
Research and implementation of multi-disease diagnosis on chest X-ray based on vision transformer
Introduction
Computer-aided diagnosis (CAD) is an advanced technology that uses computer technology and artificial intelligence methods to analyze and process medical images and data, providing diagnostic and treatment recommendations for patients. Traditional CAD methods often rely on machine-learning algorithms that require manual feature design for diagnostic tasks. In the early stages of chest X-ray disease recognition research, manually designed and extracted features were commonly used.
To address the challenges posed by manual feature extraction and the multi-class classification of chest X-ray diseases, we developed a multi-modal fusion network model based on the Vision Transformer (ViT) architecture. The ViT model, introduced by Dosovitskiy et al. (1) in 2020, represents a breakthrough in the field of visual self-attention models, and has demonstrated remarkable performance in image classification tasks. The ViT network model builds on the transformer architecture by making improvements such as discarding the decoder and using only the encoder to encode and compute image information. By leveraging the self-attention mechanism, the ViT model captures the global context of the input image, enabling it to effectively process visual information.
Currently, convolutional neural networks (CNNs) are commonly used for chest X-ray disease recognition. However, the ViT model used in this article has a number of advantages over CNNs, as it can capture global information for an entire image and learn long-range dependencies between different regions, which provides a better understanding of the image’s structure and semantics. However, the image segmentation method employed by the ViT model in the previous study may disrupt the spatial correlation between image patches and cannot focus on important regions during network training. To address these limitations, we proposed a sliding window image segmentation method to preserve the correlation between image patches. Additionally, we introduced an attention region selection module to guide the model’s focus toward crucial areas of the image. The proposed model had an average area under the curve (AUC) of 0.837 for diagnosing 14 diseases in the ChestX-ray14 data set, representing a 2.1% improvement compared to that of previous models (2-5). Notably, the most significant enhancements were observed for pneumonia and edema diseases, with increases of 7.5% and 6.8%, respectively. Further, our method demonstrated substantial improvements for diseases with a large amount of data, such as effusion and atelectasis, for which it showed increases of 6.5% and 5.7%, respectively.
In the process of diagnosing multi-diseases in chest X-rays using the ViT model, we identified the following main objectives:
- Automatic small lesion location extraction: the lesions in the chest are primarily found in the lungs and chest wall, and they often occupy a small portion of an entire X-ray image. Our first objective was to develop a method to automatically extract the location of these small lesions from X-ray images.
- Accurate identification of multiple diseases: each chest X-ray image may contain one or more diseases, and there are complex coexistence and dependency relationships among these diseases. Our second objective was to accurately identify and classify various diseases present in the chest X-ray images.
- Integration of patient metadata information: in the diagnosis of chest X-ray diseases, relying solely on imaging data may have limitations. In clinical practice, doctors often consider additional information, such as the patient’s age, gender, and medical history, to make a more comprehensive diagnosis. Our ultimate objective was to integrate patient metadata information into the diagnostic network to enhance the accuracy and reliability of the chest disease diagnosis.
By addressing these objectives, we sought to improve the efficiency and effectiveness of chest X-ray diagnosis, ultimately leading to better patient care and outcomes.
Overall, the main contributions of our work can be summarized as follows:
- We proposed an image segmentation approach based on a sliding window mechanism. This approach ensures that small lesion areas in chest X-ray images are not divided into multiple image patches, thus improving segmentation accuracy.
- We introduced a visual self-attention model with an attention region selection module. This model allows for the fine-grained identification of multiple diseases in chest X-ray images by focusing on specific regions of interest. This improves the accuracy and specificity of disease identification.
- We proposed a multi-modal fusion network model that combines patient metadata and X-ray image data. This addresses the limitations of diagnosing with single-image data, enabling the more accurate and comprehensive identification of complex diseases.
The remainder of this article is structured as follows: in Section 2, we provide a brief introduction to related works in the field of chest X-ray image analysis. In Section 3, we describe the data sets used in our experiments and explain how we split the data sets for training and evaluation. We also provide details about the architecture of our model and the implementation of its components. In Section 4, we compare our proposed model to state-of-the-art methods and present the results of ablation experiments. We also analyze the results and discuss the strengths and limitations of our approach. In Section 5, we discuss the experimental setup for the ablation experiments, including the evaluation metrics used and the computational resources employed. Finally, in Section 6, we conclude our work and highlight the key findings and contributions of our research.
Methods
Data sets and data set splitting
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The ChestXray14 data set is one of the earliest publicly available, large-scale chest X-ray data sets. It was released by the National Institutes of Health (NIH) in the United States in 2017. The data set contains a total of 112,120 chest X-ray images taken from 30,805 patients. The data set has the following 15 label categories: infiltration, effusion, atelectasis, nodule, mass, pneumothorax, consolidation, pleural thickening, cardiomegaly, emphysema, edema, fibrosis, pneumonia, hernia, and no finding. The first 14 labels represent common chest diseases, and each X-ray image may be labeled with one or more of these diseases. The “no finding” label indicates that none of the 14 diseases are present in the X-ray image.
Data set splitting
The distribution of various disease samples and their proportions in the ChestX-ray14 data set is shown in the “official partitioning” column of Table 1. There is a significant class imbalance issue among the different disease categories. For instance, diseases such as infiltration, effusion, and atelectasis have a large number of samples, accounting for over 10% of all disease samples. Conversely, diseases such as pneumonia and hernia have a smaller number of samples, comprising only around 1% of all disease samples. The ChestX-ray14 data set divides all chest X-ray images on a per-patient basis, and 80% of the data set is used for training and validation, and the remaining 20% is used for testing evaluation. This ensures that X-ray images taken from the same patient do not appear in both the training and testing sets.
Table 1
| Lesion type | Official partitioning | Past medical history partitioning | |||
|---|---|---|---|---|---|
| Sample size | Proportion of samples (%) | Sample size | Proportion of samples (%) | ||
| Infiltration | 19,894 | 24.51 | 18,339 | 24.57 | |
| Effusion | 13,317 | 16.41 | 12,606 | 16.89 | |
| Atelectasis | 11,559 | 14.24 | 10,675 | 14.30 | |
| Nodule | 6,331 | 7.80 | 5,461 | 7.32 | |
| Mass | 5,782 | 7.12 | 4,999 | 6.70 | |
| Pneumothorax | 5,302 | 6.53 | 5,109 | 6.85 | |
| Consolidation | 4,667 | 5.74 | 4,423 | 5.93 | |
| Pleural thickening | 3,385 | 4.17 | 3,013 | 4.04 | |
| Cardiomegaly | 2,776 | 3.42 | 2,442 | 3.27 | |
| Emphysema | 2,516 | 3.10 | 2,369 | 3.17 | |
| Edema | 2,303 | 2.84 | 2,253 | 3.02 | |
| Fibrosis | 1,686 | 2.08 | 1,424 | 1.91 | |
| Pneumonia | 1,431 | 1.76 | 1,336 | 1.79 | |
| Hernia | 227 | 0.28 | 181 | 0.24 | |
| Total | 81,176 | 100 | 74,630 | 100 | |
Data sets re-divided based on patient medical history
As Figure 1 shows, the ChestX-ray14 data set not only contains 110,000 chest radiographs from over 30,000 patients, but also includes the following patient-specific information for each radiograph: identification (ID) number, age, gender, and view position. The first eight digits of the ID number represent the patient’s ID, and the last three digits indicate the order in which the radiographs of the patient were taken. In addition, the proportion of patients to images in the ChestX-ray14 data set is 1:3.6, and there is a common phenomenon of multiple radiographs belonging to the same patient. Only 15.61% of patients had only one radiograph taken, while the majority of patients had 2–35 radiographs taken. Thus, there is a high degree of redundancy in the image data.
There is a correlation between chest diseases; for example, patients with atelectasis symptoms are more likely to have diseases such as infiltration and effusion (6). Therefore, based on the original official data set’s segmentation method, we re-divided the data set. The radiographs of patients with multiple images were sorted by the time of acquisition, and the disease information from the earliest radiograph was taken as the patient’s medical history information. We also ensured that the samples of the same patient did not overlap in the training, validation, and test sets. The distribution of the re-divided data set is shown in the “past medical history partitioning” column of Table 1; a total of 6,546 radiographs were selected and transformed into patient medical history information, reducing the image data by about 5%, but leaving the proportion of each disease in the total disease count basically unchanged.
Overall architecture
The overall architecture of the model based on the visual self-attention mechanism is shown in Figure 2. The model mainly consists of a serialized feature extraction module, a self-attention model encoder, an attention region selection module, and a metadata feature extraction network.
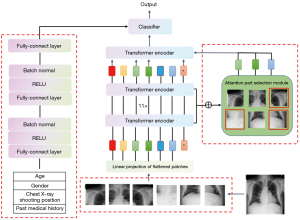
First, the input image is segmented into N equally sized image patches, which are then flattened into a one-dimensional (1D) sequence of image blocks. After incorporating the positional information of the images, the sequence is input to the encoder for encoding and computation. While the data is being computed in the encoder, the attention weight matrices from the previous encoder layers are saved to enable the attention region selection module to select key areas in the image. Once the encoding computation is completed in the encoder, the image features are generated. Finally, the image features are concatenated with the metadata features output by the patient metadata network to generate fused features, which are then input into a classifier for the final disease diagnosis.
The proposed joint network model captures the relative positional relationships between the image patches by adding an additional positional vector encoding to the vector of each image patch, forming the final input sequence. The subsequent encoder comprises 11 stacked transformer encoding modules, each of which comprises a multi-head self-attention layer, a normalization layer, and a fully connected layer. The metadata feature extraction subnetwork comprises two fully connected layers, each followed by an activation function and a normalization layer. The image feature extraction subnetwork and the metadata feature extraction subnetwork concatenate the extracted features along the last dimension, completing the fusion of image features and metadata features.
Serialized feature extraction module
As the self-attention model was originally designed to handle sequential data in the natural language processing field, it first divides the input image into multiple equally sized image blocks in order, and then flattens the divided image blocks to obtain multiple 1D image block sequences of the same length. In the original visual self-attention model, the image blocks are independent of each other, and there is a lack of connections between the adjacent image blocks, resulting in the loss of neighboring spatial information and the possibility of dividing distinctive and recognizable feature areas into multiple image blocks, leading to a decrease in classification performance. In this article, the sliding window segmentation approach adopted directly preserves the correlations between the adjacent image blocks. As the ViT network requires a large amount of training data for training, and performing convolutions on image blocks increases the number of network parameters, it becomes more challenging to train the network. Therefore, we adopted the approach of training with overlapping image blocks, which not only preserves the spatial correlations within the image blocks but also reduces the difficulty of network training and mitigates the risk of model overfitting.
To address this issue and avoid completely separating recognizable regions, while establishing connections between adjacent image blocks, we imitated the sliding convolutional kernel mechanism in CNNs and used a sliding window to divide the original image. Specifically, we let the size of the original image be H*W, the size of the sliding window be Ph*Pw, and the stride of the sliding window be S (S≤Ph, S≤Pw). Using the sliding window segmentation method, the window moves with a stride of S for segmentation, and each time a Ph*Pw image block is segmented. The number of segmented image blocks is calculated as follows:
Through this division method, all adjacent image blocks share a cross-sectional area of size (Ph−S)*Ph (top and bottom adjacent) or (Pw−S)*Pw (left and right adjacent), ensuring that small, recognizable areas appear intact in a single-image block as much as possible. As the stride S decreases, the overlap between the adjacent image blocks increases, preserving spatial correlation information more completely. However, as the stride S decreases, the number of segmented image blocks increases significantly, generating too many redundant image regions and increasing computational costs, making the training process difficult to converge.
After the image block sequence is serialized, the image block sequence is first mapped to a vector space of the same dimension through a learnable linear transformation to input it into the encoder for training. At the same time, imitating the method of adding global classification features in the original visual self-attention model, a learnable embedding sequence xc is added in the image block embedding sequence, which is of the same dimension as the image block sequence, to learn the global features of the entire image in the subsequent self-attention mechanism calculation process. In addition, to capture the relative positional relationships between the image blocks and determine their positions in the input sequence, a position encoding vector is added to each image block embedding vector. The input sequence after serialization and image block embedding can be represented as:
where Epos represents the position encoding vector, which is encoded in the same way as the position encoding in the transformer network.
As Figure 3 shows, the visual self-attention model encoder is mainly composed of alternating multi-head self-attention modules and fully connected layers, with layer normalization applied after each layer and residual connections to improve the model’s generalizability and avoid the problem of gradient disappearance. Assuming that the model encoder is stacked with L layers, the output of the l layer encoder can be represented as:
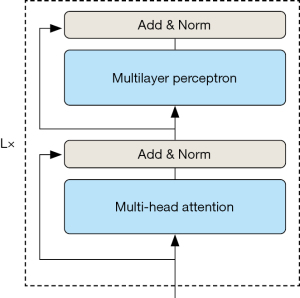
where MSA represents the multi-head self-attention layer, multi-layer perceptron (MLP) represents the fully connected layer, and Zl is the image block feature sequence after encoder operation.
Finally, in the classification module of the original visual self-attention model, only the additional global feature sequence zc is used for the final classification. Although zc learns the global features of the entire image during the encoder self-attention operation, there may still be potential information in the other feature sequences that zc has not captured. Therefore, all the image feature sequence outputs are sent by the last layer encoder together with the global feature sequence to the classifier for the final disease classification.
Attention region selection module
There is a common problem in chest X-ray images where the lesion area occupies a relatively small area of the entire chest image. To address this issue, an attention region selection module was added to the visual self-attention model (7) to select the informative regions in the image and achieve fine-grained disease recognition in chest X-ray images by focusing on specific regions of the image.
Using the inherent attention weight information in the visual self-attention model, the attention weights of all the encoders in the model are accumulated and sorted, and multiple regions with larger weights are selected as inputs to the last layer encoder. Assuming that the model is stacked with L encoders, and K-head self-attention mechanism is used in each encoder, the attention weights in the first L−1 layers of the model can be represented as:
where al represents the K-head attention weight matrix in each layer, and represents the attention weight in each attention head.
Since the weight in the attention matrix indicates the degree of attention given to the interaction between different image blocks when computing the output vector, the attention weights of all encoders is accumulated in the first L−1 layers; that is:
The final attention weight matrix afinal obtained by weight accumulation contains all the attention weight information from the previous encoder layers. In the attention region selection module, the key regions are selected based on each attention head, and the region with the highest attention weight in each head is selected. Assuming that the regions with the highest attention weights in the K attention heads of the encoder are A1, A2, …, Ak, the output of the selected encoder in the L−1 layer can be represented by Eq. [8]:
Finally, the global feature and the selected image block feature are concatenated in the last dimension as the input to the last encoder layer; that is:
A multi-disease diagnostic model based on image information and patient metadata
In clinical practice, doctors often use patient metadata information as an important reference for disease diagnosis. Doctors diagnose based on a combination of many different aspects of features, not just single information sources such as imaging data. This concept is in line with the essence of multi-information joint diagnosis in computer intelligent diagnosis, which uses the complementarity between different categories for multi-feature fusion and cross-modal relationship modeling to explore the correlation between different types of data and to more comprehensively describe and analyze data, increasing the model’s accuracy and generalization ability in decision-making and prediction tasks.
Combining patient metadata with image learning
The diagnostic decisions made by doctors are often based on the combination of features from multiple information sources. In the literature (8-10), multi-modal networks combining image features and metadata features have been extensively studied in the fields of dermatology and glaucoma. However, in the traditional CAD of chest X-ray images, patient data has long been neglected. Therefore, patient metadata is used as a supplement to image data to enhance the model’s understanding of pathological features and provide more classification evidence for model prediction.
The image feature extraction network is based on the network structure proposed in the previous section; that is, the final classifier output is removed to produce a 13×768-dimensional image feature vector. Here, 13 is composed of one global feature sequence and 12 key region sequences selected by the self-attention region selection module, while 768 is the dimension mapped by the image embedding module in the model.
Constructing a patient metadata network
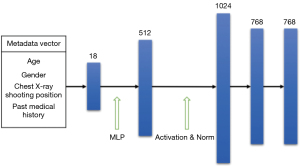
The metadata information used in this article includes the patient’s age, gender, X-ray shooting position, and past medical history. All the metadata are encoded into equally sized feature vectors using one-hot and discrete numerical encoding. The patient metadata is encoded as follows: male and female are encoded as 0 and 1, respectively, and the X-ray shooting position is encoded as 0 and 1. Patient age is normalized from [1, 97] to [0, 1]. The patient’s past medical history is encoded as a 15-dimensional vector, with the first 14 dimensions corresponding to 14 diseases in the data set. If the patient has had a certain disease, it is encoded as 1; otherwise, it is encoded as 0. If the patient does not have any of the 14 diseases, the 15th dimension is marked as 1 to indicate health; otherwise, it is marked as 0. Based on the above encoding rules, an 18-dimensional patient metadata information vector is constructed for each chest X-ray image.
The encoded metadata vector already has feature representation ability; however, the effect of each encoding feature on the classification results is different, and different weight parameters need to be assigned. Therefore, weighted calculations are needed for the metadata encoding to obtain metadata feature vectors with higher discriminability. As neural networks can map input feature vectors to a new feature space through matrix multiplication and bias addition operations, and can represent more complex and abstract features through learning weight parameters, a metadata feature extraction network was designed to further process the encoded metadata information.
The metadata feature extraction network constructed in this article consists of two fully connected layers, in addition to the activation and normalization layers after each layer. The fully connected layers are used to map low-level features to higher-level features to enhance the network’s expressive power. The activation and normalization layers introduce non-linear transformations in the network, thus improving the network’s generalization performance and reducing the risk of over fitting. Figure 4 shows the calculation process and dimensionality change of patient metadata information in the feature extraction network. After being processed by the metadata feature extraction network, the metadata vector is extended to 768 dimensions for easy feature fusion with the image feature vector. Specifically, after being encoded, the patient metadata information is first mapped to 512 dimensions through a neural network, and then normalized and activated by a rectified linear unit function before entering the second neural network layer, where it is further enhanced to 1,024 dimensions. Finally, after undergoing normalization and activation function operations, a 768-dimensional patient metadata feature vector is generated.
Joint network
The structure of the joint network model is shown in Figure 5. In the joint network, the output features of the metadata feature extraction network and the image feature extraction network are fused to generate the final classification feature vector of the model. The output features of the joint network are represented by Eq. [10] as follows:
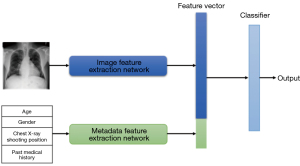
where F1 represents the image feature extraction network, which outputs features with dimensions of 13×768, F2 represents the metadata feature extraction network, which outputs features with dimensions of 1×768, and G represents the multidimensional information fusion feature vector with dimensions of 14×768, generated by concatenating the output features of the two networks on the last dimension. Finally, the fused features are fed into the classifier for the diagnosis and classification of chest X-ray diseases.
Results
Experimental environment and parameter configuration
The deep-learning framework used for this study was PyTorch, and the hardware was a graphics processing unit server with a Tesla P100-PCIE graphics card. The detailed experimental environment configuration is shown in Table 2.
Table 2
| Category | Configuration |
|---|---|
| Operating system version | Ubuntu 18.04 |
| Graphics processor | Tesla P100-PCIE |
| Central processor | Intel Xeon 2.30GHz E5-2699 v4 |
| System memory | 27G 3733MHZ LPDDR4X |
| Deep-learning framework | Pytorch 1.10.0 |
The network uses pre-trained parameters of the ViT-B/16 model. Before training, the input image size is resized to 224×224 and normalized before being fed into the network. The training batch size is set to 16, and the optimization strategy uses the stochastic gradient descent optimizer with an initial learning rate of 3e−2.
The hyperparameters of the model are set as follows: sliding window stride: 4; number of attention regions: 12; and length of the 1D sequence after unfolding the image blocks: 768. The model goes through 12 layers of transformer encoders for encoding operations. Each transformer encoder has 12 attention heads, a hidden size of 3,072 in the fully connected layer, and a dropout rate of 0.1.
Comparison to state-of-the-art and generalizability metrics
We compared our proposed method with the currently better-performing chest X-ray disease diagnosis methods. Figure 6 shows the AUC values of our method and previous methods in the diagnosis of 14 common chest diseases. The experimental results showed that our method had higher diagnostic accuracy than the previous three methods for most of the diseases in the ChestX-ray14 data set.
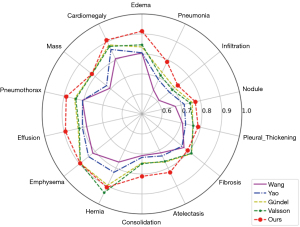
Table 3 shows the specific AUC values of the model for the diagnosis of 14 diseases and the average AUC value for all disease categories. The experimental results showed that the average AUC value of our method was 0.831, which is higher than the AUC values of Wang (0.738), Yao (0.803), Gündel (0.807), and Valsson (0.816). Specifically, the most significant improvement in the AUC values was for edema and pneumonia, which increased by 7.9 and 5.8 percentage points, respectively. Our method also showed a significant improvement in the diagnosis of diseases with small lesion areas that are difficult to identify, such as effusion and lung consolidation, which increased by 6.5 and 6.3 percentage points, respectively, which verified our method’s ability to identify small-area lesion diseases. Additionally, our method also showed a significant improvement in the diagnosis of diseases with a large number of samples, such as atelectasis and pneumothorax, which increased by 5.7 and 4.3 percentage points, respectively. As the visual self-attention mechanism-based network structure requires a large amount of data for training, the diagnostic performance of the network was slightly reduced in some disease categories with a small sample size, but it still maintained comparable diagnostic performance. For example, the AUC values for fibrosis and hernia were 0.792 and 0.907, respectively, which were slightly lower than the previous highest values of 0.818 and 0.937. Overall, these results showed the effectiveness and superiority of our proposed method in the diagnosis of common chest X-ray diseases.
Table 3
| Disease types | Diagnostic methods for chest X-ray disease | ||||
|---|---|---|---|---|---|
| Wang (2) | Yao (3) | Gündel (4) | Valsson (5) | Ours | |
| Infiltration | 0.609 | 0.675 | 0.709 | 0.694 | 0.729 |
| Effusion | 0.784 | 0.806 | 0.828 | 0.822 | 0.893 |
| Atelectasis | 0.716 | 0.733 | 0.767 | 0.763 | 0.824 |
| Nodule | 0.671 | 0.717 | 0.758 | 0.747 | 0.774 |
| Mass | 0.706 | 0.727 | 0.821 | 0.820 | 0.821 |
| Pneumothorax | 0.806 | 0.805 | 0.846 | 0.840 | 0.889 |
| Consolidation | 0.708 | 0.717 | 0.745 | 0.749 | 0.812 |
| Pleural thickening | 0.708 | 0.724 | 0.761 | 0.763 | 0.788 |
| Cardiomegaly | 0.807 | 0.858 | 0.883 | 0.875 | 0.910 |
| Emphysema | 0.815 | 0.842 | 0.895 | 0.895 | 0.896 |
| Edema | 0.805 | 0.806 | 0.835 | 0.846 | 0.914 |
| Fibrosis | 0.769 | 0.757 | 0.818 | 0.816 | 0.792 |
| Pneumonia | 0.633 | 0.690 | 0.731 | 0.714 | 0.789 |
| Hernia | 0.767 | 0.824 | 0.896 | 0.937 | 0.907 |
| AUC mean | 0.738 | 0.803 | 0.807 | 0.816 | 0.837 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
The model’s receiver operating characteristic (ROC) curves for 14 disease diagnoses on the ChestX-ray14 data sets are shown in Figure 7.
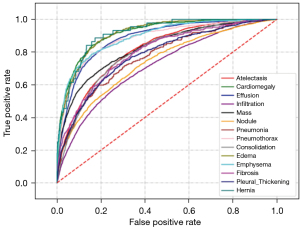
By evaluating the generalizability of the chest X-ray CAD model, we can gain a better understanding of its performance and limitations and provide guidance for further improvements and optimizations. When evaluating the generalizability of the chest X-ray model, several important metrics can be considered. First, accuracy is a crucial metric that measures a model’s overall performance in classifying normal and abnormal cases. A high accuracy indicates that a model is effective in distinguishing between the two classes. Second, sensitivity is an important metric that evaluates a model’s ability to correctly identify diseases. It measures the proportion of true positive cases identified by a model, indicating its effectiveness in detecting abnormalities. Third, specificity measures a model’s ability to correctly exclude normal cases. It represents the proportion of true negative cases correctly classified by a model, indicating its effectiveness in ruling out abnormalities when they are absent.
To assess the generalizability of our proposed model, we conducted statistical experiments on the ChestX-ray14 data set. This data set includes a diverse range of X-ray images collected from different populations, age groups, time periods, and types of X-ray devices, making it statistically significant for evaluating the model’s generalization ability.
The generalizability metrics for our proposed model on the ChestX-ray14 data set are summarized in Table 4, providing a comprehensive evaluation of its accuracy, sensitivity, and specificity. These metrics provide valuable insights into the model’s performance and its ability to generalize to unseen data.
Table 4
| Generalizability metrics | Values |
|---|---|
| Specificity | 0.821 |
| Sensitivity | 0.863 |
| Accuracy | 0.834 |
By considering these metrics and conducting rigorous evaluations, we can gain a deeper understanding of a model’s strengths and weaknesses, and identify areas for further improvement. This evaluation process is essential for enhancing a model’s performance and ensuring its reliability in real-world scenarios.
Ablation experiments
To validate the effect of different modifications to the ViT in our proposed model on the chest X-ray disease classification task, we conducted ablation experiments on the image patch segmentation method, attention region selection module, and image classification feature representation method. The detailed experimental results are shown in Table 5.
Table 5
| Disease types | Network structures | ||||
|---|---|---|---|---|---|
| ViT | ViT + overlapping segmentation methods | ViT + overlapping segmentation methods + region selection module | ViT + overlapping segmentation methods + region selection module + labeling all image blocks | ViT + overlapping segmentation methods + region selection module + labeling all image blocks + metadata information | |
| Infiltration | 0.705 | 0.707 | 0.711 | 0.715 | 0.729 |
| Effusion | 0.856 | 0.861 | 0.877 | 0.879 | 0.893 |
| Atelectasis | 0.798 | 0.804 | 0.811 | 0.812 | 0.824 |
| Nodule | 0.748 | 0.751 | 0.762 | 0.756 | 0.774 |
| Mass | 0.801 | 0.807 | 0.826 | 0.826 | 0.821 |
| Pneumothorax | 0.860 | 0.863 | 0.879 | 0.882 | 0.889 |
| Consolidation | 0.790 | 0.792 | 0.798 | 0.800 | 0.812 |
| Pleural thickening | 0.757 | 0.755 | 0.777 | 0.778 | 0.788 |
| Cardiomegaly | 0.881 | 0.881 | 0.903 | 0.912 | 0.910 |
| Emphysema | 0.875 | 0.872 | 0.899 | 0.896 | 0.896 |
| Edema | 0.897 | 0.904 | 0.909 | 0.913 | 0.914 |
| Fibrosis | 0.789 | 0.794 | 0.803 | 0.801 | 0.792 |
| Pneumonia | 0.758 | 0.766 | 0.763 | 0.771 | 0.789 |
| Hernia | 0.894 | 0.897 | 0.913 | 0.918 | 0.907 |
| AUC mean | 0.814 | 0.817 | 0.829 | 0.831 | 0.837 |
AUC, area under the ROC curve; ROC, receiver operating characteristic; ViT, Vision Transformer.
Sliding window ablation experiment
In this study, to ensure that the small lesion areas were not divided into multiple image patches, the image patch segmentation method in the original visual self-attention model was improved. A sliding window approach is used to divide the original input image, with partial overlap between adjacent image patches. This enables cross-patch connections between adjacent image patches to be established and preserves the integrity of small lesion areas. To evaluate the effect of different image patch segmentation methods on model prediction performance, the other parameters of the original visual self-attention model were left unchanged, and only the image patch segmentation method was varied in an ablation experiment. The experimental results, indicate that the sliding window segmentation method used in this study significantly improved the diagnostic performance of the model compared to the non-overlapping segmentation method (Table 6).
Table 6
| Overlapping segmentation | AUC mean |
|---|---|
| × | 0.814 |
| √ | 0.817 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
The sliding window stride values were set to 14, 12, and 8, respectively. The experimental results are shown in Table 7. As the stride decreased, the overlap area between the adjacent image blocks increased, and the AUC values for diseases with small lesion areas, such as atelectasis, infiltration, mass, and nodule, increased partially.
Table 7
| Disease types | Sliding window stride | ||
|---|---|---|---|
| 14 | 12 | 8 | |
| Atelectasis | 0.807 | 0.812 | 0.816 |
| Cardiomegaly | 0.911 | 0.912 | 0.907 |
| Effusion | 0.877 | 0.879 | 0.868 |
| Infiltration | 0.715 | 0.715 | 0.712 |
| Mass | 0.829 | 0.826 | 0.831 |
| Nodule | 0.750 | 0.756 | 0.758 |
| Pneumonia | 0.768 | 0.771 | 0.767 |
| Pneumothorax | 0.884 | 0.882 | 0.879 |
| Consolidation | 0.801 | 0.800 | 0.796 |
| Edema | 0.911 | 0.913 | 0.911 |
| Emphysema | 0.899 | 0.896 | 0.893 |
| Fibrosis | 0.807 | 0.801 | 0.789 |
| Pleural thickening | 0.768 | 0.778 | 0.774 |
| Hernia | 0.916 | 0.918 | 0.911 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
Selected key regions ablation experiment
To address the problem of small and difficult-to-identify lesion areas in chest X-ray images, this study introduced improvements to the original visual self-attention model’s image embedding feature representation. The improvements mainly comprised two parts: (I) introducing a region selection module based on the inherent self-attention mechanism of the visual self-attention model to highlight the regions of interest in the image to focus the network’ attention on lesion areas; (II) inputting all the feature sequences into a classifier for the final disease classification to retain all the feature information of the highlighted regions. The experimental results indicated that the proposed improvements in image embedding feature representation significantly enhanced the model’s predictive performance for diseases (Table 8). The AUC value for chest disease diagnosis increased from 0.814 to 0.831, demonstrating the effectiveness of the proposed improvements.
Table 8
| Labeling classifier image blocks | Labeling all image blocks | Region selection module | AUC mean |
|---|---|---|---|
| √ | × | × | 0.813 |
| × | √ | × | 0.814 |
| √ | × | √ | 0.829 |
| × | √ | √ | 0.831 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
The number of regions selected by each attention head was set to three different values (i.e., 1, 2, and 3). The experimental results are shown in Table 9. The results showed that the choice of the number of key regions had little effect on disease diagnosis. As the number of key regions increased, there was a slight decrease in the AUC values for different diseases, but the overall difference was not significant.
Table 9
| Disease types | Number of key regions | ||
|---|---|---|---|
| 1×12 | 2×12 | 3×12 | |
| Atelectasis | 0.812 | 0.812 | 0.809 |
| Cardiomegaly | 0.912 | 0.913 | 0.907 |
| Effusion | 0.879 | 0.882 | 0.880 |
| Infiltration | 0.715 | 0.711 | 0.711 |
| Mass | 0.826 | 0.823 | 0.828 |
| Nodule | 0.756 | 0.752 | 0.756 |
| Pneumonia | 0.771 | 0.766 | 0.767 |
| Pneumothorax | 0.882 | 0.878 | 0.880 |
| Consolidation | 0.800 | 0.803 | 0.796 |
| Edema | 0.913 | 0.909 | 0.916 |
| Emphysema | 0.896 | 0.896 | 0.892 |
| Fibrosis | 0.801 | 0.794 | 0.789 |
| Pleural thickening | 0.778 | 0.799 | 0.787 |
| Hernia | 0.918 | 0.919 | 0.915 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
Metadata network ablation experiments
Metadata information ablation experiments
Due to the inherent limitations of individual image data, it is often challenging to include all the necessary information for disease diagnosis. Introducing patient semantic information can extend and supplement the image data, providing additional classification evidence for model diagnosis. Therefore, in this study, a parallel patient metadata feature extraction network was further constructed based on the image feature extraction network. The patient metadata features were fused with the image features and jointly used for disease classification. The experimental results indicate that compared to the sole image features, the introduction of patient metadata information in the model increased the average AUC value for disease diagnosis from 0.831 to 0.837 (Table 10). This suggests that the constructed metadata feature extraction network in this study effectively extracted patient metadata information and supplemented the pathological information of the image data.
Table 10
| Metadata information | AUC mean |
|---|---|
| × | 0.831 |
| √ | 0.837 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
For patient gender, age, and X-ray shooting position information, we used a method based on image feature extraction network to predict non-image data to verify their effectiveness in disease diagnosis. Specifically, the image feature extraction network trained on chest X-ray image data was first used to extract image feature vectors, and the image information was then used to train classifiers to predict patient gender, age, and chest X-ray shooting position information (Figure 8).
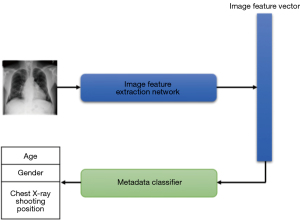
The patient metadata network was trained for five epochs on the three parts of metadata information; Table 11 shows the network’s prediction results for the patient metadata. The experimental results showed that patient gender and chest X-ray shooting position information were accurately predicted using the image feature network, indicating that these two parts of information are not well-complementary with image information and cannot effectively supplement image information. The average absolute error of age information predicted by the network was 7.92 years, which was higher than the predicted value of the bone age evaluation system for adolescent clinical applications (11), which indicates that age cannot be accurately predicted solely through image data.
Table 11
| Metadata information | Accuracy/mean absolute error |
|---|---|
| Chest X-ray shooting position (%) | 99.51 |
| Gender (%) | 94.17 |
| Age (years) | 7.92 |
Past medical history information ablation experiments
We conducted ablation experiments to verify the effectiveness of past medical history information; the experimental results are set out in Table 12. The results showed that most of the improvement of the multi-modal information fusion network in diagnosing chest diseases was due to the introduction of past medical history information, while gender, age, and X-ray shooting position information only improved the network by 0.1 percentage points, which shows the effectiveness of past medical history information in supplementing image information.
Table 12
| Gender + age + shooting position | Past medical history | AUC mean |
|---|---|---|
| × | × | 0.831 |
| √ | × | 0.831 |
| × | √ | 0.836 |
| √ | √ | 0.837 |
AUC, area under the ROC curve; ROC, receiver operating characteristic.
Discussion
Based on the findings from the above ablation experiments, a number of observations can be made. First, in the sliding window ablation experiment setup, the image blocks were divided in an overlapping manner, and the setting of the sliding window stride determined the size of the overlap area between the adjacent image blocks. The image block size was set to the default value of 16 in the original ViT model, and the sliding window stride values were set to 14, 12, and 8, respectively, corresponding to overlap areas of 1/8, 1/4, and 1/2 of the entire image block size. The adjacent overlap area of the image increased as the sliding window stride value decreased, resulting in an increase in the number of generated image blocks. This resulted in higher computational costs and challenges in achieving training convergence. In contrast to previous methods (12), we proposed a flexible adjustment to the stride of the sliding window, taking into account the size of the lesion region. This adjustment was crucial, as it determined the model’s ability to converge quickly.
Second, in the selected key regions ablation experiment setup, the number of heads in the multi-head self-attention module was set to 12. This setting was chosen because previous studies (13-16) have shown that different heads in the multi-head self-attention mechanism represent semantic information in different dimensions. Therefore, in the key region selection module, for each attention head, the regions with the highest attention weights were selected as the key regions. We conducted experiments on the number of selected key regions for each attention head, with the number of selected key regions set to 1, 2, and 3, corresponding to 12, 24, and 36 key regions, respectively. The choice of the number of key regions might only have had a small effect on disease diagnosis because most of the images in the ChestX-ray14 data set contain one to three diseases; thus, selecting 12 key regions covers most diseases in the images. However, selecting fewer key regions can increase the model’s focus on key regions. Thus, it is more appropriate to select 12 key regions for the ChestX-ray14 data set. If a data set is being used with more disease types, the selection of more key regions may achieve better diagnostic performance.
In the metadata information ablation experiment setup, to explore the effectiveness of each part of the patient metadata, we further investigated the correlation between image information and patient metadata information. The three parts of metadata information were predicted separately. The gender and chest X-ray shooting position were converted into binary classification problems, and the labels of the two parts of information were encoded using binary encoding. For patient age, as it is a number that falls within the range of 1 to 95, this section used a regression prediction method and used the mean absolute error as the evaluation index, which reflects the actual age difference between network predicted value and label value. The calculation method of mean absolute error is expressed in Eq. [11] as follows:
where y represents the patient’s true age, and y' represents the network predicted age.
If image features can effectively predict patient metadata information, it indicates that this category of metadata has poor complementarity with image data, which means that this information can be learned from image feature data and has poor complementarity with image feature data. If image features cannot effectively predict patient metadata information, it indicates that this metadata information has strong complementarity with image data, and they can therefore serve as an effective supplement to image data. Due to the large average absolute error of age information predicted from the image network, age information has strong complementarity with image information and can effectively supplement image information. Related studies have shown (17) that as age increases, the incidence of chest diseases in the elderly population also shows a growing trend compared to younger age groups. Therefore, fusing patient age information with image features can provide more decision evidence for the system to diagnose chest diseases and improve the system’s classification ability.
Conclusions
We focused our research on chest X-ray images and identified several limitations that needed to be addressed (i.e., the presence of small lesion areas, numerous and complex diseases, and the challenge of incorporating all necessary information for accurate disease diagnosis into a single image). To address these limitations, we proposed a multi-modal fusion network model that combines patient metadata and X-ray image data. To address the problem of small lesion areas, we employed a sliding window–based image segmentation approach. This allows us to extract relevant regions of interest and focus on important lesion areas using an attention region selection module. By doing so, we ensure that the model pays attention to the most crucial parts of the image for accurate diagnosis.
In addition to image data, we also incorporated patient metadata into our model. This metadata provides valuable contextual information that can enhance the classification performance. By fusing patient metadata and image information, we are able to improve the model’s ability to predict multiple diseases in chest X-rays.
We chose the ViT model for the task of multi-label classification of chest X-ray images. Through extensive experiments on the ChestX-ray14 data set, our approach achieved an average AUC of 0.837. This represents a 2.7% improvement in the average AUC compared to previous CNN-based methods. These results demonstrate the effectiveness of our proposed approach in accurately diagnosing chest diseases. To further validate our improvements, we conducted ablation experiments. These experiments confirmed the effectiveness of our modifications to the ViT model and the introduction of patient metadata.
However, we acknowledge that the available patient metadata types are limited and metadata information may lack information about strong correlation among chest diseases, such as smoking history or family medical history. In future research, we plan to use real-world hospital data sets to collect more comprehensive patient metadata information. This will allow us to further improve the performance of our model and enhance its applicability in real-world clinical settings.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1280/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, Dehghani M, Minderer M, Heigold G, Gelly S, Uszkoreit J, Houlsby N. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv:2010.11929. [Preprint]. 2020. Available online: https://doi.org/
10.48550/arXiv.2010.11929 - Wang W, Xie E, Li X, Fan DP, Shao L. Pyramid Vision Transformer: A Versatile Backbone for Dense Prediction without Convolutions. arXiv.2102.12122 [Preprint]. 2021. Available online: https://doi.org/
10.48550/arXiv.2010.11929 - Yao L, Prosky J, Poblenz E, Covington B, Lyman K. Weakly Supervised Medical Diagnosis and Localization from Multiple Resolutions. arXiv:1803.07703. [Preprint]. 2018. Available online: https://doi.org/
10.48550/arXiv.1803.07703 - Gündel S, Grbic S, Georgescu B, Liu S, Maier A, Gomaniciu D. Learning to Recognize Abnormalities in Chest X-Rays with Location-Aware Dense Networks. In: Vera-Rodriguez R, Fierrez J, Morales A. editors. Progress in Pattern Recognition, Image Analysis, Computer Vision, and Applications. CIARP 2018. Lecture Notes in Computer Science, vol 11401. Springer, Cham; 2019. doi:
10.1007/978-3-030-13469-3_88 . - Valsson S, Arandjelovic O. Nuances of Interpreting X-ray Analysis by Deep Learning and Lessons for Reporting Experimental Findings. Sci 2022;4:3.
- Bustos A, Pertusa A, Salinas JM, de la Iglesia-Vayá M. PadChest: A large chest x-ray image dataset with multi-label annotated reports. Med Image Anal 2020;66:101797. [Crossref] [PubMed]
- Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser L, Polosukhin I. Attention Is All You Need. arXiv.1706.03762 [Preprint]. 2017. Available online: https://doi.org/
10.48550/arXiv.1803.07703 - Thomas SA. Combining Image Features and Patient Metadata to Enhance Transfer Learning. Annu Int Conf IEEE Eng Med Biol Soc 2021;2021:2660-3. [Crossref] [PubMed]
- Pacheco AGC, Krohling RA. An Attention-Based Mechanism to Combine Images and Metadata in Deep Learning Models Applied to Skin Cancer Classification. IEEE J Biomed Health Inform 2021;25:3554-63. [Crossref] [PubMed]
- Li Y, Han Y, Li Z, et al. A transfer learning-based multimodal neural network combining metadata and multiple medical images for glaucoma type diagnosis. Sci Rep 2023;13:12076. [Crossref] [PubMed]
- van Rijn RR, Thodberg HH. Bone age assessment: automated techniques coming of age? Acta Radiol 2013;54:1024-9. [Crossref] [PubMed]
- Li G, Xu D, Cheng X, Si L, Zheng C. SimViT: Exploring a Simple Vision Transformer with sliding windows. arXiv.2112.13085 [Preprint]. 2021. Available online: https://doi.org/
10.48550/arXiv.2112.13085 - Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019;25:954-61. [Crossref] [PubMed]
- Zheng J, Wang H, Deng Z, et al. Structure and biological activities of glycoproteins and their metabolites in maintaining intestinal health. Crit Rev Food Sci Nutr 2023;63:3346-61. [Crossref] [PubMed]
- Ren J, Chen L, Xu H, et al. A Bi-LSTM and multihead attention-based model incorporating radiomics signatures and radiological features for differentiating the main subtypes of lung adenocarcinoma. Quant Imaging Med Surg 2023;13:4245-56. [Crossref] [PubMed]
- Yang D, Ren G, Ni R, et al. Deep learning attention-guided radiomics for COVID-19 chest radiograph classification. Quant Imaging Med Surg 2023;13:572-84. [Crossref] [PubMed]
- Aker MB, Taha AS, Zyoud SH, et al. Estimation of 10-year probability bone fracture in a selected sample of Palestinian people using fracture risk assessment tool. BMC Musculoskelet Disord 2013;14:284. [Crossref] [PubMed]


