Automated anatomical landmark detection on 3D facial images using U-NET-based deep learning algorithm
Introduction
Anthropometry on 3-dimensional (3D) images, or 3D photogrammetry, is a powerful tool for the study of facial morphology. It outperforms direct measurement for its convenience and efficiency (1). It is also considered a better alternative to 2-dimensional (2D) photogrammetry because it has higher precision and allows for volume and surface area measurement (2). Previous studies have confirmed 3D photogrammetry as the best method for the analysis of facial soft tissue morphology (3,4). In recent years, 3D imaging, and facial anthropometric analysis have been widely used in many research fields including ethnic study, facial aging study, facial attractiveness study, preoperative consultation, and postoperative follow-up (5-10).
However, 3D photogrammetry is not without limitations. One constraint is that manually localizing landmarks onto the 3D meshes can be a time-consuming process. There is a trend to include an increasing number of 3D facial images for analysis and to plot more anthropometric landmarks for detailed meticulous measurements, calling for the need for automated landmark localization algorithms. Nair and Cavallaro proposed a landmark detection framework based on a 3D point distribution model (11). The study mainly tested the detection accuracy of 5 key landmarks. The absolute mean error in the detection of the endocanthion, exocanthion, and pronasale was about 12, 20, and 9 mm, respectively. There was a non-negligible percentage of detection failure. Liang et al. introduced an improved method to achieve automatic landmark detection (12). They located a set of landmarks on each 3D mesh by geometric techniques and created a dense correspondence between the individual mesh and the template mesh using a deformable transformation algorithm. The study had the advantage of detecting a set of 20 facial landmarks automatically, surpassing the previous studies. Moreover, the study showed a remarkable improvement in detection accuracy. The average distance between the automatic landmarks and the ground truth was only 2.64 mm. Baksi et al. invented another automated landmark identification algorithm, which innovatively added texture information into the landmark detection process (13). 3D meshes underwent initialization, alignment, and elastic deformation. The mean Euclidean difference between automated identified landmarks and manually plotted landmarks was 3.2±1.64 mm. The algorithm would be more convincing if tested on a larger dataset.
U-NET, a convolutional network architecture proposed by Ronneberger et al. in 2015 (14), has gained popularity in medical image processing as it largely increases the efficiency of using annotated samples and yields excellent results in medical image recognition. A common use of U-NET in the medical field is the automated segmentation of tumor lesions and other structures of interest. It is theoretically possible to use U-NET for the segmentation of landmarks in 3D facial images. One of the difficulties in 3D facial image processing is the limited sample size. U-NET has the potential to perform accurate image segmentation using highly limited training samples, making it an ideal network architecture for developing automated landmark localization algorithms. This study aimed to develop a novel automated algorithm based on U-NET for facial landmark detection and test its accuracy on different case groups.
Methods
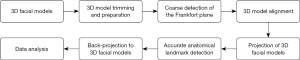
The landmark detection process is briefly outlined in Figure 1. The main steps involved 3D image acquisition, coarse detection of the Frankfort plane, 3D model alignment, 3D model projection, automated landmark detection, back-projection to 3D model, and data analysis.
Study sample and development tools
This study enrolled 200 3D facial images including 160 from healthy individuals, 20 from acromegaly patients, and 20 from localized scleroderma patients (82 males and 118 females, mean age: 37 years old). The healthy cases and localized scleroderma patients were recruited from the Department of Plastic and Aesthetic Surgery, Peking Union Medical College Hospital (PUMCH). The acromegaly patients were recruited from the Department of Endocrinology, PUMCH. All the 3D images were acquired by Vectra H1 handheld 3D camera (Canfield Scientific, Inc., Parsippany, NJ, USA). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of PUMCH. All volunteers signed the informed consent form and agreed on their images and anthropometric data to be used for analysis.
TensorFlow 1.8 (Google, Mountain View, CA, USA) was used as the deep learning framework, and NVIDIA RTX3060 Ti (NVIDIA, Santa Clara, CA, USA) was used as the computing hardware. The training process took about 70 epochs to finish with a batch size of 8, we applied Adam optimizer and a learning rate of 3e-5 in the training process. The landmark detection process is briefly outlined in Figure 1.
Anatomical landmark selection
We selected 20 facial anthropometric landmarks in this study to train the automated landmark detection algorithm. In the periorbital region, the algorithm was trained to automatically localize 8 landmarks including the right endocanthion (enR), left endocanthion (enL), right exocanthion (exR), left exocanthion (exL), right palpebrale superiori (psR), left palpebrale superiori (psL), right palpebrale inferioris (piR), and left palpebrale inferioris (piL). In the nasal region, the algorithm was trained to automatically localize 5 landmarks including nasion (n), pronasale (prn), subnasale (sn), right alare (alR), and left alare (alL). In the orolabial region, the algorithm was trained to automatically localize 7 landmarks including the right crista philtri (cphR), left crista philtri (cphL), right chelion (chR), left chelion (chL), labiale superius (ls), stomion (sto), and labiale inferius (li).
Manual landmark annotation
The 20 landmarks above were manually plotted onto the 200 3D facial images using Geomagic Wrap 2017 (Geomagic, Inc., Research Triangle Park, NC, USA) by the first author with years of experience in stereophotogrammetry. To ensure the accuracy of the manual annotation, 5 3D images were randomly selected from each group to test for intra-observer reliability and another 5 3D images were randomly selected from each group for inter-observer reliability. A mean distance between the 2 sets of landmarks of less than 2 mm was considered clinically acceptable (15-17).
Network architecture and design
The main structure of our network stacks 2 U-NETs. The U-NET architecture, as shown in Figure 2, consisted of a contracting path (left) and an expansive path (right). In the contracting path, we used 3×3 convolution kernel (padded convolutions) and doubled the number of feature channels. Each convolution layer was followed by a rectified linear unit (ReLU) activation function and a 2×2 max pooling operation for down-sampling. The above steps were repeated 3 times. After contracting, the image size became 1/8 of the original input size, and the number of feature channels became 256. In the contracting path, we used transpose convolution that halved the number of the feature channels. Then, there was a concatenation with the corresponding feature map from the contracting path, and 2 3×3 convolutions, each followed by a ReLU. The above steps were also repeated 3 times. At the final layer, a 1×1 convolution was used to correspond each feature vector to the intermediate feature with the same size to generate the final heat map. Also, we embedded drop-out layers in the network to relieve overfit effect during the training process. The outputs of U-NET blocks were heat maps, as shown in Figure 2. The pixel value represented the probability of the target point lying on this pixel position.
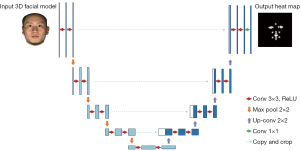
We fed output heat maps of both U-NET blocks into loss function during the training process for intermediate supervision. The loss function was defined as follows:
was the output heat map of the first U-NET block; was the output heat map of the second U-NET block; was the output heat map of the ground truth.
Evaluation metrics and results
Percentage of correct key points (PCK) and normalized mean error (NME) were used to evaluate facial landmark detection accuracy. PCK calculated the fraction of successfully predicted results, of which the distance to ground truth was smaller than a specified threshold, which was usually set as 2 mm, the clinically acceptable threshold used in previous studies (15-17). NME calculated the distance between ground-truth points and model prediction result, then divided it by a normalized factor, as defined below.
was the normalized factor and was set to 1 in this study; was the predicted result; is the ground truth point.
Results
The intra- and inter-observer reliability is shown in Table 1. The mean distances were all less than 2 mm, indicating that manual landmark annotation had good reliability and could be used as the ground truth.
Table 1
| Landmark | Intra-observer variability | Inter-observer variability | |||||
|---|---|---|---|---|---|---|---|
| Healthy cases | Acromegaly patients | LS patients | Healthy cases | Acromegaly patients | LS patients | ||
| Periorbital region | |||||||
| enR | 0.29 | 0.64 | 0.16 | 0.56 | 0.70 | 0.83 | |
| enL | 0.26 | 0.35 | 0.28 | 0.30 | 0.46 | 0.55 | |
| exR | 0.79 | 1.09* | 0.62 | 1.41* | 1.33* | 0.74 | |
| exL | 0.66 | 0.48 | 0.31 | 0.61 | 1.36* | 0.46 | |
| psR | 0.98 | 1.32* | 1.36* | 1.71* | 1.44* | 1.61* | |
| psL | 0.74 | 1.67* | 1.43* | 1.54* | 1.06* | 1.26* | |
| piR | 1.40* | 1.31* | 1.49* | 1.43* | 1.66* | 1.65* | |
| piL | 1.17* | 1.36* | 0.92 | 1.93* | 1.71* | 1.67* | |
| Nasal region | |||||||
| n | 0.82 | 0.73 | 0.58 | 0.49 | 0.77 | 0.57 | |
| prn | 0.84 | 0.56 | 0.93 | 0.23 | 0.81 | 0.35 | |
| sn | 0.58 | 1.13* | 0.59 | 0.54 | 0.43 | 0.62 | |
| alR | 0.96 | 1.49* | 1.45* | 1.10* | 1.51* | 1.81* | |
| alL | 1.14* | 1.50* | 1.06* | 1.48* | 1.32* | 1.88* | |
| Orolabial region | |||||||
| cphR | 0.50 | 0.86 | 0.59 | 0.49 | 0.34 | 0.24 | |
| cphL | 0.66 | 0.87 | 0.66 | 0.39 | 0.23 | 0.22 | |
| chR | 1.04* | 0.74 | 0.71 | 0.40 | 0.53 | 0.28 | |
| chL | 0.68 | 0.63 | 0.44 | 0.77 | 0.43 | 0.31 | |
| ls | 0.35 | 0.71 | 0.37 | 0.46 | 0.41 | 0.27 | |
| sto | 0.30 | 0.78 | 0.29 | 0.50 | 0.37 | 0.42 | |
| li | 0.59 | 1.28* | 0.57 | 0.80 | 0.66 | 0.69 | |
*, values between 1.0 and 2.0 mm. LS, localized scleroderma; enR, right endocanthion; enL, left endocanthion; exR, right exocanthion; exL, left exocanthion; psR, right palpebrale superiori; psL, left palpebrale superiori; piR, right palpebrale inferioris; piL, left palpebrale inferioris; n, nasion; prn, pronasale; sn, subnasale; alR, right alare; alL, left alare; cphR, right crista philtri; cphL, left crista philtri; chR, right chelion; chL, left chelion; ls, labiale superius; sto, stomion; li, labiale inferius.
The detection accuracy on different participant groups is shown in Table 2. Among normal healthy cases, the average NME of the 20 landmarks was 1.4 mm; the NME of 90% (18/20) landmarks were within the clinical acceptance of 2.0 mm. Among the acromegaly patients, the average NME of the 20 landmarks was 2.8 mm; among localized scleroderma patients, the average NME of the 20 landmarks was 2.2 mm. The output of automated landmark detection on 3D facial images is shown in Figure 3.
Table 2
| Landmark | Healthy cases | Acromegaly patients | LS patients |
|---|---|---|---|
| Periorbital region | |||
| enR | 1.03 | 1.52 | 1.01 |
| enL | 1.11 | 2.02* | 1.42 |
| exR | 1.01 | 1.95 | 1.72 |
| exL | 1.15 | 2.10* | 1.42 |
| psR | 1.39 | 1.92 | 1.33 |
| psL | 1.18 | 2.07* | 1.44 |
| piR | 2.04* | 3.52* | 2.07* |
| piL | 1.54 | 2.15* | 1.79 |
| Nasal region | |||
| n | 2.76* | 6.31** | 5.55** |
| prn | 1.51 | 4.4* | 2.58* |
| sn | 0.83 | 1.73 | 1.67 |
| alR | 1.53 | 1.82 | 1.94 |
| alL | 1.02 | 1.57 | 1.57 |
| Orolabial region | |||
| cphR | 1.56 | 4.01* | 2.84* |
| cphL | 1.69 | 3.21* | 1.97 |
| chR | 1.46 | 1.80 | 1.70 |
| chL | 1.61 | 2.95* | 2.28* |
| ls | 1.09 | 3.49* | 2.51* |
| sto | 1.39 | 5.07** | 3.61* |
| li | 1.53 | 3.31* | 3.41* |
*, values between 2.0 and 5.0 mm; **, values higher than 5.0 mm. LS, localized scleroderma; enR, right endocanthion; enL, left endocanthion; exR, right exocanthion; exL, left exocanthion; psR, right palpebrale superiori; psL, left palpebrale superiori; piR, right palpebrale inferioris; piL, left palpebrale inferioris; n, nasion; prn, pronasale; sn, subnasale; alR, right alare; alL, left alare; cphR, right crista philtri; cphL, left crista philtri; chR, right chelion; chL, left chelion; ls, labiale superius; sto, stomion; li, labiale inferius.

PCK revealed the percentage of landmarks that were accurately detected. When the threshold was set to the clinically acceptable limit of 2 mm, the PCK among normal healthy cases, acromegaly patients, and the localized scleroderma patients was 90%, 35%, and 60%, respectively; if the threshold was set to 3 mm, the PCK among normal healthy cases, acromegaly patients, and the localized scleroderma patients was 100%, 60%, and 85%, respectively; if the threshold was set to 5 mm, the PCK among the 3 groups of participants was 100%, 90%, and 95%, respectively (Table 3).
Table 3
| Threshold | Healthy cases | Acromegaly patients | LS patients |
|---|---|---|---|
| 2 mm | 90% | 35% | 60% |
| 3 mm | 100% | 60% | 85% |
| 5 mm | 100% | 90% | 95% |
LS, localized scleroderma.
Discussion
This study proposed a novel U-NET based algorithm to achieve automated landmark detection on 3D facial images. The algorithm was tested on 3 groups of participants. The algorithm achieved accurate landmark detection on normal healthy cases. The average NME on normal cases was only 1.4 mm, whereas the average NME on acromegaly patients and localized scleroderma patients was 2.2 and 2.8 mm, respectively.
Currently, deep learning techniques are widely applied in clinical areas to increase the diagnosis and treatment efficiency of surgeons. For example, deep neural networks have been applied to automatically identify and segment lung cancer regions (18,19). Shen et al. used a Visual Geometry Group (VGG)-structured network to localize and extract suspicious breast cancer areas on full-field digital mammography images (20). Orthopedic doctors utilized deep learning-related technology in recent years to accurately segment bones for disease evaluation and for pre-surgical planning to intervene clinical treatment process (21-23).
Attempts to develop algorithms for landmark detection on 3D models are not new. Abu et al. automated the craniofacial landmark detection on 3D images and validated it on 8 inter-landmark distances (24). Baksi et al. developed an algorithm and tested on 30 3D facial images and achieved a mean Euclidean difference of 3.2 mm (13). In comparison, this study achieved an average NME of 1.4 mm among normal cases, which was significantly smaller than previous results. In this study, we mainly used U-NET structure as our backbone network, as U-NET has displayed high robustness and stability in other medical research fields. The network was down-sampled to extract high-level features and then up-sampled to restore original input size, and down-sampled layers would be concatenated to up-sampled layers for feature enhancement and derivative maintenance. Besides, we stacked 2 U-NET structures and made an intermediate supervision instead of building up a bigger and deeper neural network, so that the network architecture would be more light-weighted and easily converged during the training process.
3D imaging and facial anthropometric analysis have gained increasing popularity among plastic surgeons. It gradually replaces traditional 2D photogrammetry to be the most reliable tool for morphologic analysis. A previous meta-analysis demonstrated that common 3D facial optical instruments were reliable in linear distance measurement and suitable for research and clinical use (25). As 3D images accumulate, surgeons tend to spend increasingly more time plotting anatomical landmarks and doing measurements. Although some commercial software such as Vectra Analytical Module (VAM) supports automatic landmark detection, its accuracy is debatable, and it is usually bundled with hardware for sale. This study proposed a novel algorithm for landmark localization. It automatically plots 20 landmarks on a 3D image in seconds with high accuracy and reproducibility. This algorithm spares surgeons from the laborious and time-consuming process of plotting landmarks.
Additionally, manual landmark identification is subject to human error. Different researchers, or even the same researcher at different times, reliably yield inconsistent results. Testing for intra- and inter-observer reliability reveals whether the bias is acceptable but does not deny its existence. Using an automated algorithm to detect landmarks ensures the placement of the landmarks is always consistent so that the measurement is more reliable and no further statistical testing is needed.
The algorithm showed different prediction accuracy in different groups of cases. Landmark automatic localization on normal healthy humans and localized scleroderma patients was reliable, whereas the test on acromegaly patients did not yield clinically acceptable results. It is reasonable that prediction accuracy decreased when there were local deformities. For example, acromegaly patients usually have soft tissue changes in the nasolabial region. The algorithm had poor prediction accuracy on landmarks in this region such as prn, cph, ls, and sto. This study innovatively trained the algorithm with 3D facial images from patients with facial abnormalities to improve its prediction accuracy on 3D images with local deformities. There are possible ways to improve the prediction accuracy. Apart from enlarging the sample size for algorithm training, prediction accuracy can be improved by adding attention gates to highlight salient features that passed through the skip connections and using context fuse modules to aggregate contextual information at multiple scales. In the future, the algorithm will be improved to be more accurate and applied to a wider range of diseases with facial abnormalities.
Conclusions
Automated landmark detection on 3D facial images has the potential to improve the efficiency and the accuracy of 3D photogrammetry. This study developed a U-NET-based algorithm for automated facial landmark detection on 3D models. The algorithm was validated in 3 different participant groups. It achieved great landmark localization accuracy on normal healthy cases. The average NME of the 20 landmarks was 1.4 mm. The prediction error of 90% of the landmarks was within the clinically acceptable range (<2 mm). The algorithm achieved good landmark localization accuracy on localized scleroderma patients. The average NME was 2.2 mm. The accuracy on acromegaly patients needs improvement, for whom the average NME was 2.8 mm. Possible improvements to the algorithm include adding attention gates and context fuse modules.
Acknowledgments
Funding: This study was funded by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1108/coif). Y.A. and Q.H. report that they were full-time employees of Beijing Li-Med Medical Technology Co., Ltd. during the conduct of the study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Peking Union Medical College Hospital. All volunteers signed the informed consent form and agreed on their images and anthropometric data to be used for analysis.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wong JY, Oh AK, Ohta E, Hunt AT, Rogers GF, Mulliken JB, Deutsch CK. Validity and reliability of craniofacial anthropometric measurement of 3D digital photogrammetric images. Cleft Palate Craniofac J 2008;45:232-9. [Crossref] [PubMed]
- Farkas LG, Bryson W, Klotz J. Is photogrammetry of the face reliable? Plast Reconstr Surg 1980;66:346-55. [Crossref] [PubMed]
- Ghoddousi H, Edler R, Haers P, Wertheim D, Greenhill D. Comparison of three methods of facial measurement. Int J Oral Maxillofac Surg 2007;36:250-8. [Crossref] [PubMed]
- Weissler JM, Stern CS, Schreiber JE, Amirlak B, Tepper OM. The Evolution of Photography and Three-Dimensional Imaging in Plastic Surgery. Plast Reconstr Surg 2017;139:761-9. [Crossref] [PubMed]
- Celebi AA, Kau CH, Femiano F, Bucci L, Perillo L. A Three-Dimensional Anthropometric Evaluation of Facial Morphology. J Craniofac Surg 2018;29:304-8. [Crossref] [PubMed]
- Chong Y, Li J, Liu X, Wang X, Huang J, Yu N, Long X. Three-dimensional anthropometric analysis of eyelid aging among Chinese women. J Plast Reconstr Aesthet Surg 2021;74:135-42. [Crossref] [PubMed]
- Kim YC, Kwon JG, Kim SC, Huh CH, Kim HJ, Oh TS, Koh KS, Choi JW, Jeong WS. Comparison of Periorbital Anthropometry Between Beauty Pageant Contestants and Ordinary Young Women with Korean Ethnicity: A Three-Dimensional Photogrammetric Analysis. Aesthetic Plast Surg 2018;42:479-90. [Crossref] [PubMed]
- Modabber A, Peters F, Galster H, Möhlhenrich SC, Bock A, Heitzer M, Hölzle F, Kniha K. Three-dimensional evaluation of important surgical landmarks of the face during aging. Ann Anat 2020;228:151435. [Crossref] [PubMed]
- Chong Y, Dong R, Liu X, Wang X, Yu N, Long X. Stereophotogrammetry to reveal age-related changes of labial morphology among Chinese women aging from 20 to 60. Skin Res Technol 2021;27:41-8. [Crossref] [PubMed]
- Miller TR. Long-term 3-Dimensional Volume Assessment After Fat Repositioning Lower Blepharoplasty. JAMA Facial Plast Surg 2016;18:108-13. [Crossref] [PubMed]
- Nair P, Cavallaro A. 3-D Face Detection, Landmark Localization, and Registration Using a Point Distribution Model. IEEE Transactions on Multimedia 2009;11:611-23. [Crossref]
- Liang S, Wu J, Weinberg SM, Shapiro LG. Improved detection of landmarks on 3D human face data. Annu Int Conf IEEE Eng Med Biol Soc 2013;2013:6482-5. [PubMed]
- Baksi S, Freezer S, Matsumoto T, Dreyer C. Accuracy of an automated method of 3D soft tissue landmark detection. Eur J Orthod 2021;43:622-30. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention; Cham: Springer, 2015:234-41.
- Dindaroğlu F, Kutlu P, Duran GS, Görgülü S, Aslan E. Accuracy and reliability of 3D stereophotogrammetry: A comparison to direct anthropometry and 2D photogrammetry. Angle Orthod 2016;86:487-94. [Crossref] [PubMed]
- Othman SA, Saffai L, Wan Hassan WN. Validity and reproducibility of the 3D VECTRA photogrammetric surface imaging system for the maxillofacial anthropometric measurement on cleft patients. Clin Oral Investig 2020;24:2853-66. [Crossref] [PubMed]
- Weinberg SM, Naidoo S, Govier DP, Martin RA, Kane AA, Marazita ML. Anthropometric precision and accuracy of digital three-dimensional photogrammetry: comparing the Genex and 3dMD imaging systems with one another and with direct anthropometry. J Craniofac Surg 2006;17:477-83. [Crossref] [PubMed]
- Kalaivani N, Manimaran N, Sophia D, Devi D. Deep Learning Based Lung Cancer Detection and Classification. IOP Conf Ser: Mater Sci Eng 2020;994:012026.
- Kadir T, Gleeson F. Lung cancer prediction using machine learning and advanced imaging techniques. Transl Lung Cancer Res 2018;7:304-12. [Crossref] [PubMed]
- Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci Rep 2019;9:12495. [Crossref] [PubMed]
- Gangwar T, Calder J, Takahashi T, Bechtold JE, Schillinger D. Robust variational segmentation of 3D bone CT data with thin cartilage interfaces. Med Image Anal 2018;47:95-110. [Crossref] [PubMed]
- Chen F, Liu J, Zhao Z, Zhu M, Liao H. Three-Dimensional Feature-Enhanced Network for Automatic Femur Segmentation. IEEE J Biomed Health Inform 2019;23:243-52. [Crossref] [PubMed]
- Chen H, Zhao N, Tan T, Kang Y, Sun C, Xie G, Verdonschot N, Sprengers A. Knee Bone and Cartilage Segmentation Based on a 3D Deep Neural Network Using Adversarial Loss for Prior Shape Constraint. Front Med (Lausanne) 2022;9:792900. [Crossref] [PubMed]
- Abu A, Ngo CG, Abu-Hassan NIA, Othman SA. Automated craniofacial landmarks detection on 3D image using geometry characteristics information. BMC Bioinformatics 2019;19:548. [Crossref] [PubMed]
- Gibelli D, Dolci C, Cappella A, Sforza C. Reliability of optical devices for three-dimensional facial anatomy description: a systematic review and meta-analysis. Int J Oral Maxillofac Surg 2020;49:1092-106. [Crossref] [PubMed]