
A fusion model integrating magnetic resonance imaging radiomics and deep learning features for predicting alpha-thalassemia X-linked intellectual disability mutation status in isocitrate dehydrogenase–mutant high-grade astrocytoma: a multicenter study
Introduction
Gliomas are common primary malignant tumors in the brain and can be categorized into different subtypes based on their histopathological characteristics (1). The presence of the same genetic alterations in patients with different pathological histological classifications of glioma suggests that they may have similar biological behavior and prognosis (2). Therefore, isocitrate dehydrogenase (IDH)–mutant astrocytoma is classified as a distinct type in the 2021 World Health Organization (WHO) Central Nervous System (CNS) Tumor Classification Criteria. It is classified into 3 grades, WHO CNS grades 2 to 4, based on histological morphology and features (3,4). High-grade glioma (grades 3 and 4) involves a poor prognosis and a low cure rate due to the lack of effective treatments (1). However, patients with glioblastoma with alpha-thalassemia X-linked intellectual disability (ATRX) deletion experience a longer overall survival time and benefit more from temozolomide (TMZ) treatment (5). The combination therapy of TMZ and multitargeted receptor tyrosine kinase inhibitors (RTKis) may expand the therapeutic window for patients with high-grade gliomas carrying ATRX mutations (1). There are significant differences in the treatment approach and prognosis between high- and low-grade IDH-mutant astrocytoma (6). Therefore, knowledge of the mutational status of ATRX is important for both the prognostic assessment and treatment options in high-grade IDH-mutant astrocytoma.
The most common methods for detecting ATRX mutation status are based on sequencing or immunohistochemistry after biopsy or surgical excision (7,8). However, brain biopsy is often hampered by factors such as the patient’s poor health condition and tumor location or patient’s refusal to undergo invasive tests. Additionally, the accuracy of gene detection can be compromised by limited tissue samples, and biopsies involve certain risks, such as brain swelling, bleeding, and other neurological issues (9). Therefore, noninvasively predicting the ATRX mutation status of IDH-mutant high-grade astrocytoma could have considerable clinical value.
Imaging techniques have the advantage over standard pathological examination of being able to analyze the invasive, non-resected components of gliomas and thus capture and characterize the status of the tumor as a whole. To facilitate a consistent and standardized analysis of qualitative magnetic resonance imaging (MRI) features, the Visually Accessible Rembrandt Images (VASARI) terminology was developed (10). Previous studies have shown that VASARI features are biologically relevant to glioblastoma (11). Radiomics can extract high-throughput quantitative features that reveal tumor information from MRI images, and mathematical models based on these quantitative features can predict tumor phenotypes (12). As a common type of artificial neural network in deep learning (DL), convolutional neural networks (CNNs) have been proven capable of performing well in both image recognition and segmentation (13,14). DL and radiomics based on conventional and functional MRI have been widely used for preoperative differential diagnosis, grading, genotyping, and prognosis of gliomas (15-17). They have demonstrated good performance in predicting O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation and IDH mutation in diffuse glioma (18,19). A radiomics approach based on multiparametric MRI can noninvasively determine the molecular status of IDH1 and ATRX in patients with low-grade glioma (LGG) (20). In a previous study (21), a clinical radiomics–integrated model based on 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) and multimodal MRI successfully predicted the ATRX mutation status of patients with IDH-mutant LGG. Although the findings of these studies are promising, the primary focus has been on LGG, and the noninvasive prediction of ATRX mutational status in high-grade IDH-mutant astrocytoma has not yet been examined. Since functional MRI and PET imaging are not as widely available as is conventional MRI (cMRI), it is necessary to thoroughly investigate the potential of cMRI in predicting ATRX mutation status in patients with IDH-mutant high-grade astrocytoma.
In this study, we aimed to combine the quantitative and qualitative features derived from cMRI and clinical variables to build a fusion model to predict ATRX mutation status in IDH-mutant high-grade astrocytoma. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-807/rc).
Methods
Patients
From January 2017 to June 2022, this study analyzed data from 3 different institutions: the First Affiliated Hospital of Chongqing Medical University, Sichuan Cancer Hospital, and Chongqing United Medical Imaging Center. The data and pathological information were obtained from a total of 234 patients with IDH-mutant astrocytoma classified as WHO CNS grades 3 or 4 according to the 2021 WHO criteria. Of these patients, 120 had ATRX mutations.
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the institutional review boards of Chongqing Medical University, Sichuan Cancer Hospital, and the United Medical Imaging Center. All participating institutions were formally informed and agreed to the protocol of the study. Given the retrospective nature of the design, the requirements for informed consent from patients was waived. The inclusion criteria were patients with pathologically confirmed astrocytoma, IDH-mutant status, WHO CNS grades 3 or 4, available MRI with conventional sequences including T2 fluid-attenuated inversion recovery (T2f) and contrast-enhanced T1-weighted (T1c) images, available information regarding ATRX mutation status, and available clinical features including gender and age. The exclusion criteria included images with severe artifacts; previous treatment with radiotherapy, stereotactic radiosurgery, anti-vascular therapy, or surgery; and unknown ATRX mutation status. The patient screening process is shown in Figure 1.
Detection of ATRX
The tumor samples were preserved in a 10% formaldehyde solution at room temperature for a full day, encased in paraffin, and then sliced into sections 3.5-µm thick. The primary antibodies were applied for immunohistochemistry following the guidelines provided by the manufacturer (Cell Signaling Technology, Boston, USA). Each tissue section was treated with a 3% hydrogen peroxide solution at 37 ℃ for 10 min, which was followed by an overnight incubation with the primary antibody at 4 ℃. Finally, sections were exposed to goat anti-mouse/rabbit immunoglobin G (IgG) antibodies for half an hour at room temperature using a 1:100 dilution. The staining with the DAB Detection Kit (ZSGB-BIO, Beijing, China) was observed using a Nikon microscope (Nikon Corporation, Tokyo, Japan; magnification 40×).
Assessment of qualitative clinical variables
The VASARI feature set consists of 30 categorical variables, such as tumor location, proportion enhancing, and proportion of edema (additional details about the VASARI scoring standard can be found in Table S1). The VASARI features were assessed by 2 radiologists with 5- and 10-year experience, respectively, under a double-blind method, and any disagreements were resolved by a neuroradiologist with 15-year experience. For each case, a VASARI score was constructed from the VASARI feature set and considered as a clinical variable along with age, gender, and WHO grading, to differentiate it from the imaging score calculated based on radiomics features, which is described in a later section.
Image preprocessing and region of interest (ROI) segmentation
MR images, including T1c and T2f, were acquired from various 3.0T MRI scanners using different acquisition parameters. The specific acquisition protocols can be found in Table S2. To minimize differences in imaging parameters across devices, all images underwent preprocessing steps such as registration, bias correction, intensity normalization, and resampling. Additional information regarding the image preprocessing can be found in Appendix 1 (22,23).
The segmentation of the 3-dimensional ROI was performed by 2 radiologists with 5 and 10 years of experience, respectively, using 3D-slicer software (version 4.3; https://www.slicer.org) (24). They manually segmented the ROIs of the overall lesion (enhancing tumor + edema) and enhancing tumor area from T2f and T1C, respectively, to obtain the ROI of edema habitat determination (25). If the difference between the ROIs obtained by the 2 radiologists was less than 5%, the final ROI was determined as the overlapping region of the 2 ROIs. Otherwise, it was determined by the neuroradiologist with 15 years of experience. None of these 3 experts knew of the final diagnosis or ATRX mutation status. The overall process of the experiment after image preprocessing is shown in Figure 2.

Feature extraction
The extraction of radiomics features was performed using the open-source software Pyradiomics (version 3.0.1; https://www.radiomics.io/index.html). The radiomics features derived from the ROI of edema habitat (edema), enhancing tumor (tumor), and overall lesion (overall) were extracted in both T1c and T2f images. A total of 1111 radiomics features were extracted for each ROI. For more information regarding these features, please refer to Appendix 1.
Deep features were extracted from pretrained residual network 34 (ResNet34) using transfer learning. The Pytorch (version 1.9.0; https://pytorch.org) framework was used for CNN network construction and feature extraction. The CNN network uses the well-known ResNet34, which inputs a 224×224×3 pixels natural image, and after multiple consecutive convolutional layers and pooling layers, it can output a 1,000-dimensional vector, which we regarded as the depth feature extracted from the image. Weights were pretrained using the open-source dataset ImageNet-1k (https://www.image-net.org/download.php). The slices with the largest edema area and tumor area were selected from T1c and T2f sequences, and then the ROI region was cut out. Following this, the image was enlarged to 224×224 using the bilinear interpolation algorithm, which was copied into 3 channels for input into the ResNet34. After inputting the ROI image were input into the model, a 1,000-dimensional depth feature was extracted from each ROI region of each sequence. Deep features and radiomics features were combined for subsequent filtering.
Feature selection
The feature data extracted from different ROIs were standardized with z scores. To improve the generalization performance of the model, the independent samples t-test was first used for the preliminary filtering of features, significant features were selected (P<0.05), the least absolute shrinkage and selection operator (LASSO) was applied for further dimension reduction, and the area under curve (AUC) was used as the evaluation index. The optimal parameter λ was determined through 10-fold cross-validation, and features with a nonzero coefficient were selected. If there were still many features after LASSO filtering (according to rule of thumb, the sample size needs to cover 10–15 observations per predictor variable to yield a stable estimate; in our study, the sample size was 194, so we aimed to keep the number of features below 20), the Akaike information criterion (AIC) was used as the evaluation index, and the optimal feature subset was obtained using the backward step search algorithm.
Imaging model building and signature building
We utilized filtered features to construct logistic regression (LR), random forest (RF), and support vector machine (SVM) models. LR was chosen as the classifier for subsequent model building due to its superior generalization performance. Based on the combination of 3 types of features (radiomics, DL, and radiomics + DL as the hybrid feature) in different regions, 9 imaging models were constructed to verify the prediction effect of different types of features and ROI regions on ATRX mutation status, and the image models were named according to the combination of feature types (i.e., radiomics, DL, hybrid) and feature source (i.e., edema, tumor, overall). The optimal imaging model was selected according to the average AUC of the models with 5-fold cross-validation.
Based on the optimal imaging model, an imaging signature was constructed using a linear combination of coefficients weighted features (i.e., first feature coefficient × first feature value + … + nth feature coefficient × nth feature value). The imaging score for each patient was then calculated. The formula for calculating the imaging score of imaging features can be found in Appendix 2, and patients were divided into high-risk group and low-risk group according to a cutoff value of 0.48.
Clinical model building
Clinical variables included age, sex, WHO grade, and VASARI score. More detailed information on the clinical variables can be found in Table S3. A multivariate logical regression model was constructed using clinical variables.
Fusion model building and performance evaluation
A fusion predictive model was constructed by combining clinical variables and image scores obtained from the optimal imaging model. To assess the performance and utility of the fusion model, several methods were employed, including receiver operating characteristic (ROC) analysis, nomogram construction, calibration curve analysis, decision curve analysis, and clinical application curve analysis. These evaluations helped to determine the accuracy and applicability of the model in clinical settings.
Statistical analysis
All statistical analyses were performed using R software (version 4.2.0; https://www.r-project.org). The t test or Mann-Whitney test was used for continuous variables, the chi-squared test was used for classifying variables, and the Delong test was used to evaluate the differences between ROC curves. All statistical tests were 2-sided with a statistical significance threshold of P<0.05.
Results
Construction of image models
A total of 234 patients (120 ATRX mutant type, 114 ATRX wild type) were included in this study. The clinical characteristics of these patients are shown in Table 1. There were no significant differences in clinical characteristics except for WHO grade in cohort A and age in cohort C. Cohort A and cohort B data were randomly sampled based on gender and WHO grade and were split into a training set (N=155) and validation set (N=39) at a ratio of 4:1 for model development according to the practice of previous machine learning research (the ratio of training set to verification set is generally maintained between 4:1 and 3:1) (26). Cohort C data (N=40) was used as an independent test set for external validation of the model. Through multivariate LR, 9 imaging models were constructed and 5-fold cross-validation was performed.
Table 1
| Clinical characteristics | Cohort A (N=82) | Cohort B (N=112) | Cohort C (N=40) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATRX (+) | ATRX (−) | P (intra) | ATRX (+) | ATRX (−) | P (intra) | ATRX (+) | ATRX (–) | P (intra) | |||
| Gender | 0.142 | 0.504 | 0.859 | ||||||||
| Male | 29 | 18 | 32 | 33 | 9 | 14 | |||||
| Female | 15 | 20 | 27 | 20 | 8 | 9 | |||||
| WHO | 0.001 | 0.107 | 0.712 | ||||||||
| III | 2 | 17 | 29 | 35 | 7 | 7 | |||||
| IV | 42 | 21 | 30 | 18 | 10 | 16 | |||||
| Age (years), mean ±SD | 52.7±12.3 | 53.6±11.4 | 0.740 | 53.6±14.3 | 54.4±10.9 | 0.743 | 45.4±12.4 | 58.3±15.2 | 0.001 | ||
| VASARI, mean ±SD | 71.7±5.5 | 73.5±7.2 | 0.221 | 70.7±5.9 | 72.8±6.2 | 0.073 | 71.5±9.3 | 71.5±6.2 | 0.984 | ||
ATRX, alpha-thalassemia X-linked intellectual disability; WHO, World Health Organization; VASARI, Visually Accessible Rembrandt Images.
Selection of the optimal imaging model
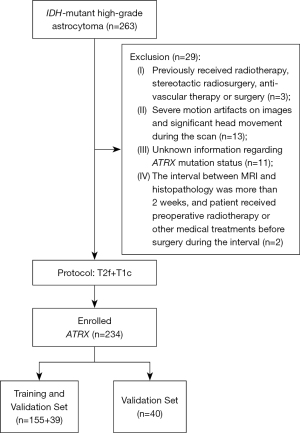
Table 2 summarizes the results of 5-fold cross-validation on the training and validation sets for 9 imaging models, and Table 3 shows the results of the models on the external test set; all the models shown good predictive performance (AUC >0.75). Figure 3A,3B depict the performance of the 6 models constructed from single-type features (radiomics or DL) of 3 ROI regions (edema, tumor, overall), and the models based on overall had a higher AUC relative to the models based on edema and tumor area (overall DL model on the validation set: AUC =0.910, 95% CI: 0.833–0.999; overall radiomics model on the test set: AUC =0.916, 95% CI: 0.819–1.000). In the 3 models based on hybrid features (Figure 3C,3D), the model derived from the overall had the best predictive performance (validation set: AUC =0.916, 95% CI: 0.822–0.999; test set: AUC =0.936, 95% CI: 0.859–1.000). After the Delong test, there was no statistical difference between the ROC curves of the 6 models constructed from single-type features (P>0.05; Figure 4A,4B). Finally, the overall hybrid model was identified as the optimal imaging model due to it having the highest AUC among all the imaging models. The Delong test of the ROC comparison between the 9 image models can be found in Table S4.
Table 2
| Model | Training cohort (N=155) | Validation cohort (N=39) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC (95% CI) | Sensitivity | Specificity | Accuracy | AUC (95% CI) | ||
| Edema DL | 0.869 | 0.819 | 0.845 | 0.903 (0.869−0.958) | 0.798 | 0.770 | 0.789 | 0.834 (0.645−0.989) | |
| Edema radiomics | 0.852 | 0.802 | 0.829 | 0.910 (0.870−0.958) | 0.847 | 0.755 | 0.804 | 0.896 (0.774−0.990) | |
| Edema hybrid | 0.833 | 0.804 | 0.820 | 0.915 (0.869−0.960) | 0.837 | 0.792 | 0.814 | 0.914 (0.842−1.000) | |
| Tumor DL | 0.832 | 0.777 | 0.807 | 0.900 (0.849−0.945) | 0.814 | 0.749 | 0.783 | 0.880 (0.804−0.996) | |
| Tumor radiomics | 0.828 | 0.779 | 0.805 | 0.882 (0.829−0.932) | 0.798 | 0.747 | 0.773 | 0.834 (0.672−0.963) | |
| Tumor hybrid | 0.872 | 0.819 | 0.847 | 0.929 (0.886−0.965) | 0.844 | 0.798 | 0.824 | 0.903 (0.796−0.998) | |
| Overall DL | 0.874 | 0,857 | 0.866 | 0.945 (0.904−0.979) | 0.859 | 0.762 | 0.819 | 0.910 (0.833−0.999) | |
| Overall radiomics | 0.861 | 0.827 | 0.845 | 0.917 (0.879−0.962) | 0.829 | 0.804 | 0.815 | 0.898 (0.751−1.000) | |
| Overall hybrid | 0.915 | 0.876 | 0.897 | 0.966 (0.948−0.991) | 0.852 | 0.862 | 0.861 | 0.916 (0.822−0.999) | |
| Clinical | 0.705 | 0.571 | 0.643 | 0.677 (0.586−0.766) | 0.658 | 0.546 | 0.604 | 0.641 (0.489−0.796) | |
| Fusion | 0.920 | 0.881 | 0.902 | 0.969 (0.964−0.997) | 0.925 | 0.860 | 0.900 | 0.956 (0.878−1.000) | |
AUC, area under curve; CI, confidence interval; edema DL, edema deep learning feature model; edema radiomics, edema radiomics model; edema hybrid, edema radiomics and deep learning feature model; tumor DL, tumor deep learning feature model; tumor radiomics, tumor radiomic model; tumor hybrid, tumor deep learning feature model; overall DL, overall lesion region deep learning feature model; overall radiomics, overall lesion region radiomic model; overall hybrid, overall lesion region radiomics and deep learning feature model; clinical, clinical model; fusion, fusion model.
Table 3
| Models | Sensitivity | Specificity | Accuracy | AUC (95% CI) |
|---|---|---|---|---|
| Edema DL | 0.647 | 0.783 | 0.725 | 0.775 (0.629−0.921) |
| Edema radiomics | 0.765 | 0.870 | 0.825 | 0.875 (0.757−0.992) |
| Edema hybrid | 0.765 | 0.957 | 0.875 | 0.893 (0.789−0.996) |
| Tumor DL | 0.706 | 0.826 | 0.775 | 0.887 (0.788−0.987) |
| Tumor radiomics | 0.647 | 0.696 | 0.675 | 0.818 (0.687−0.950) |
| Tumor hybrid | 0.774 | 0.869 | 0.835 | 0.923 (0.845−1.000) |
| Overall DL | 0.705 | 0.783 | 0.750 | 0.903 (0.814−0.992) |
| Overall radiomics | 0.760 | 0.827 | 0.800 | 0.916 (0.819−1.000) |
| Overall hybrid | 0.824 | 0.870 | 0.850 | 0.936 (0.859−1.000) |
| Clinical | 0.471 | 0.783 | 0.650 | 0.772 (0.624−0.920) |
| Fusion | 0.824 | 0.913 | 0.875 | 0.949 (0.890−1.000) |
AUC, area under curve; CI, confidence interval; edema DL, edema deep learning feature model; edema radiomics, edema radiomics model; edema hybrid, edema radiomics and deep learning feature model; tumor DL, tumor deep learning feature model; tumor radiomics, tumor radiomic model; tumor hybrid, tumor deep learning feature model; overall DL, overall lesion region deep learning feature model; overall radiomics, overall lesion region radiomic model; overall hybrid, overall lesion region radiomics and deep learning feature model; clinical, clinical model; fusion, fusion model.
Predictive performance of the overall hybrid model
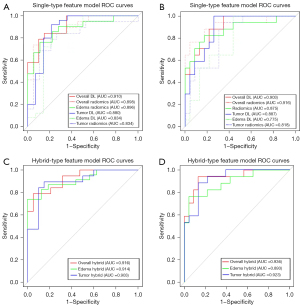
The overall hybrid model was constructed using 16 imaging features, 10 of which were deep features and 6 radiomics features. These features were selected from a total of 8,444 features in the overall lesion region. The selection process involved filtering via t test, LASSO, and the backward step search algorithm to prevent overfitting. The resulting features were found to have low correlation with each other (Figure S1), which could indicate that they complemented each other in the model. The classification performance of the model was evaluated in the validation and test sets, as shown in Figure S2. The model demonstrated good performance in accurately classifying lesions, indicating its potential as a diagnostic tool.
Construction and evaluation of the fusion model
The imaging signature was constructed with the overall hybrid model to calculate the imaging score of each patient (see Appendix 2 for details). The imaging score was then combined with WHO grade, age, sex, and VASARI score to develop a fusion model. The AUC of the fusion model on the training, validation, and test sets were 0.969, 0.956, and 0.949, respectively; the sensitivity was 0.920, 0.925, and 0.824, respectively; the specificity was 0.881, 0.860, and 0.913, respectively; and the accuracy was 0.902, 0.900, and 0.875, respectively. In the training, validation, and test set, the fusion model had the highest AUC value compared to the overall hybrid model and the clinical model, and the overall hybrid model had a higher AUC than did the clinical model (Tables 2,3). With the Delong test (Figure 4C,4D), there was no significant difference between the fusion model and the overall hybrid model, and both the fusion model and the overall model had significant differences compared to the clinical model, regardless of whether the validation set or a test set was considered.
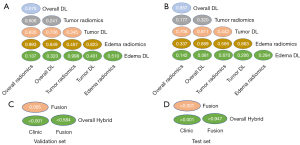
A clinical nomogram was established using the fusion model to show the value of combining imaging scores and clinical variables to predict the ATRX mutation status (Figure 5A). The linear predictive value and risk probability of the patient can be obtained from the clinical nomogram. For example, for a 34-year-old female patient, with WHO CNS grade 4, VASARI 77, and imaging score −4, the score of each item is first determined on the Points line according to the patient’s information, which is added up to a total score and is further transformed into the linear predictive value and risk probability according to the total score in the Total Points line. In this example, the linear predictive value of the patient is −1, indicating that her risk is relatively low, with a risk probability of 0.25 below the risk threshold and an expected ATRX mutation–negative status. The calibration curves showed good agreement between predictions and observations on the test sets (Figure 5B). It was found that the fusion model and the overall hybrid model both had a higher net benefit compared to the clinical model (Figure 5C). After the fusion model was simulated in a sample that scaled up to 1,000 for risk stratification, it could be surmised from the clinical impact curve that the fusion model predictions were in good agreement with the actual true-positive results (Figure 5D).
Discussion
In this study, we developed a fusion model that integrated the radiomics and DL features derived from cMRI and clinical variables for the noninvasive prediction of ATRX mutation status in patients with IDH-mutant high-grade astrocytoma. Compared with the imaging models and the clinical model, the fusion model had the best performance. In addition, all imaging models performed better than did the clinic model.
ATRX loss drives glioma-related biological behaviors by directly regulating chromatin structure and composition (27) and favors the malignant progression of gliomas (28). ATRX mutations can be used to determine prognosis and even indicate clinicopathological grading (29). The heterogeneity of tumor biological behavior can be captured by MRI image features. Preoperative MRI features have been used to predict ATRX mutations in previous studies, but mainly in LGG. Li et al. (30) built a T2-weighted imaging-based radiomics model to determine ATRX mutations in LGGs and achieved the highest AUC of 0.94. Wu et al. (31) predicted ATRX mutations in LGGs by combining age, gender, and radiomic features, with a concordance index of 0.863 and 0.840 for the training and test sets, respectively. Calabrese et al. (32) assessed 9 genetic biomarkers including ATRX in 400 adults with WHO grade 4 gliomas using a radiomic signature, CNN, and a combination of the 2, and the AUC value of ATRX reached as high as 0.97. Compared with previous studies, our study focused more on the ATRX mutation of high-grade IDH-mutant astrocytoma, which is more meaningful for clinical treatment. Unlike Calabrese et al. (32), who averaged the 2 output probabilities (1 from the CNN limb and 1 from the radiomics limb) to create a final combined model probability, we used CNN to extract deep features of each patient and built an LR model together with traditional radiomics features. Before constructing the model, we fused the features, which allowed us to screen a larger number and variety of features, enhancing the model’s flexibility. Additionally, we explored the possibility of using the fused features to predict the status of ATRX mutations, which enriches the existing prediction models. In addition, we added qualitative features such as VASARI, which improved the predictive performance.
Advancements in DL methods have shown superior performance over traditional machine learning methods in predicting tumor genetics and molecular biology based on MRI data (33). The combination of DL and radiomics features has stronger differential ability and is more robust (34). In this study, rather than constructing a direct end-to-end DL model, we extracted highly abstracted semantic features as DL features for the model, and their prediction performance was comparable to, or even surpassed, that of the radiomics features. Out of the 16 image features screened for the optimal image model construction, 10 were derived from DL features. Additionally, the correlation coefficient plots indicated that the final 16 selected features exhibited a low interfeature correlation but had a high correlation with the outcome variables. This suggests that DL features, unlike radiomics features, can effectively capture the information related to tumor genetics and molecular biology. Hybrid models combining radiomics and DL features outperformed those relying solely on either DL or radiomics features across the training, validation, and test sets. This indicates that DL features play a crucial role in the predictive performance of image models and that radiomics and DL features have complementary roles in the model. While the clinical model incorporating the VASARI score showed limited predictive performance, the fusion model that integrated clinical features with radiomics and DL features demonstrated the best predictive performance. This implies the VASARI score can contributed to predicting ATRX mutation status in high-grade IDH-mutant astrocytoma.
MRI is the preferred method for the in vivo investigation of most brain diseases (35). MRI data analysis techniques enable the exploration of associations between image features and diverse molecular phenotypes. This facilitates a more profound investigation of specific molecular variations and the biological behaviors of gliomas. In this study, using cMRI sequences (T2f and T1c) alone was sufficient to achieve good predictive performance. This can be explained by the ability of T2f to effectively distinguish between different components of tumors and the ability of T1c to reveal important information regarding tumor blood supply and internal features. Furthermore, we found that models based on the overall lesion features outperformed those based solely on edema and enhancing tumor area. This suggests that the combination of T1c and T2f can characterize the glioma-specific changes caused by ATRX mutations.
In this study, we fully leveraged the information garnered from cMRI and innovatively used it to predict the ATRX mutation status of patients with high-grade IDH-mutant astrocytoma, achieving promising results. Nevertheless, this study had several limitations that should be addressed and improved upon. First, although the model we developed was based on a multicenter study and showed good performance in the external validation, the sample size was not sufficiently large. Larger sample sizes and prospective studies are still required to validate this model. Second, although our model based on cMRI showed promising potential, further research should explore whether more interpretable qualitative or semiquantitative features from functional MRI sequences could enhance the prediction of ATRX status in IDH-mutant high-grade gliomas. Finally, the manual segmentation methods used to obtain the ROIs were highly time-consuming. Therefore, future research should focus on developing semiautomatic or automatic segmentation methods to obtain ROIs, which could also potentially improve the prediction accuracy.
Conclusions
We developed a multicenter clinical trait-imaging fusion model that combines MRI radiomics and DL features with clinical variables, including VASARI features. The model could effectively predict the ATRX mutation status of patients with IDH-mutant high-grade astrocytoma based on cMRI and may thus aid in the development of more targeted and effective treatment strategies.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-807/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-807/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the institutional review boards of Chongqing Medical University, Sichuan Cancer Hospital, and the United Medical Imaging Center. All participating institutions were formally informed of and agreed to the study protocol. Given the retrospective nature of the design, the requirements for informed consent from patients was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Pladevall-Morera D, Castejón-Griñán M, Aguilera P, Gaardahl K, Ingham A, Brosnan-Cashman JA, Meeker AK, Lopez-Contreras AJ. ATRX-Deficient High-Grade Glioma Cells Exhibit Increased Sensitivity to RTK and PDGFR Inhibitors. Cancers (Basel) 2022;14:1790. [Crossref] [PubMed]
- Reardon DA, Wen PY. Glioma in 2014: unravelling tumour heterogeneity-implications for therapy. Nat Rev Clin Oncol 2015;12:69-70. [Crossref] [PubMed]
- Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, Hawkins C, Ng HK, Pfister SM, Reifenberger G, Soffietti R, von Deimling A, Ellison DW. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021;23:1231-51. [Crossref] [PubMed]
- Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol 2016;131:803-20. [Crossref] [PubMed]
- Han B, Cai J, Gao W, Meng X, Gao F, Wu P, Duan C, Wang R, Dinislam M, Lin L, Kang C, Jiang C. Loss of ATRX suppresses ATM dependent DNA damage repair by modulating H3K9me3 to enhance temozolomide sensitivity in glioma. Cancer Lett 2018;419:280-90. [Crossref] [PubMed]
- Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol 2021;18:170-86. [Crossref] [PubMed]
- Ceccarelli M, Barthel FP, Malta TM, Sabedot TS, Salama SR, Murray BA, et al. Molecular Profiling Reveals Biologically Discrete Subsets and Pathways of Progression in Diffuse Glioma. Cell 2016;164:550-63. [Crossref] [PubMed]
- Purkait S, Miller CA, Kumar A, Sharma V, Pathak P, Jha P, Sharma MC, Suri V, Suri A, Sharma BS, Fulton RS, Kale SS, Dahiya S, Sarkar C. ATRX in Diffuse Gliomas With its Mosaic/Heterogeneous Expression in a Subset. Brain Pathol 2017;27:138-45. [Crossref] [PubMed]
- Di Bonaventura R, Montano N, Giordano M, Gessi M, Gaudino S, Izzo A, Mattogno PP, Stumpo V, Caccavella VM, Giordano C, Lauretti L, Colosimo C, D'Alessandris QG, Pallini R, Olivi A. Reassessing the Role of Brain Tumor Biopsy in the Era of Advanced Surgical, Molecular, and Imaging Techniques-A Single-Center Experience with Long-Term Follow-Up. J Pers Med 2021;11:909. [Crossref] [PubMed]
- Wangaryattawanich P, Hatami M, Wang J, Thomas G, Flanders A, Kirby J, Wintermark M, Huang ES, Bakhtiari AS, Luedi MM, Hashmi SS, Rubin DL, Chen JY, Hwang SN, Freymann J, Holder CA, Zinn PO, Colen RR. Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: imaging predictors of overall and progression-free survival. Neuro Oncol 2015;17:1525-37. [Crossref] [PubMed]
- Gevaert O, Mitchell LA, Achrol AS, Xu J, Echegaray S, Steinberg GK, Cheshier SH, Napel S, Zaharchuk G, Plevritis SK. Glioblastoma Multiforme: Exploratory Radiogenomic Analysis by Using Quantitative Image Features. Radiology 2015;276:313. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. Las Vegas, NV, USA: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); IEEE, 2016.
- Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic Image Segmentation with Deep Convolutional Nets, Atrous Convolution, and Fully Connected CRFs. IEEE Trans Pattern Anal Mach Intell 2018;40:834-48. [Crossref] [PubMed]
- Buda M, AlBadawy EA, Saha A, Mazurowski MA. Deep Radiogenomics of Lower-Grade Gliomas: Convolutional Neural Networks Predict Tumor Genomic Subtypes Using MR Images. Radiol Artif Intell 2020;2:e180050. [Crossref] [PubMed]
- Matsui Y, Maruyama T, Nitta M, Saito T, Tsuzuki S, Tamura M, Kusuda K, Fukuya Y, Asano H, Kawamata T, Masamune K, Muragaki Y. Prediction of lower-grade glioma molecular subtypes using deep learning. J Neurooncol 2020;146:321-7. [Crossref] [PubMed]
- Li G, Li L, Li Y, Qian Z, Wu F, He Y, Jiang H, Li R, Wang D, Zhai Y, Wang Z, Jiang T, Zhang J, Zhang W. An MRI radiomics approach to predict survival and tumour-infiltrating macrophages in gliomas. Brain 2022;145:1151-61. [Crossref] [PubMed]
- Chen S, Xu Y, Ye M, Li Y, Sun Y, Liang J, Lu J, Wang Z, Zhu Z, Zhang X, Zhang B. Predicting MGMT Promoter Methylation in Diffuse Gliomas Using Deep Learning with Radiomics. J Clin Med 2022;11:3445. [Crossref] [PubMed]
- Choi YS, Bae S, Chang JH, Kang SG, Kim SH, Kim J, Rim TH, Choi SH, Jain R, Lee SK. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro Oncol 2021;23:304-13. [Crossref] [PubMed]
- Ren Y, Zhang X, Rui W, Pang H, Qiu T, Wang J, Xie Q, Jin T, Zhang H, Chen H, Zhang Y, Lu H, Yao Z, Zhang J, Feng X. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J Magn Reson Imaging 2019;49:808-17. [Crossref] [PubMed]
- Zhang L, Pan H, Liu Z, Gao J, Xu X, Wang L, Wang J, Tang Y, Cao X, Kan Y, Wen Z, Chen J, Huang D, Chen S, Li Y. Multicenter clinical radiomics-integrated model based on [18F]FDG PET and multi-modal MRI predict ATRX mutation status in IDH-mutant lower-grade gliomas. Eur Radiol 2023;33:872-83.
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033-44. [Crossref] [PubMed]
- Shinohara RT, Sweeney EM, Goldsmith J, Shiee N, Mateen FJ, Calabresi PA, Jarso S, Pham DL, Reich DS, Crainiceanu CMAustralian Imaging Biomarkers Lifestyle Flagship Study of Ageing. Alzheimer's Disease Neuroimaging Initiative. Statistical normalization techniques for magnetic resonance imaging. Neuroimage Clin. 2014;6:9-19. Erratum in: Neuroimage Clin 2015;7:848.
- Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin JC, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward S, Miller JV, Pieper S, Kikinis R. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012;30:1323-41. [Crossref] [PubMed]
- Wei J, Yang G, Hao X, Gu D, Tan Y, Wang X, Dong D, Zhang S, Wang L, Zhang H, Tian J. A multi-sequence and habitat-based MRI radiomics signature for preoperative prediction of MGMT promoter methylation in astrocytomas with prognostic implication. Eur Radiol 2019;29:877-88. [Crossref] [PubMed]
- Raschka S. Model Evaluation, Model Selection, and Algorithm Selection in Machine Learning. doi:
10.48550 /arXiv.1811.12808. - Danussi C, Bose P, Parthasarathy PT, Silberman PC, Van Arnam JS, Vitucci M, Tang OY, Heguy A, Wang Y, Chan TA, Riggins GJ, Sulman EP, Lang FF, Creighton CJ, Deneen B, Miller CR, Picketts DJ, Kannan K, Huse JT. Atrx inactivation drives disease-defining phenotypes in glioma cells of origin through global epigenomic remodeling. Nat Commun 2018;9:1057. [Crossref] [PubMed]
- Wang Y, Yang J, Wild AT, Wu WH, Shah R, Danussi C, Riggins GJ, Kannan K, Sulman EP, Chan TA, Huse JT. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat Commun 2019;10:943. [Crossref] [PubMed]
- Xie Y, Tan Y, Yang C, Zhang X, Xu C, Qiao X, Xu J, Tian S, Fang C, Kang C. Omics-based integrated analysis identified ATRX as a biomarker associated with glioma diagnosis and prognosis. Cancer Biol Med 2019;16:784-96. [Crossref] [PubMed]
- Li Y, Liu X, Qian Z, Sun Z, Xu K, Wang K, Fan X, Zhang Z, Li S, Wang Y, Jiang T. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol 2018;28:2960-8. [Crossref] [PubMed]
- Wu S, Zhang X, Rui W, Sheng Y, Yu Y, Zhang Y, Yao Z, Qiu T, Ren Y. A nomogram strategy for identifying the subclassification of IDH mutation and ATRX expression loss in lower-grade gliomas. Eur Radiol 2022;32:3187-98. [Crossref] [PubMed]
- Calabrese E, Rudie JD, Rauschecker AM, Villanueva-Meyer JE, Clarke JL, Solomon DA, Cha S. Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma. Neurooncol Adv 2022;4:vdac060. [Crossref] [PubMed]
- Bangalore Yogananda CG, Shah BR, Vejdani-Jahromi M, Nalawade SS, Murugesan GK, Yu FF, Pinho MC, Wagner BC, Mickey B, Patel TR, Fei B, Madhuranthakam AJ, Maldjian JA. A novel fully automated MRI-based deep-learning method for classification of IDH mutation status in brain gliomas. Neuro Oncol 2020;22:402-11. [Crossref] [PubMed]
- Ding J, Zhao R, Qiu Q, Chen J, Duan J, Cao X, Yin Y. Developing and validating a deep learning and radiomic model for glioma grading using multiplanar reconstructed magnetic resonance contrast-enhanced T1-weighted imaging: a robust, multi-institutional study. Quant Imaging Med Surg 2022;12:1517-28. [Crossref] [PubMed]
- Esmaeili M, Vettukattil R, Banitalebi H, Krogh NR, Geitung JT. Explainable Artificial Intelligence for Human-Machine Interaction in Brain Tumor Localization. J Pers Med 2021;11:1213. [Crossref] [PubMed]


