:
where MSE[•, 1] is the mean square error between the discriminator map of the sVMI and a unit mask.
The total loss
The total loss function of the model consists of the generator loss and the discriminator loss, defined as:
where λ(cyc+syn) is the coefficient of the cycle and synthetic consistency loss. We aim to solve:
GSECT−VMI, GVMI-SECT are updated to minimize this objective, whereas DVMI, DSECT attempt to maximize it.
Implementation and configuration of parameters
The model was implemented in Python 3.6 (37) and Tensorflow r1.3 (38) framework. The version of used CUDA was 10.0.
Hyperparameter values in the loss function were set as 10 for , 1 for , 1 for , 1 for , 1 for λMPL, 1 for λGML, 1 for λWLL, and 1 for λ(cyc+syn). The initial learning rate was set to be 0.0002 for both the generator and the descriminator. The Adam optimizer with beta1=0.5 and beta2=0.999 was employed to optimize both the generator and the descriminator.
Experiment design
Radiomics feature extraction
In this study, radiomics extraction included 4 steps: (I) collect the images; (II) delineate and segment the ROIs; (III) preprocessing; (IV) feature extraction.
All radiomics features were extracted using the PyRadiomics package (39). For each patient, conventional CT images and various VMIs were reconstructed and obtained. The extracted features included 18 first order features, 22 gray-level co-occurrence matrix (GLCM) features (40), 14 gray-level difference matrix (GLDM) features, 16 gray-level run-length matrix (GLRLM) features (41), 16 gray-level size-zone matrix (GLSZM) features (42), and 5 neighborhood gray-tone difference matrix (NGTDM) features (43). Since ROI contour in various monoenergetic images for each patient is the same, the shape features were not considered. The settings of imaging for radiomics extraction are listed as Table 2.
Table 2
| Setting item | Value |
|---|---|
| Scanner type | IQon Spectral CT System (Vendor: Philips Healthcare, Cleveland) |
| Acquisition protocol | SD Chest + ABD + C/Thorax |
| Tube voltage, kVp | 120 |
| Tube current, mA | 359 |
| Pixel spacing, mm | 0.86 |
| Image slice thickness, mm | 3 |
| Convolution kernel | B |
| Exposure, ms | 493 |
| Interpolation method | BSpline |
| Discretization: width of the bins, gray level | 25 |
| CM symmetry | Symmetry |
| CM distance, pixel | 1 |
| SZM linkage distance, pixel | 1 |
| NGTDM distance, pixel | 1 |
| Software availability | PyRadiomics version: 3.0.1 |
CM, co-occurrence matrix; SZM, size-zone matrix; NGTDM, neighborhood gray-tone difference matrix; kVp, kilovoltage peak.
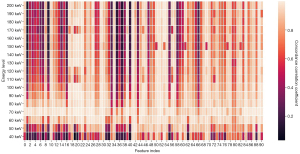
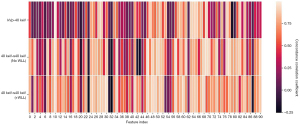
Figure 3 shows the concordance correlation coefficient between conventional CT images and different VMIs. Each cell represents the concordance correlation coefficient between radiomics features of conventional CT images and VMIs.
Evaluation metrics
Our proposed method was compared with the pix2pix (44) model and the CycleGAN model using several evaluation metrics. After training, we evaluated the performance of the model through comparing the radiomics feature values extracted of synthetic images and source images. The first order, GLCM, GLDM, GLRLM, GLSZM, NGTDM, and wavelet from the whole body and ROI of conventional CT images, VMIs, and synthetic virtual monoenergetic CT images were compared.
For quantitative evaluation, the error of each feature was defined as:
The concordance correlation coefficient (CCC) was defined as:
where µx and µy are the means for the two variables and σx2 and σy2 are the corresponding variances. ρ is the correlation coefficient between the 2 variables.
Results
Image evaluation
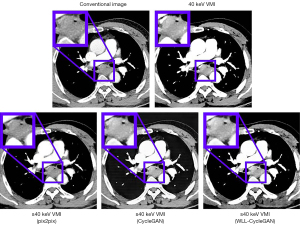
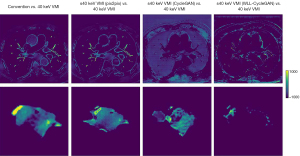
Conventional CT image, synthetic 40 keV VMIs, and 40 keV monoenergetic images are displayed in Figure 4. The results indicate that the CycleGAN model can synthesize approximately 40 keV monoenergetic images with high contrast and less streak artifacts from corresponding conventional kVp images. The difference images of conventional CT image versus 40 keV image and synthetic 40 keV image versus 40 keV image are displayed in Figure 5. The top line shows the result of the whole body, whereas the bottom line shows the result of the ROI of the tumor. These results suggests that the deep learning model converts original conventional CT image pixel values to 40 keV pixel values in both ROI and whole body accurately.
Standard evaluation measures were produced for image quality comparison with within ROIs and entire body contour. The results, including the mean values and standard deviations of measures, were shown in Table 3 and Table 4. The synthetic 40 keV VMIs generated from the 120 kVp conventional CT images using the proposed CycleGAN-based method were similar to the 40-keV VMIs obtained from the DECT device. Our proposed wavelet loss enhancement significantly improved the quality of the VMIs synthesized by the CycleGAN model.
Table 3
| Comparison item | RMSE | PSNR | SSIM |
|---|---|---|---|
| Conv image vs. 40 keV | 54.75±11.9 | 12.94±2.1 | 0.625±8.3 |
| s40 keV (pix2pix) vs. 40 keV | 43.61±6.1 | 13.58±1.1 | 0.716±4.5 |
| s40 keV (CycleGAN) vs. 40 keV | 38.46±4.8 | 30.41±1.1 | 0.831±2.6 |
| s40 keV (WLL-CycleGAN) vs. 40 keV | 32.03±4.7 | 32.03±1.4 | 0.906±3.1 |
Data format: mean ± standard deviation. CT, computed tomography; VMI, virtual monoenergetic CT images; RMSE, root mean square error; PSNR, peak signal-to-noise ratio; SSIM, structure similarity index measure; s40 keV, synthetic 40 keV; GAN, generative adversarial network; WLL, wavelet loss.
Table 4
| Comparison item | RMSE | PSNR | SSIM |
|---|---|---|---|
| Conv image vs. 40 keV | 92.67±36.1 | 23.78±4.9 | 0.994±0.1 |
| s40 keV (pix2pix) vs. 40 keV | 36.74±3.5 | 30.77±0.8 | 0.997±0.2 |
| s40 keV (CycleGAN) vs. 40 keV | 23.12±1.3 | 34.77±0.5 | 0.998±0.1 |
| s40 keV (WLL-CycleGAN) vs. 40 keV | 21.90±0.7 | 35.23±0.3 | 0.999±0.1 |
Data format: mean ± standard deviation. CT, computed tomography; VMI, virtual monoenergetic CT images; RMSE, root mean square error; PSNR, peak signal-to-noise ratio; SSIM, structure similarity index measure; s40 keV, synthetic 40 keV; GAN, generative adversarial network; WLL, wavelet loss.
The line intensity histograms of conventional CT images, VMI, and sVMIs using various methods are shown in Figure 6. The results suggested that the CycleGAN model can calibrate the Hounsfield unit (HU) values distribution in conventional CT images nearing that of actual 40 keV monoenergetic images. The CT HU distribution of VMI synthesized by the proposed method closely reflects the actual VMI. The histogram line of our method is closer to that of the 40 keV VMI than other methods.
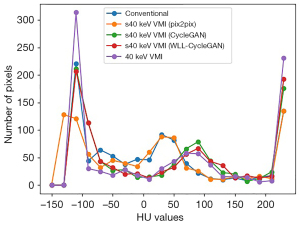
Quantitative evaluation for the comparison of histograms was also produced. We used the root mean square error to evaluate the difference between 2 histograms. The results (Table 5) indicated that the proposed method produced the best effect among all models to improve the reproducibility of radiomics features.
Table 5
| Conventional vs. VMI | s40 keV VMI pix2pix | s40 keV VMI CycleGAN | s40 keV VMI WLL-CycleGAN | |
|---|---|---|---|---|
| RMSE | 58.59 | 83.49 | 28.59 | 28.32 |
GAN, generative adversarial network; s40 keV VMI, synthetic 40 keV virtual monoenergetic image; WLL, wavelet loss; RMSE, root mean square errors.
Radiomics evaluations
The results of CCC passing rate comparison are shown in Table 6. Alternative CCC thresholds were configured as 0.8 and 0.85. The proposed WLL-CycleGAN produced the best results in 4 feature classes (GLCM, GLDM, GLRLM, and wavelet) at both thresholds, whereas CycleGAN obtained the best result in GLSZM at both thresholds and in the first-order feature class at a threshold of 0.85. The heatmap for CCC passing rate comparison is shown in Figure 7. The results suggest that our proposed WLL-CycleGAN outperforms other models.
Table 6
| Feature class | Conv-40 keV | 40 keV-s40 keV (pix2pix) | 40 keV-s40 keV (CycleGAN) | 40 keV-s40 keV (WLL-CycleGAN) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.8 | 0.85 | 0.8 | 0.85 | 0.8 | 0.85 | 0.8 | 0.85 | ||||
| First order | 1 [5.6] | 1 [5.6] | 3 [16.7] | 1 [5.6] | 6 [33.3] | 4 [22] | 6 [33.3] | 2 [11.1] | |||
| GLCM | 2 [9] | 2 [9] | 3 [13.6] | 1 [4.5] | 7 [31.8] | 3 [13] | 9 [40.9] | 8 [36.4] | |||
| GLDM | 2 [14.3] | 2 [14.3] | 0 [0] | 0 [0] | 5 [35.7] | 4 [28.6] | 7 [50] | 7 [50] | |||
| GLRLM | 8 [50] | 5 [31.2] | 0 [0] | 0 [0] | 6 [37.5] | 5 [31.2] | 8 [50] | 7 [43.8] | |||
| GLSZM | 4 [25] | 3 [18.8] | 5 [31.25] | 3 [18.8] | 7 [43.8] | 4 [25] | 4 [25] | 3 [18.8] | |||
| NGTDM | 0 [0] | 0 [0] | 0 [0] | 0 [0] | 1 [20] | 1 [20] | 1 [20] | 1 [20] | |||
| Wavelet | 268 [36.8] | 225 [31] | 130 [17.9] | 123[16.9] | 274 [37.6] | 203 [27.9] | 324 [44.5] | 277 [38] | |||
(I) Numbers are count of radiomic features. Numbers in square brackets are percentages; (II) CCC are used as thresholds. s40 keV, synthetic 40 keV; GAN, generative adversarial network; WLL, wavelet loss; GLCM, gray-level co-occurrence matrix; GLDM, gray-level difference matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size-zone matrix; NGTDM, neighborhood gray-tone difference matrix; CCC, concordance correlation coefficient.
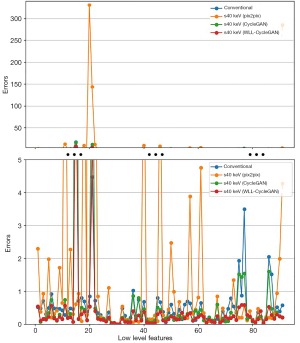
The comparisons of feature value error were revealed in tables and figures. Figure 8 represents the error comparison of low-level texture features and Figure 9 represents error comparison of wavelet-based features. The magnitude of the errors is significantly dependent on the feature. The results of various methods including pix2pix, CycleGAN, and proposed WLL-CycleGAN are displayed in these figures.
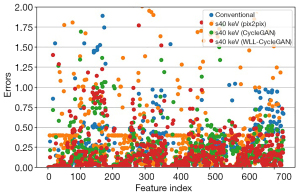
Evaluations for the conversion from conventional CT image to VMIs
Inverse conversion, which involves synthesizing conventional CT images from VMIs, was also conducted in this work. The same image evaluation and radiomics feature evaluation were employed in this scheme.
The standard image evaluations of results of different methods were shown in several tables and figures. Image comparisons with standard evaluation measures are displayed in Table 7. Image and ROI comparison are displayed in Figure 10. Figure 11 shows the image difference comparison among all methods. Histograms of HU values for each case for comparison are displayed in Figure 12 and Table 8 shows the quantitively metrics of the intensity histogram.
Table 7
| Comparison item | RMSE | PSNR | SSIM |
|---|---|---|---|
| 40 keV vs. Conv image | 54.75±11.9 | 12.94±2.1 | 0.625±8.3 |
| sConv (pix2pix) vs. Conv image | 47.80±41.9 | 31.17±6.4 | 0.703±3.1 |
| sConv (CycleGAN) vs. Conv image | 30.44±0.5 | 32.37±0.1 | 0.850±4.5 |
| sConv (WLL-CycleGAN) vs. Conv image | 23.66±0.5 | 34.56±0.2 | 0.894±2.3 |
Data format: mean ± standard deviation. CT, computed tomography; RMSE, root mean square error; PSNR, peak signal-to-noise ratio; SSIM, structure similarity index measure; sConv, synthetic conventional CT image; GAN, generative adversarial network; WLL, wavelet loss.
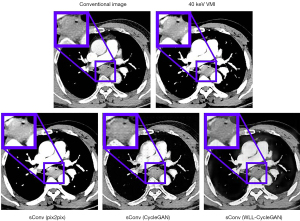
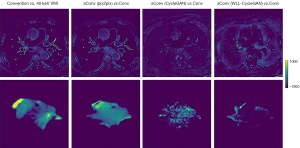
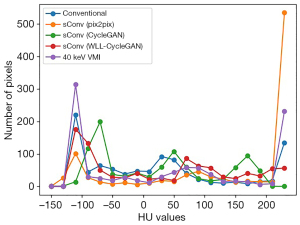
Table 8
| Conventional vs. VMI | sConv VMI pix2pix | sConv VMI CycleGAN | sConv VMI WLL-CycleGAN | |
|---|---|---|---|---|
| RMSE | 58.59 | 98.71 | 70.16 | 42.04 |
VMI, virtual monoenergetic image; sConv, synthetic conventional CT image; GAN, generative adversarial network; WLL, wavelet loss; CT, computed tomography; RMSE, root mean square errors.
The radiomics evaluations are also represented in tables and figures. Figure 13 represents the error comparison of low-level texture features, whereas Figure 14 represents the comparison of wavelet-based features. Table 9 shows the CCC passing rate comparison among methods. The proposed WLL-CycleGAN produced the best results in 3 feature classes (first order, GLCM, and GLSZM) at both thresholds, whereas CycleGAN obtained the best result in GLDM at a threshold of 0.8. All methods performed worse than the baseline level in GLRLM and wavelet at a threshold of 0.8.
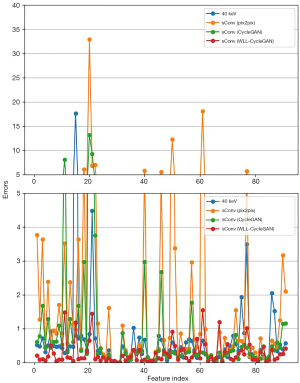
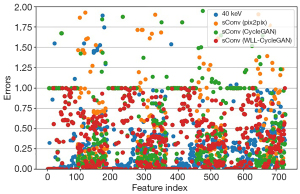
Table 9
| Feature class | Conv-40 keV | Conv-sConv (pix2pix) | Conv-sConv (CycleGAN) | Conv-sConv (WLL-CycleGAN) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.8 | 0.85 | 0.8 | 0.85 | 0.8 | 0.85 | 0.8 | 0.85 | ||||
| First order | 1 [5.6] | 1 [5.6] | 2 [11.1] | 2 [11.1] | 1 [5.6] | 1 [5.5] | 8 [44.4] | 6 [33.3] | |||
| GLCM | 2 [9] | 2 [9] | 3 [13.6] | 2 [9] | 3 [13.6] | 1 [4.5] | 7 [31.8] | 4 [18.2] | |||
| GLDM | 2 [14.3] | 2 [14.3] | 2 [14.3] | 2 [14.3] | 6 [42.9] | 4 [28.6] | 4 [28.6] | 3 [21.4] | |||
| GLRLM | 8 [50] | 5 [31.2] | 4 [25] | 4 [25] | 3 [18.8] | 3 [18.8] | 5 [31.2] | 5 [31.3] | |||
| GLSZM | 4 [25] | 3 [18.8] | 4 [25] | 4 [25] | 4 [25] | 3 [18.8] | 7 [43.8] | 6 [37.5] | |||
| NGTDM | 0 [0] | 0 [0] | 0[0] | 0[0] | 1 [20] | 0 [0] | 2 [40] | 0 [0] | |||
| Wavelet | 268 [36.8] | 225 [31] | 20 [8] | 13 [6] | 212 [30] | 196 [26.9] | 249 [34.2] | 229 [1.5] | |||
(I) Numbers are count of radiomic features. Numbers in square brackets are percentages; (II) CCC are used as thresholds. sConv, synthetic conventional CT images; GAN, generative adversarial network; WLL, wavelet loss; GLCM, gray level co-occurrence matrix; GLDM, gray-level difference matrix; GLRLM, gray-level run-length matrix; GLSZM, gray-level size-zone matrix; NGTDM, neighborhood gray-tone difference matrix; CCC, concordance correlation coefficient; CT, computed tomography.
Discussion
Radiomic analysis has been widely applied to diagnosis and outcome prediction for cancers. Robustness and reproducibility of radiomic features is crucial in radiomics and has attracted the attention of researchers recently.
To address the issue of radiomics reproducibility, several compensation methods have been proposed to improve reproducibility of individual radiomics features (45,46). In contrast, the advantage of deep learning-based image synthesis is to work at an image level. This method can directly correct biases for original pixels. GAN-based deep learning methods are potential techniques for generating medical images from existing ones. Recently, many medical image translation studies have been reported: positron emission tomography (PET)-CT translation, correction of magnetic resonance motion artifacts, CBCT-to-CT translation, PET denoising, metal artifact reduction, and noise reduction from low dose CT (47-51).
In clinical practice, improved contrast attenuation with virtual monoenergetic (VM) imaging at lower kiloelectron volt levels enables better delineation and diagnostic accuracy in the detection of various vascular or oncologic abnormalities. Since providing more information of tissue than single polyenergetic image, monoenergetic images have been employed for radiomics analysis for diagnosis and outcome prediction recently (20-22).
The VMIs produced by DECT can provide different image contrasts: low energy (40–60 keV) for high soft tissue contrast and iodine attenuation; high energy (120–200 keV) to depress beam-hardening and metal artifacts (23-25). Therefore, different levels of VMIs obtained from 1 scan may provide different values of the same radiomics feature from conventional CT images. This facilitates the extraction of different dimensions of information from the same image, thus contributing to radiomics analysis. In other hand, data missing and inconsistency have become barriers in retrospective, longitudinal, or multicenter clinical studies. Deep learning-based image synthesis (translation or conversion) approaches may help to alleviate the above problems.
Conventional CT images, namely, kVp images, are reconstructed from the acquired project data by DECT devices using the conventional reconstruction algorithm. Meanwhile, VMIs are reconstructed from the same project data using the reconstruction algorithm with material decomposition (34). Since the difference between conventional CT images and VMIs is generated by the reconstruction algorithm with material decomposition, different parameters will result in VMIs at varying energy levels.
All VMIs are reconstructed from one dual-energy project data by the 2-layer detector in the same way as the conventional CT image through material decomposition. Therefore, all VMIs and conventional CT images are pixel-wise consistent without position shift. The image registration is not necessary before feeding data into the network (23,52-54). Therefore, the CycleGAN-based model is suitable for these data since the paired reference images are ready-made.
In this paper, we first investigated the impact of VMIs on radiomics features. The radiomics features extracted from both conventional CT images and VMIs were compared. Secondly, we defined the wavelet loss of 2 images to evaluate the high-frequency detailed differences. The proposed wavelet loss was incorporated into the CycleGAN model to synthesize VMIs from conventional CT images. Finally, we evaluated the performance of image conversion using the proposed method and assessed whether it could improve the reproducibility of radiomic features between conventional CT images and VMIs.
Radiomics analysis can obtain internal features of images, whereas DWT can provide structure information of multi-level frequency in original images. The results indicate that the proposed model reduces errors for 84% of low-level features, whereas the original CycleGAN reduces errors for 73% of low-level features, and the pix2pix model reduces errors for 20%. The proposed model reduces errors for 72% of wavelet features, whereas the original CycleGAN reduces errors for 40%, and the pix2pix model reduces errors for 6%. Due to the limitation of computational resource, a 2-dimensional (2D) network model and 2D-DWT were employed for image translating. With the improvement of the image quality of each slice, the robustness of 3D radiomics features are improved. In the future, we can try to use a 3D network model with 3D-DWT to translate image blocks directly.
Although the CycleGAN network was originally designed for unpaired data and unsupervised learning tasks, in several studies, training images are still paired by registration to preserve quantitative pixel values and remove large geometric mismatch to allow the network to focus on mapping details and accelerate training (32,55). Due to the inverse path favoring a one-to-one correspondence between the input and output, the training of the GAN is less affected by mode collapse (55). The cycle consistency may help to reduce the space of possible mapping functions and allows for higher accuracy because the model is doubly constrained (31,32). The above reasons may help to understand the result that the CycleGAN-based model outperforms pix2pix-GAN model.
As shown in Figures 4,5, CycleGAN produces artifacts in synthetic images such as streaking lines, possibly due to the cyclic loss term and some form of mode collapse. Our proposed WLL-CycleGAN method avoids these artifacts by incorporating paired loss terms in addition to the cycle-consistency loss.
There are a few of limitations of this work. First, only the dual-layer spectral DECT of 1 manufacturer (Philips) was considered. Different mechanisms for obtaining VMIs may affect the reproducibility of radiomics features, and the influence should be studied. Second, only the conversion between conventional CT images and VMIs were performed. In the future, the performance for improving radiomics features’ reproducibility of deep learning models translating CT images at any monoenergetic level to another can be evaluated. Third, the effectiveness of the deep learning model for improving radiomics features was validated through only value errors and CCC. The actual effect of synthetic images for radiomics analysis tasks, such as disease prognosis and outcome prediction, should be evaluated.
In the future, we can also investigate the performance of the deep learning model for improving radiomics feature reproducibility in other types of images, such as multi-sequence MRI or PET. We can extend our proposed wavelet-based loss paired CycleGAN to a 3D model.
Conclusions
In this paper, we first explored the reproducibility of radiomic features extracted from conventional CT images and VMIs. The results indicated that radiomics features obtained from VMIs may differ from those obtained from conventional CT images. Secondly, by exploiting the wavelet loss to evaluate the high-frequency detailed differences, the WLL-CycleGAN based deep learning method was developed to translate conventional CT images to VMIs for improving the reproducibility of radiomics features. Our proposed method was shown to be effective for this addressing issue. Compared to pix2pix and CycleGAN, our proposed WLL-CycleGAN produced better performance.
Acknowledgments
Funding: This paper was supported in part by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-922/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shandong Cancer Hospital and Institute, and the requirement for individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Nie K, Al-Hallaq H, Li XA, Benedict SH, Sohn JW, Moran JM, Fan Y, Huang M, Knopp MV, Michalski JM, Monroe J, Obcemea C, Tsien CI, Solberg T, Wu J, Xia P, Xiao Y, El Naqa I. NCTN Assessment on Current Applications of Radiomics in Oncology. Int J Radiat Oncol Biol Phys 2019;104:302-15. [Crossref] [PubMed]
- Touati R, Le WT, Kadoury S. A feature invariant generative adversarial network for head and neck MRI/CT image synthesis. Phys Med Biol 2021; [Crossref]
- Dalmaz O, Yurt M, Cukur T. ResViT: Residual Vision Transformers for Multimodal Medical Image Synthesis. IEEE Trans Med Imaging 2022;41:2598-614. [Crossref] [PubMed]
- Touati R, Kadoury S. Bidirectional feature matching based on deep pairwise contrastive learning for multiparametric MRI image synthesis. Phys Med Biol 2023; [Crossref]
- Touati R, Kadoury S. A least square generative network based on invariant contrastive feature pair learning for multimodal MR image synthesis. Int J Comput Assist Radiol Surg 2023;18:971-9. [Crossref] [PubMed]
- Shan H, Padole A, Homayounieh F, Kruger U, Khera RD, Nitiwarangkul C, Kalra MK, Wang G. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat Mach Intell 2019;1:269-76. [Crossref] [PubMed]
- Kyong Hwan Jin. McCann MT, Froustey E, Unser M. Deep Convolutional Neural Network for Inverse Problems in Imaging. IEEE Trans Image Process 2017;26:4509-22. [Crossref] [PubMed]
- Zhao T, McNitt-Gray M, Ruan D. A convolutional neural network for ultra-low-dose CT denoising and emphysema screening. Med Phys 2019;46:3941-50.
- Traverso A, Wee L, Dekker A, Gillies R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys 2018;102:1143-58. [Crossref] [PubMed]
- Jahanshahi A, Soleymani Y, Fazel Ghaziani M, Khezerloo D. Radiomics reproducibility challenge in computed tomography imaging as a nuisance to clinical generalization: a mini-review. Egypt J Radiol Nucl Med 2023;83:54.
- Choe J, Lee SM, Do KH, Lee G, Lee JG, Lee SM, Seo JB. Deep Learning-based Image Conversion of CT Reconstruction Kernels Improves Radiomics Reproducibility for Pulmonary Nodules or Masses. Radiology 2019;292:365-73. [Crossref] [PubMed]
- Marcadent S, Hofmeister J, Preti MG, Martin SP, Van De Ville D, Montet X. Generative Adversarial Networks Improve the Reproducibility and Discriminative Power of Radiomic Features. Radiol Artif Intell 2020;2:e190035. [Crossref] [PubMed]
- Li Y, Han G, Wu X, Li ZH, Zhao K, Zhang Z, Liu Z, Liang C. Normalization of multicenter CT radiomics by a generative adversarial network method. Phys Med Biol 2021; [Crossref]
- Zhang Z, Huang M, Jiang Z, Chang Y, Torok J, Yin FF, Ren L. 4D radiomics: impact of 4D-CBCT image quality on radiomic analysis. Phys Med Biol 2021;66:045023. [Crossref] [PubMed]
- Park S, Lee SM, Do KH, Lee JG, Bae W, Park H, Jung KH, Seo JB. Deep Learning Algorithm for Reducing CT Slice Thickness: Effect on Reproducibility of Radiomic Features in Lung Cancer. Korean J Radiol 2019;20:1431-40. [Crossref] [PubMed]
- Booz C, Nöske J, Martin SS, Albrecht MH, Yel I, Lenga L, Gruber-Rouh T, Eichler K, D'Angelo T, Vogl TJ, Wichmann JL. Virtual Noncalcium Dual-Energy CT: Detection of Lumbar Disk Herniation in Comparison with Standard Gray-scale CT. Radiology 2019;290:446-55. [Crossref] [PubMed]
- Euler A, Laqua FC, Cester D, Lohaus N, Sartoretti T, Pinto Dos Santos D, Alkadhi H, Baessler B. Virtual Monoenergetic Images of Dual-Energy CT-Impact on Repeatability, Reproducibility, and Classification in Radiomics. Cancers (Basel) 2021;13:4710. [Crossref] [PubMed]
- Han D, Yu Y, He T, Yu N, Dang S, Wu H, Ren J, Duan X. Effect of radiomics from different virtual monochromatic images in dual-energy spectral CT on the WHO/ISUP classification of clear cell renal cell carcinoma. Clin Radiol 2021;76:627.e23-627.e29. [Crossref] [PubMed]
- An C, Li D, Li S, Li W, Tong T, Liu L, Jiang D, Jiang L, Ruan G, Hai N, Fu Y, Wang K, Zhuo S, Tian J. Deep learning radiomics of dual-energy computed tomography for predicting lymph node metastases of pancreatic ductal adenocarcinoma. Eur J Nucl Med Mol Imaging 2022;49:1187-99. [Crossref] [PubMed]
- Albrecht MH, Vogl TJ, Martin SS, Nance JW, Duguay TM, Wichmann JL, De Cecco CN, Varga-Szemes A, van Assen M, Tesche C, Schoepf UJ. Review of Clinical Applications for Virtual Monoenergetic Dual-Energy CT. Radiology 2019;293:260-71. [Crossref] [PubMed]
- Siegel MJ, Kaza RK, Bolus DN, Boll DT, Rofsky NM, De Cecco CN, Foley WD, Morgan DE, Schoepf UJ, Sahani DV, Shuman WP, Vrtiska TJ, Yeh BM, Berland LL. White Paper of the Society of Computed Body Tomography and Magnetic Resonance on Dual-Energy CT, Part 1: Technology and Terminology. J Comput Assist Tomogr 2016;40:841-5.
- Paganetti H, Beltran C, Both S, Dong L, Flanz J, Furutani K, Grassberger C, Grosshans DR, Knopf AC, Langendijk JA, Nystrom H, Parodi K, Raaymakers BW, Richter C, Sawakuchi GO, Schippers M, Shaitelman SF, Teo BKK, Unkelbach J, Wohlfahrt P, Lomax T. Roadmap: proton therapy physics and biology. Phys Med Biol 2021; [Crossref]
- Cong W, Xi Y, Fitzgerald P, De Man B, Wang G. Virtual Monoenergetic CT Imaging via Deep Learning. Patterns (N Y) 2020;1:100128. [Crossref] [PubMed]
- Funama Y, Oda S, Kidoh M, Nagayama Y, Goto M, Sakabe D, Nakaura T. Conditional generative adversarial networks to generate pseudo low monoenergetic CT image from a single-tube voltage CT scanner. Phys Med 2021;83:46-51. [Crossref] [PubMed]
- Kawahara D, Saito A, Ozawa S, Nagata Y. Image synthesis with deep convolutional generative adversarial networks for material decomposition in dual-energy CT from a kilovoltage CT. Comput Biol Med 2021;128:104111. [Crossref] [PubMed]
- Lyu T, Zhao W, Zhu Y, Wu Z, Zhang Y, Chen Y, Luo L, Li S, Xing L. Estimating dual-energy CT imaging from single-energy CT data with material decomposition convolutional neural network. Med Image Anal 2021;70:102001. [Crossref] [PubMed]
- Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In: Jolesz F. editor. Intraoperative Imaging and Image-Guided Therapy. Springer, New York, NY, 2024.
- Zhu JY, Park T, Isola P, Efros AA. Unpaired image-to-image translation using cycle-consistent adversarial networks. IEEE International Conference on Computer Vision (ICCV), Venice, Italy, 2017:2223-32.
- Harms J, Lei Y, Wang T, Zhang R, Zhou J, Tang X, Curran WJ, Liu T, Yang X. Paired cycle-GAN-based image correction for quantitative cone-beam computed tomography. Med Phys 2019;46:3998-4009. [Crossref] [PubMed]
- Liu Y, Lei Y, Wang T, Fu Y, Tang X, Curran WJ, Liu T, Patel P, Yang X. CBCT-based synthetic CT generation using deep-attention cycleGAN for pancreatic adaptive radiotherapy. Med Phys 2020;47:2472-83. [Crossref] [PubMed]
- Philips, “Philips Healthcare Spectral CT.” 2023. Available online: https://www.philips.com.tw/healthcare/sites/spectral-ct-learning-center
- Chen KM. Principles and clinical applications of spectral CT imaging. Science 2012. (in Chinese). Available online: https://book.sciencereading.cn/shop/book/Booksimple/show.do?id=B9759E7063FB34D34A47A514F466D5789000
- Johnson TR. Dual-energy CT: general principles. AJR Am J Roentgenol 2012;199:S3-8. [Crossref] [PubMed]
Foundation P. S. 2021 . Available online: https://www.python.org- Google, Tensorflow, 2021. Available online: https://www.tensorflow.org
- Pyradiomics,
2020 . Available online: https://www.radiomics.io/pyradiomics.html - Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. IEEE Transactions on Systems, Man, and Cybernetics 1973;6:610-21.
- Galloway MM. Texture analysis using grey level run lengths. STIN 1974;75:18555.
- Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng 2014;61:630-7. [Crossref] [PubMed]
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Transactions on Systems, Man, and Cybernetics 1989;19:1264-74.
- Isola P, Zhu JY, Zhou T, Efros AA. Image-to-image translation with conditional adversarial networks. IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 2017:1125-34.
- O'Connor JP, Aboagye EO, Adams JE, Aerts HJ, Barrington SF, Beer AJ, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017;14:169-86. [Crossref] [PubMed]
- Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, Soussan M, Frouin F, Frouin V, Buvat I. A Postreconstruction Harmonization Method for Multicenter Radiomic Studies in PET. J Nucl Med 2018;59:1321-8. [Crossref] [PubMed]
- Yi X, Walia E, Babyn P. Generative adversarial network in medical imaging: A review. Med Image Anal 2019;58:101552.
- Dar SU, Yurt M, Karacan L, Erdem A, Erdem E, Cukur T. Image Synthesis in Multi-Contrast MRI With Conditional Generative Adversarial Networks. IEEE Trans Med Imaging 2019;38:2375-88. [Crossref] [PubMed]
- Armanious K, Jiang C, Fischer M, Küstner T, Hepp T, Nikolaou K, Gatidis S, Yang B. MedGAN: Medical image translation using GANs. Comput Med Imaging Graph 2020;79:101684. [Crossref] [PubMed]
- Chen L, Liang X, Shen C, Nguyen D, Jiang S, Wang J. Synthetic CT generation from CBCT images via unsupervised deep learning. Phys Med Biol 2021; [Crossref]
- Zhang Y, Yue N, Su MY, Liu B, Ding Y, Zhou Y, Wang H, Kuang Y, Nie K. Improving CBCT quality to CT level using deep learning with generative adversarial network. Med Phys 2021;48:2816-26. [Crossref] [PubMed]
- Grant KL, Flohr TG, Krauss B, Sedlmair M, Thomas C, Schmidt B. Assessment of an advanced image-based technique to calculate virtual monoenergetic computed tomographic images from a dual-energy examination to improve contrast-to-noise ratio in examinations using iodinated contrast media. Invest Radiol 2014;49:586-92. [Crossref] [PubMed]
- Albrecht MH, Trommer J, Wichmann JL, Scholtz JE, Martin SS, Lehnert T, Vogl TJ, Bodelle B. Comprehensive Comparison of Virtual Monoenergetic and Linearly Blended Reconstruction Techniques in Third-Generation Dual-Source Dual-Energy Computed Tomography Angiography of the Thorax and Abdomen. Invest Radiol 2016;51:582-90.
- Albrecht MH, Scholtz JE, Hüsers K, Beeres M, Bucher AM, Kaup M, Martin SS, Fischer S, Bodelle B, Bauer RW, Lehnert T, Vogl TJ, Wichmann JL. Advanced image-based virtual monoenergetic dual-energy CT angiography of the abdomen: optimization of kiloelectron volt settings to improve image contrast. Eur Radiol 2016;26:1863-70. [Crossref] [PubMed]
- Zhou L, Schaefferkoetter JD, Tham IWK, Huang G, Yan J. Supervised learning with cyclegan for low-dose FDG PET image denoising. Med Image Anal 2020;65:101770. [Crossref] [PubMed]


