MCSE-U-Net: multi-convolution blocks and squeeze and excitation blocks for vessel segmentation
Introduction
Atherosclerosis, diabetic retinopathy, and hypertension are just a few examples of the many eye-related disorders that can be diagnosed using retinal vascular segmentation (1,2). The morphological characteristics of retinal blood vessels, such as their thickness, curvature, and density, can serve as key indicators for the early detection and diagnosis of various illnesses. Therefore, ophthalmologists frequently evaluate these morphological characteristics, which serve as crucial diagnostic markers for a range of ophthalmological illnesses, in retinal fundus pictures to determine the clinical status of the retinal blood vessels. Key markers can be used to diagnose many ocular illnesses. However, the automatic segmentation of retinal vessels is crucial, as the manual labeling of retinal vessels in such images is time consuming, difficult, and requires a high level of clinical knowledge (3).
Traditional segmentation algorithms rely heavily on hand-labeled features for training to improve performance and are mostly based on computer vision techniques (4). It is challenging to accurately and completely describe a particular region of interest for a small number of hand-produced elements. In recent years, image segmentation and other image processing methods have repeatedly shown that convolutional neural networks (CNNs) outperform conventional computer vision algorithms (5). Performance advantages are achieved by stacking convolutional layers and extracting millions of features. These successive layers produce intricate feature mappings that enable the CNNs to automatically “learn” and “organize” the picture data in relation to the segmentation goal. The U-net CNN architecture comprises a symmetric encoder-decoder backbone with hopping links (2). It is frequently used for automatic image segmentation and has been shown to perform well, which has led to a continual stream of advancements based on the U-net (6).
To address the information imbalance between the U-net’s high and low levels and enhance the network’s capacity for generalization, Jiang et al. proposed the addition of a multilevel attention module (7). To improve the U-net’s ability to focus on structures of interest while suppressing background noise, Gao et al. included attention gates to improve the ability of the U-net architecture to deal with spatial disparity and loss of information (8). Beeche et al. further simplified the U-net structure and suppressed the overfitting phenomenon by using a fused up-sampling module and a dynamic sensory field module (9). To further simplify the U-net structure and address the issue of overfitting, Hu et al. developed the minimal U-net (10).
In the above methods, the accuracy (acc) of fine blood vessel segmentation remains problematic. Thus, inspired by the model architectures of Xin et al. and Beeche et al. (9,11), we proposed a new and improved U-net based on multi-convolution (MC) blocks and squeeze and excitation (SE) modules; that is, the multi-convolution block and squeeze and excitation based on the U-shape network (MCSE-U-net) model. The main contributions of the model are as follows:
- The method performs exceptionally well in retinal blood vessel segmentation, and it could be extended to other medical image segmentation tasks in the future.
- The addition of the MC module to the U-net framework improves the overall segmentation performance of the model.
- The addition of the SE module to the network model improves the fine vessel segmentation performance of the model.
In terms of retinal vascular segmentation, our experimental findings and computational comparisons showed that our enhanced model performed slightly better than already existing models.
Methods
Network architecture
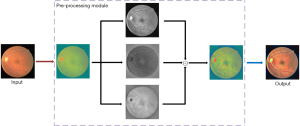
This section explains in detail how the created module units were introduced to the U-net benchmark model using the U-net as the reference model. Pre-processing, the MC module and the SE module comprise the bulk of the MCSE-U-net module. Figure 1 shows the structure formed by connecting these three modules. The data set image is first processed using the pre-processing module, and the resulting image is then fed into the MCSE-U-net model. This improvement to the U-net model uses the MC module rather than the two 3×3 convolutions in the original U-net model. Finally, in the encoder path, the features obtained by passing through the MC block then enter into the SE module to change the number of feature channels in the image, resulting in a maximum pooling size of 2×2. The number of feature channels obtained in the decoder route is cut in half, as the features are first up-sampled using a bilinear interpolation operation in each layer before being fed through the MC module. Through a skip connection between the encoder and decoder, the shallow, low-level features present in the encoder part are simultaneously combined with the high-level abstract features present in the decoder part, allowing the image to retain a sizable amount of spatial information for improved localization.
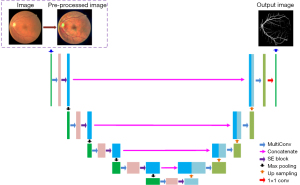
The following section provides further specifics on the pre-processing module, MC module, and SE module.
Pre-processing module
The photographs in the data set need to be treated to improve the contrast between the blood vessels and the background because it is poor, which negatively affects the effectiveness of blood vessel segmentation (12). This study employed the luminance, chrominance-blue, chrominance-red (YCbCr) color space conversion method to process the data sets. This method reduces color information interference for the segmentation task, enhancing the contrast and revealing the image edges and details more clearly. By creating a YCbCr color space, chromaticity components (Cb and Cr) can be adjusted to correct color distribution, thereby reducing color differences between the images and improving the model’s generalization ability under various lighting and sensor conditions.
However, the enhancing process of the YCbCr image processing method significantly affects the ability of model to differentiate between vascular structure and the background. To improve the image, gamma correction and contrast limited adaptive histogram equalization (CLAHE) are employed. If there are not enough training data, multi-scale equalized sampling is used to prevent overfitting.
Figure 2 shows the pre-processing procedure, which comprises four primary steps. First, the data set image is converted from the RGB color space to the YCbCr color space. Second, it is divided into three channels of Y, Cb, and Cr within the YCbCr color space. Third, the three channels are combined using CLAHE, and the images of the three channels are merged. Fourth, the merged image is converted back to the RGB color space. As Figure 2 shows, the blood vessel pixels in the output image are clearer than those in the input image.
MC module
Two consecutive 3×3 convolutions are done to extract features using the U-net. Picture features can be extracted; however, this method is not very useful for segmenting features with a great deal of variance, such as the minute blood veins in the retina. This is due to the fact that this feature extraction technique can only extract features that are 3×3 in size, which leads to the inadequate model segmentation of small blood vessels.
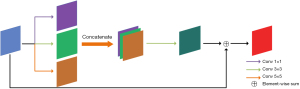
To improve the retinal image segmentation effect on the small blood vessels, we changed the U-net feature extraction method of fixed-size convolution. Specifically, we use three convolutions of different kernel sizes to extract and then fuse multi-scale feature information. This significantly improves the model’s ability to extract microvascular features. Figure 3 shows the MC module’s structure. First, the MC block performs one 1×1 convolution, 3×3 convolution, and 5×5 convolution on the input feature maps to obtain three feature maps of the same size representing different scale information. Second, these three feature maps are spliced according to the channels, and the spliced image is subjected to one 3×3 convolution so that the number of channels is the same as that of the input feature maps. Finally, the obtained features and input features are added together with input feature mapping, and the overall feature extraction results of MC block are obtained. MC block extracts multi-scale features by convolutions of three different kernel sizes and then fuses them. The input is added to the multi-scale feature mapping through shortcuts to form the residual structure, which makes the model easy to train. The improved feature extraction module can extract multi-scale features more efficiently than the traditional structure of two consecutive 3×3 convolutions, and the segmentation of small blood vessels is significantly improved.
SE module
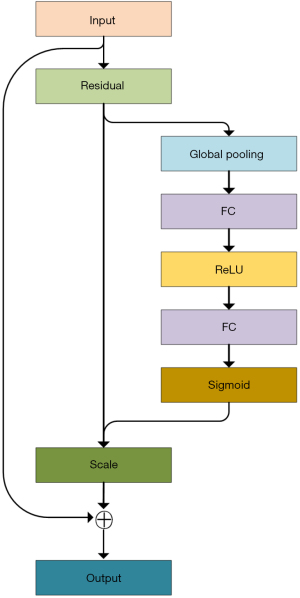
In deep-learning image segmentation tasks, the SE module can improve a model’s performance, generalization ability, and long-range dependency modeling capabilities (9). The SE module improves the model’s capacity to represent various targets in an image by assisting it to adaptively learn the relevance weights of various channels in the feature map during the image segmentation task. This helps to direct the model’s attention to features that are meaningful for a specific target and reduces the attention paid to irrelevant information. This attentional mechanism improves the discriminative and generalization abilities of the model, thus enhancing its performance in image segmentation tasks.
SE are the two primary stages of the SE module. Figure 4 shows the SE module’s structure. After passing through a residual convolution block, the input features are first subjected to global average pooling, which is the fusion of feature information from each channel to encode the spatial features on each channel into a global feature. Next, the significance of each channel is predicted through a fully connected layers. Again, following a rectified linear unit (ReLU) activation function, the obtained feature map is then subjected to a global average pooling and a Sigmoid activation function. Finally, the SE module performs a global average pooling and a Sigmoid activation function by means of a scale operation, which is the multiplication of channel weights. Once again, after a ReLU activation function, global average pooling and a Sigmoid activation function are performed on the obtained feature map. Finally, through a scale operation (i.e., channel weight multiplication), the weight values of each channel computed by the SE module are multiplied by each two-dimensional matrix of the corresponding channels of the original feature map, to obtain the output feature map.
The experimental environments
The overall experiments and all the ablation experiments were performed using Pytorch version 3.8 with an AMD Ryzen 5 5600 H Radeon graphics processor, a Nvidia GeForce RTX 2070 S (8GB) graphics card, and 16GB of RAM.
Description of data sets
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The new methodology was tested on the following three public data sets of vessels: Digital Retinal Images for Vessel Extraction (DRIVE) (13); Structured Analysis of the Retina (STARE) (14), and Child Heart and Health Study in England (CHASE_DB1) (15). The DRIVE data set, which was obtained from a diabetic retinopathy screening program in the Netherlands, has a total of 40 color fundus images of patients. The images were acquired using a Canon CR5 astigmatism-free 3 CCD camera with a field of view of 45 degrees. The resolution size of the images is 565×584 with 8 bits per color channel. The data set has been divided into a training set and a test set, each set consists of 20 images. Each image corresponds to a mask, and a vessel image that has been manually labeled by two experts. In this experiment, we used the already established training and test DRIVE data sets to train and validate the model; the image labeled by the first expert was used by default as the ground truth of the experiment.
The STARE data set, which was created by the University of Florida Eye Research Center, is a public data set that comprises high-resolution images captured by digital photography with a resolution of 700×605. Each image corresponds to the manual segmentation results of two experts and is commonly used for fundus image analysis and ocular disease diagnosis. However, this data set does not include any mask images and need to be set up manually. To validate the baseline U-net model and the CEDMU-U-net model, we manually extracted the mask corresponding to each image. The data set contains 20 images, of which 12 are used for training in experiments, and eight images are used as the validation set for testing.
The CHASE_DB1 data set is a subset of multi-ethnic children’s retinal images from the United Kingdom’s Children’s Heart and Health Study. It comprises 28 retinal images obtained from the left and right eyes of 14 children, each with a resolution size of 999×960. Each image similarly corresponds to the results of two manual segmentations performed by experts. Typically, the first 20 images are used for training, and the remaining eight images are used for testing.
Both data sets underwent pre-processing through CLAHE and normalization.
Data augmentation
As the vascular data set used was only moderately sized, we enriched the data for the experiment. After pre-processing, the minimum and maximum size of the image scaling were 0.5 times and 1.2 times that of the size of the image of the input model. We performed random horizontal and vertical flipping of the image. Finally, we used random cropping to expand the data set image.
Parameter settings
Based on the properties of the eye image itself, we found that cropping the image uniformly to a size of 480×480 provided better results than other sizes with the initial learning rate set to 0.001. Due to the performance settings of the server, we set the epoch number to 200, the batch size to 2, the optimizer to the default parameter Adam, the momentum to 0.9, and the network’s input layer channel to 3. We extended the first convolutional layer channel to 16 and set the output layer channel to 1. The MCSE-U-net model was trained from scratch using the training set of each data set, and the weights obtained were used to make predictions for the data in each test set.
Evaluation metrics
We used a confusion matrix to calculate the assessment metrics, including the Dice coefficient, sensitivity (sen), specificity (spe), acc, and mean Intersection over Union (mIoU), and to quantitatively assess the correctness of the approach for segmenting the retinal vessels presented in this study. In terms of vascular segmentation, the Dice coefficient illustrates the relationship between sen and acc It assesses the degree of overlap between the predicted and ground truth and is a crucial measure in segmentation work. Sen measures how well the model can distinguish between different types of vessels by representing the percentage of pixels properly identified as vessels compared to all blood vessel pixels. Spe measures how well a model can distinguish between the background and foreground by comparing the percentage of successfully segmented background pixels to the total number of real background pixels. Acc refers to the proportion of accurately segmented blood vessel pixels compared to the total number of segmented blood vessel pixels. The mIoU is frequently employed to rate image segmentation operations. The mIoU for each category is determined, and the IoU for all categories is then averaged.
The formulas for the assessment metrics are expressed as follows:
where true positive (TP) represents a correctly classified vessel pixel; true negative (TN) represents a correctly classified background pixel; false positive (FP) represents a pixel where the background was misclassified as a vessel; and false negative (FN) represents a pixel where the vessel was mislabeled as the background. In Eq. [5], the k represents the number of categories, and the value of k in vessel segmentation is 1, while k+1 represents the number of categories after adding the background.
Cross verification
In this experiment, given the small sample size of each data set, K-fold cross-validation was used in training to avoid large experimental errors and overfitting to enhance the robustness of the analysis (16). In the K-fold cross-validation, the entire data set was divided into K equally sized parts. Each partition was called a “fold”. Therefore, as we had K parts, we called it a k-fold. One-fold was used as the verification set, and the remaining K-1 folds were used as the training set. This validation was repeated K times until each fold was used as a validation set and the remaining folds were used as a training set, and the final acc of the model was calculated by taking the average acc of the K-model’s validation data.
Results
We used cross-validation methods on all three data sets and compared the results. Tables 1-3 set out the quantitative results for the retinal vessel segmentation using the DRIVE, STARE, and CHASE_DB1 data sets, respectively. Table 1 compares the segmentation results obtained by the relevant advanced models using the DRIVE data set. As Tables 1-3 show, our MCSE-U-net method performed better across all five evaluation metrics on the DRIVE, STARE, and CHASE_DB1 data sets.
Table 1
| Method | Year | Dice | Sen | Spe | Acc | mIoU |
|---|---|---|---|---|---|---|
| Multi-scale CNN (17) | 2018 | – | 0.7772 | 0.9794 | 0.9533 | – |
| Laddernet (18) | 2018 | – | 0.7856 | 0.9810 | 0.9561 | – |
| DU-net (19) | 2019 | – | – | – | 0.9697 | – |
| AD-U-net (20) | 2019 | – | 0.8075 | 0.9814 | 0.9663 | – |
| Vessel-net (21) | 2019 | – | 0.8038 | 0.9802 | 0.9578 | – |
| FA-FCN (22) | 2019 | – | 0.7940 | 0.9820 | 0.9579 | – |
| BA-Transform (23) | 2019 | – | 0.7940 | 0.9816 | 0.9567 | – |
| SE-GAN (24) | 2020 | – | 0.8135 | 0.9768 | 0.9560 | – |
| Octave-U-net (25) | 2019 | – | 0.8374 | 0.9790 | 0.9664 | – |
| BEFD (26) | 2020 | – | 0.8215 | 0.9845 | 0.9701 | – |
| FAE-Segmentation (27) | 2021 | – | 0.8448 | 0.9900 | 0.9819 | – |
| SA-U-net (3) | 2021 | – | 0.8212 | 0.9840 | 0.9698 | – |
| MCPANet (7) | 2022 | 0.8315 | 0.8356 | 0.9836 | 0.9705 | – |
| MR-U-net (11) | 2022 | – | 0.8058 | 0.9863 | 0.9705 | – |
| MC-U-net (28) | 2022 | – | 0.8100 | 0.9879 | 0.9678 | – |
| BCR-U-net (29) | 2022 | – | 0.8183 | 0.9840 | 0.9695 | – |
| SDAU-net (30) | 2023 | – | 0.7955 | 0.9848 | 0.9682 | – |
| MRC-net (31) | 2023 | 0.8270 | 0.8250 | 0.9837 | 0.9698 | – |
| U-net | 2023 | 0.8122 | 0.8130 | 0.9840 | 0.9664 | 0.8172 |
| MCSE-U-net | 2023 | 0.8430 | 0.8752 | 0.9902 | 0.9725 | 0.8473 |
MCSE-U-net, multi-convolution block and squeeze and excitation based on the U-shape network; DRIVE, Digital Retinal Images for Vessel Extraction; sen, sensitivity; spe, specificity; acc, accuracy; mIoU, mean intersection over union; CNN, convolutional neural networks; DU, deformable U-net; AD, attention densenet; FA-FCN, Separable Spatial and Channel Flow and Densely Adjacent and Fully Convolutional Network; BA, Bilinear attentional; SE-GAN, Squeeze and Excitation and Generative Adversarial Network; BEFD, Boundary Enhancement and Feature Denoising; SA, spatial attention; MCPANet, Multiscale Cross-Position Attention Network; MR, Multi-scale and Residual convolutions; MC, Multimodule Concatenation; BCR, Bi-directional ConvLSTM Residual; SDA, Series Deformable convolution and Lightweight Attention; MRC, Multi-resolution Contextual.
Table 2
| Method | Dice | Sen | Spe | Acc | mIoU |
|---|---|---|---|---|---|
| U-net | 0.7918 | 0.8603 | 0.9660 | 0.9569 | 0.8081 |
| MCSE-U-net | 0.8108 | 0.8997 | 0.9738 | 0.9636 | 0.8200 |
MCSE-U-net, multi-convolution block and squeeze and excitation based on the U-shape network; STARE, Structured Analysis of the Retinal; sen, sensitivity; spe, specificity; acc, accuracy; mIoU, mean intersection over union.
Table 3
| Method | Dice | Sen | Spe | Acc | mIoU |
|---|---|---|---|---|---|
| U-net | 0.6841 | 0.7877 | 0.9763 | 0.9611 | 0.7391 |
| MCSE-U-net | 0.7085 | 0.7975 | 0.9799 | 0.9653 | 0.7550 |
MCSE-U-net, multi-convolution block and squeeze and excitation based on the U-shape network; CHASE_DB1, Child Heart and Health Study in England; sen, sensitivity; spe, specificity; acc, accuracy; mIoU, mean intersection over union.
As Table 1 shows, in relation to the DRIVE data set, our MCSE-U-net model performed well on all the metrics overall. Notably, it had a Dice coefficient of 0.8430, a sen of 0.8752, a spe of 0.9902, an acc of 0.9725, and a mIoU of 0.8473. The baseline U-net model was compared to the MCSE-U-net model, and the Dice coefficient improved significantly from 0.8122 to 0.8430, which represents an improvement of 3.08%, the sen improved from 0.8130 to 0.8752, the spe improved from 0.9840 to 0.9902, the acc improved from 0.9664 to 0.9725, and the mIoU improved significantly from 0.8172 to 0.8473. We also compared the results listed in Table 1 for the DRIVE data set with the results for the most recent models on the DRIVE data set, and found that the MCSE-U-net model ranked first across all the metrics except acc. More specifically, it achieved the highest Dice coefficient (1.15% higher than the second result), the best sen (0.02% higher than the previous highest score), the highest spe (3.04% higher than the previous highest score), and the best mIoU (3.01% higher than the next best result). Notably, the acc (0.9725) of our method was close to the best score achieved (0.9819). The above comparison shows the power of our model in handling semantic segmentation.
As Table 2 shows, in relation to the STARE data set, the MCSE-U-net model was more stable than the baseline model and performed better across all five performance indicators. Specifically, the Dice coefficient increased from 0.7918 to 0.8108, the sen increased from 0.8603 to 0.8997, the spe increased from 0.9660 to 0.9738, the acc increased from 0.9569 to 0.9636, and the mIoU increased from 0.8081 to 0.8200.
As Table 3 shows, in relation to the CHASE_DB1 data set, the MCSE-U-net model demonstrated superior stability than the baseline model, and exhibited better performance across all five evaluation metrics. Specifically, the Dice coefficient increased from 0.6841 to 0.7085, the sen increased from 0.7877 to 0.7975, the spe increased from 0.9763 to 0.9799, the acc increased from 0.9611 to 0.9653, and the mIoU increased from 0.7391 to 0.7550.
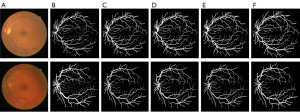
Figure 5 shows blood vessel images extracted from the DRIVE data set images; Figure 5A shows the original image; Figure 5B shows the ground truth; Figure 5C shows the segmentation results of the baseline U-net model; Figure 5D displays the segmentation results of the baseline SA-U-net model; Figure 5E displays the segmentation results of the baseline MR-U-net model; and Figure 5F presents the results obtained according to the algorithm of this article. As Figure 5 shows, both the U-net model and the MCSE-U-net model could detect the coarse blood vessels, but the MCSE-U-net model could detect some fine blood vessels that the U-net model could not detect (Figure 5).
In addition, to verify the stability of the model’s performance, we used the weights obtained from the training of the DRIVE data set to verify the data in the STARE data set and CHASE_DB1 data set, and compared the results (Tables 2,3) obtained from the training and prediction of the STARE data set and CHASE_DB1 data set. As Table 4 shows, the proposed model had better generalization performance.
Table 4
| Data set | Dice | Sen | Spe | Acc | mIoU |
|---|---|---|---|---|---|
| STARE | 0.7903 | 0.8694 | 0.9653 | 0.9503 | 0.8264 |
| CHASE_DB1 | 0.7102 | 0.7998 | 0.9777 | 0.9667 | 0.7538 |
MCSE-U-net, multi-convolution block and squeeze and excitation based on the U-shape network; sen, sensitivity; spe, specificity; acc, accuracy; mIoU, mean intersection over union; STARE, Structured Analysis of the Retina; CHASE_DB1, Child Heart and Health Study in England.
Discussion
To show that each module added in this article enhanced the performance of the model, ablation experiments were conducted on each module to verify its effect and the related data are presented in Table 5. We performed a set of ablation experiments on the data set to compare the baseline U-net model with the model to which a pre-processing module had been added. The results of the quantitative evaluation are shown in Table 5. Notably, in terms of the five kinds of segmentation metrics, the model to which the pre-processing module was added performed better than the model with no pre-processing module. The Dice coefficient increased from 0.7863 to 0.8545, the sen increased from 0.9165 to 0.9212, the spe increased from 0.9700 to 0.9898, the acc increased from 0.9676 to 0.9740, and the mIoU increased from 0.8067 to 0.8589. Thus, we can conclude that adding the pre-processing module to the whole model made it more effective.
Table 5
| DRIVE | Dice | Sen | Spe | Acc | mIoU |
|---|---|---|---|---|---|
| U-net | 0.7863 | 0.9165 | 0.9700 | 0.9676 | 0.8067 |
| U-net + Pre | 0.8545 | 0.9212 | 0.9898 | 0.9740 | 0.8589 |
| U-net + MC | 0.8496 | 0.9206 | 0.9877 | 0.9730 | 0.8547 |
| U-net + SE | 0.8065 | 0.9206 | 0.9804 | 0.9705 | 0.8221 |
| U-net + Pre + MC | 0.8560 | 0.9078 | 0.9907 | 0.9747 | 0.8604 |
| U-net + Pre + MC + SE + post-processing | 0.8255 | 0.8550 | 0.9837 | 0.9688 | 0.8347 |
| U-net + Pre + MC + SE | 0.8573 | 0.9122 | 0.9904 | 0.9748 | 0.8615 |
DRIVE, Digital Retinal Images for Vessel Extraction; sen, sensitivity; spe, specificity; acc, accuracy; mIoU, mean intersection over union; U-net + Pre, U-net and pre-processing; U-net + MC, U-net and multi-convolution block; U-net + SE, U-net and Squeeze and Excitation; U-net + Pre + MC, U-net and pre-processing and Multi-convolution block; U-net + Pre + MC + SE + post-processing, U-net and pre-processing and multi-convolution block and post-processing; U-net + Pre + MC + SE, U-net and pre-processing and multi-convolution block.
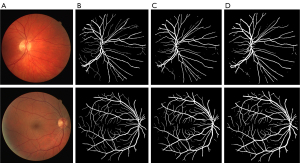
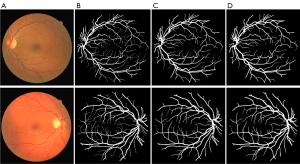
In addition, we also compared the baseline model and the image after the addition of the pre-processing module. Figure 6A shows the original image; Figure 6B shows the ground truth; Figure 6C shows the segmentation results of the baseline U-net model; Figure 6D shows the segmentation results of the U-net + Pre model. As these figures show, compared to the baseline model, the addition of the pre-processing module improved the segmentation effect. As Figure 6 shows, the addition of the pre-processing module enhanced the ability of the model to inspect blood vessels.
We performed two sets of ablation experiments on the DRIVE data set, the first of which compared the benchmark U-net model with the benchmark U-net model after the addition of the multi-scale module alone. The quantitative evaluation results are shown in Table 5. As Table 5 shows, for the five kinds of segmentation metrics, the model to which the multi-scale framework had been added performed better than the benchmark model, and the metrics for each segmentation improved. Specifically, the Dice coefficient improved from 0.7863 to 0.8496, the sen improved from 0.9165 to 0.9206, the spe improved from 0.9700 to 0.9877, the acc improved from 0.9676 to 0.9730, and the mIoU from 0.8067 to 0.8547.
The second group of ablation experiments was based on the first group of ablation experiments; however, a pre-processing module was added to the model for the two experiments. The quantitative evaluation results are shown in Table 5. As Table 5 shows, compared to the first group of ablation experiments, the results changed. The addition of the pre-processing module improved the experimental effect of the second group compared to the first group. A multi-scale module was also added in addition to the pre-processing module. However, in terms of the sen, the performance of the model that included both the pre-processing module and the multi-scale module was slightly lower than that of the benchmark model. However, after the addition of the pre-processing module, the overall performance of the model was still good. Specifically, the Dice coefficient increased from 0.8545 to 0.8560, the spe increased from 0.9898 to 0.9907, the acc increased from 0.9740 to 0.9747, and the mIoU increased from 0.8589 to 0.8604. These results show that the model’s overall performance was better than that of model to which only the multi-scale module had been added.
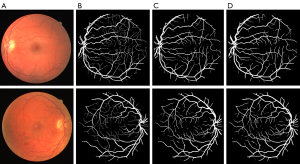
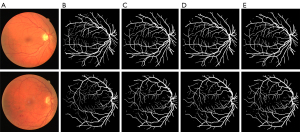
Table 5 shows the importance of the multi-scale framework in improving the performance of the models. Additionally, the overall performance of the model continued to improve after the addition of the pre-processing module. Thus, the pre-processing module is also very important in the multi-scale framework. We also generated graphs and diagrams to show the effects of the multi-scale framework on the segmentation effect. Figure 7A shows the original image; Figure 7B shows the ground truth; Figure 7C shows the segmentation results of the baseline U-net model; Figure 7D shows the segmentation results of the U-net + MC model. Figure 8A shows the original image; Figure 8B shows the ground truth; Figure 8C shows the segmentation results of the benchmark U-net model; Figure 8D shows the segmentation results of the U-net + Pre model; Figure 8E shows the segmentation results of the U-net + Pre + MC model. As these figures show, adding the multi-scale module increased the ability of the model to accurately detect the fine blood vessels compared to the benchmark model (i.e., the ability of the model to inspect blood vessels was enhanced by the addition of the MC module).
To evaluate the effectiveness of the SE module for retinal vessels, we conducted two sets of ablation experiments using the DRIVE data set. The first set of ablation experiments compared the baseline U-net model to a model to which the SE module had been added. The quantitative evaluation of the results are shown in Table 5. As Table 5 shows, in terms of segmentation, the model to which the SE module had been added performed better than the U-net model. Specifically, the Dice coefficient increased from 0.7865 to 0.8065, the sen increased from 0.9165 to 0.9206, the spe increased from 0.9700 to 0.9804, the acc increased from 0.9676 to 0.9705, and the mIoU increased from 0.8067 to 0.8221.
The second group of ablation experiments was based on the first group of ablation experiments. A multi-scale framework and pre-processing module were added to two experimental models. The quantitative evaluation results are shown in Table 5, which compares the results of the second group of ablation experiments to the first group of ablation experiments. The results showed certain changes. The overall model effect improved. Specifically, the Dice coefficient increased from 0.8560 to 0.8573, the sen increased from 0.9078 to 0.9122, the acc increased from 0.9747 to 0.9748, and the mIoU increased from 0.8604 to 0.8615, while the spe decreased by 0.03% from 0.9907 to 0.9904.
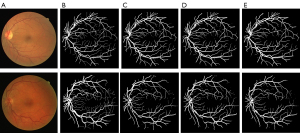
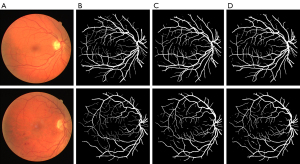
In addition, we evaluated the effect of the SE module on segmentation (Figures 9,10). Figure 9A shows the original image; Figure 9B shows the ground truth; Figure 9C shows the segmentation results of the baseline U-net model; and Figure 9D shows the segmentation results of the U-net + SE model. Figure10A shows the original image; Figure 10B shows the ground truth; Figure 10C shows the segmentation results of the benchmark model U-net; Figure 10D shows the segmentation results of U-net + Pre + MC model; and Figure 10E shows the segmentation results of the U-net + Pre + MC + SE model. As Figures 9 and 10 show, the model to which the SE module had been added was better able to detect fine blood vessels than the baseline model to which the pre-processing module and the MC module had been added.
In addition, inspired by Liu et al. (32), we introduced a post-processing method of double threshold iteration in the ablation experiment to improve the acc of the segmentation results. As Table 5 shows, after the addition of this post-processing method, the acc of the MCSE-U-net model decreased. Specifically, the Dice coefficient decreased from 0.8573 to 0.8255, the sen decreased from 0.9122 to 0.8550, the spe decreased from 0.9904 to 0.9837, the acc decreased from 0.9748 to 0.9688, and the mIoU decreased from 0.8615 to 0.8347. However, as Figure 11 shows, the continuity of the blood vessels was enhanced. Therefore, we will continue to study segmentation models that include this post-processing method in the future to increase segmentation acc.
Conclusions
In this article, a novel MCSE module was proposed to improve the recognition and segmentation of fine blood vessels in retinal vascular images. The comparison and ablation experiment results showed that the segmentation performance of the MSCE-U-net model was superior to the baseline U-net model and most state-of-the-art methods, proving the superiority of the proposed method.
Acknowledgments
The authors would like to thank Wang Biao and Hu Ming for their helpful advice.
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-1454/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wu H, Wang W, Zhong J, Lei B, Wen Z, Qin J. SCS-Net: A Scale and Context Sensitive Network for Retinal Vessel Segmentation. Med Image Anal 2021;70:102025. [Crossref] [PubMed]
- Ahn E, Kumar A, Fulham M, Feng D, Kim J. Convolutional sparse kernel network for unsupervised medical image analysis. Med Image Anal 2019;56:140-51. [Crossref] [PubMed]
- Guo C, Szemenyei M, Yi Y, Wang W, Chen B, Fan C. SA-UNet: Spatial Attention U-Net for Retinal Vessel Segmentation. 2020 25th International Conference on Pattern Recognition (ICPR) 2021:1236-42.
- Khandouzi A, Ariafar A, Mashayekhpour Z, Pazira M, Baleghi Y. Retinal Vessel Segmentation, a Review of Classic and Deep Methods. Ann Biomed Eng 2022;50:1292-314. [Crossref] [PubMed]
- Yun J, Ning T, Tingting P, Hai Z. Retinal Vessels Segmentation Based on Dilated Multi-Scale Convolutional Neural Network. IEEE Access 2019;7:76342-52.
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Lecture Notes in Computer Science, Springer, Cham, 2015:9351.
- Jiang Y, Liang J, Cheng T, Zhang Y, Lin X, Dong J. MCPANet: Multiscale Cross-Position Attention Network for Retinal Vessel Image Segmentation. Symmetry 2022;14:1357-76.
- Gao G, Li J, Yang L, Liu Y. A multi-scale global attention network for blood vessel segmentation from fundus images. Measurement 2023;222:113553.
- Beeche C, Singh JP, Leader JK, Gezer S, Oruwari AP, Dansingani KK, Chhablani J, Pu J. Super U-Net: a modularized generalizable architecture. Pattern Recognit 2022;128:108669. [Crossref] [PubMed]
- Hu J, Wang H, Gao S, Bao M, Liu T, Wang Y, Zhang J. S-UNet: A Bridge-Style U-Net Framework With a Saliency Mechanism for Retinal Vessel Segmentation. IEEE Access 2019;7:174167-77.
- Xin Y, Li L, Tao L. MR‐UNet: An UNet model using multi‐scale and residual convolutions for retinal vessel segmentation. Int J Imaging Syst Technol 2022;32:1588-603.
- Sooyeon Lee YK. Youn Jin Kim, SeHyeok Park, and Jaehyun Kim. Contrast preserved chroma enhancement technique using YCbCr color space. IEEE Transactions on Consumer Electronics 2012;58:641-5.
- Available online: https://drive.grand-challenge.org/
- Available online: http://cecas.clemson.edu/ahoover/stare/
- Available online: https://blogs.kingston.ac.uk/retinal/chasedb1/
- Arlot S, Celisse A. Segmentation of the mean of heteroscedastic data via cross-validation. Stat Comput 2011;21:613-32.
- Hu K, Zhang Z, Niu X, Zhang Y, Cao C, Xiao F, Gao X. Retinal vessel segmentation of color fundus images using multiscale convolutional neural network with an improved cross-entropy loss function. Neurocomputing 2018; [Crossref]
Zhuang J. LadderNet: Multi-path networks based on U-Net for medical image segmentation. arXiv: 1810.07810.- Jin Q, Meng Z, Pham TD, Chen Q, Wei L, Su R. DUNet: A deformable network for retinal vessel segmentation. Knowledge-Based Systems 2019;178:149-62.
- Luo Z, Zhang Y, Zhou L, Zhang B, Luo J, Wu H. Micro-Vessel Image Segmentation Based on the AD-UNet Model. IEEE Access 2019;7:143402-11.
- Yue K, Zou B, Chen Z, Liu Q. Retinal vessel segmentation using dense U-net with multiscale inputs. J Med Imaging (Bellingham) 2019;6:034004. [Crossref] [PubMed]
- Lyu J, Cheng P, Tang X. Fundus Image Based Retinal Vessel Segmentation Utilizing a Fast and Accurate Fully Convolutional Network. In: Fu H, Garvin M, MacGillivray T, Xu Y, Zheng Y. editors. Ophthalmic Medical Image Analysis. OMIA 2019. Lecture Notes in Computer Science, vol 11855. Springer, Cham, 2019:112-20.
- Chi L, Yuan Z, Mu Y, Wang C. Non-local neural networks with grouped bilinear attentional transforms. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Seattle, WA, USA, 2020:11801-10.
Zhou Y Chen Z Shen H Peng P Zeng Z Zheng X. A Symmetric Equilibrium Generative Adversarial Network with Attention Refine Block for Retinal Vessel Segmentation. arXiv: 1909.11936.Fan Z Mo J Qiu B. Accurate Retinal Vessel Segmentation via Octave Convolution Neural Network. arXiv: 1906.12193- Zhang M, Yu F, Zhao J, Zhang L, Li Q. BEFD: Boundary Enhancement and Feature Denoising for Vessel Segmentation. In: Martel AL, Abolmaesumi P, Stoyanov D, Mateus D, Zuluaga ZA, Zhou SK, Racoceanu D, Joskowicz L. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science, vol 12265. Springer, Cham, 2020.
- Boudegga H, Elloumi Y, Akil M, Hedi Bedoui M, Kachouri R, Abdallah AB. Fast and efficient retinal blood vessel segmentation method based on deep learning network. Comput Med Imaging Graph 2021;90:101902. [Crossref] [PubMed]
- Li J, Zhang T, Zhao Y, Chen N, Zhou H, Xu H, Guan Z, Xue L, Yang C, Chen R, Wei L. MC-UNet: Multimodule Concatenation Based on U-Shape Network for Retinal Blood Vessels Segmentation. Comput Intell Neurosci 2022;2022:9917691. [Crossref] [PubMed]
- Yi Y, Guo C, Hu Y, Zhou W, Wang W. BCR-UNet: Bi-directional ConvLSTM residual U-Net for retinal blood vessel segmentation. Front Public Health 2022;10:1056226. [Crossref] [PubMed]
- Sun K, Chao Y, Chen Y. A retinal vessel segmentation method based improved U-Net model. Biomedical Signal Processing and Control 2023;82:104574-86.
- Khan TM, Naqvi SS, Robles-Kelly A, Razzak I. Retinal vessel segmentation via a Multi-resolution Contextual Network and adversarial learning. Neural Netw 2023;165:310-20. [Crossref] [PubMed]
- Liu W, Yang H, Tian T, Cao Z, Pan X, Xu W, Jin Y, Gao F. Full-Resolution Network and Dual-Threshold Iteration for Retinal Vessel and Coronary Angiograph Segmentation. IEEE J Biomed Health Inform 2022;26:4623-34. [Crossref] [PubMed]