
Artificial intelligence for segmentation and classification of lobar, lobular, and interstitial pneumonia using case-specific CT information
Introduction
Pneumonia is an inflammatory and infectious condition of the lungs caused by various pathogens such as bacteria, viruses, fungi, or other microorganisms (1,2). It is a significant public health issue, leading to high rates of morbidity and mortality worldwide, especially in young children, the elderly, and individuals with compromised immune systems (3,4). Timely detection and appropriate treatment are vital to mitigate pneumonia’s public health impact. In this context, medical imaging techniques, particularly chest X-rays and computed tomography (CT) scans, are crucial tools in diagnosing pneumonia and monitoring treatment progress (5,6).
Interpreting CT images can be challenging due to the intra-lesion variability and inter-lesion similarity in different types of pneumonia. Furthermore, manual diagnosis of CT scans is a time-consuming and subjective process that can vary significantly among physicians with different levels of expertise. Hence, there is a critical need for automated, objective methods for pneumonia segmentation and classification. One potential solution is the development of an automated system using advanced deep learning techniques for pneumonia segmentation and type interpretation. This approach could improve the efficiency of pneumonia diagnosis.
Artificial intelligence (AI), particularly convolutional neural networks (CNNs), has shown immense potential in improving pneumonia detection, classification, and segmentation in medical imaging (7,8). Research has indicated that CNNs can effectively differentiate coronavirus disease 2019 (COVID-19) from other types of pneumonia using CT images (9). U-Net and its variants have shown significant potential in segmenting pneumonia lesions from medical images (10-12). The no new U-Net (nnU-Net), a self-configuring method, automatically adapts the U-Net architecture based on input data, enhancing performance across various biomedical image segmentation tasks (13,14).
Although AI has achieved notable diagnostic outcomes in past studies, most of the current research focuses primarily on differentiating between one or a limited number of pneumonia types (15-17). In real-world clinical scenarios, multiple pneumonia types may be detected during CT screenings; as such, a multi-class pneumonia identification solution would be advantageous for clinical use. However, given the numerous pneumonia pathogens, achieving simultaneous identification of a wide array of pneumonia types remains a formidable challenge.
Pneumonia can be anatomically classified into lobar, lobular, and interstitial types, each associated with different pathogens (18-22). Lobar pneumonia is characterized by the involvement of an entire lobe of the lung, usually caused by bacterial pathogens, such as Streptococcus pneumoniae, Klebsiella pneumoniae, and Legionella species (19). Bronchopneumonia, also known as lobular pneumonia, is characterized by patchy consolidation involving multiple bronchopulmonary segments, often caused by bacteria such as Haemophilus influenzae or Staphylococcus aureus (20). Interstitial pneumonia is characterized by inflammation and consolidation in the interstitium of the lung, which can be caused by viral infections (21) such as influenza or COVID-19 (22), as well as other non-infectious conditions. Utilizing AI to determine the anatomical classifications of pneumonia and assist in refining the differential diagnosis may offer a more viable and clinically relevant solution.
Our study aimed to develop a multi-classification model capable of detecting pneumonia in patients using case-specific CT data, classifying the type of pneumonia (lobar, lobular, or interstitial), and segmenting associated lesions. To achieve this, we assembled a multiclass chest CT dataset encompassing three types of pneumonia: lobar, lobular, and interstitial. To our knowledge, this is the first multiclass CT dataset of pneumonia based on anatomical classification. Each CT scan and image were meticulously re-examined by experienced radiologists. Utilizing a 3-dimensional (3D) model and a multi-task learning approach, we endeavored to fully leverage spatial data and improve the speed of model processing data and providing results, to increase the model’s stability. Therefore, the current study aimed to provide radiologists with valuable decision-making supporting tools for more efficient or accurate detection and diagnosis pneumonia. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-945/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Board of the Third Hospital of Peking University (No. M2022854). As this was a retrospective study, the need for signed informed consent was waived.
Study participants
A total of 977 patients were identified by searching chest CT reports from June 2020 to May 2022 in our picture archiving and communication systems (PACS) database for the following keyword: pneumonia. The inclusion criteria were as follows: (I) chest CT scans showed patchy infiltrative shadows or interstitial changes in the lungs before antibiotic therapy; (II) patients met at least 1 of the following conditions: (i) newly developed or worsening cough with sputum production, possibly with chest pain, (ii) fever, (iii) moist rales identified upon lung auscultation, or (iv) peripheral blood leukocyte count greater than 10×109/L or less than 4×109/L. Among the total of 613 patients who met the inclusion criteria, 432 were excluded from this study for the following reasons: (I) severe tuberculosis, lung tumor, non-infectious interstitial lung disease, pulmonary edema, pulmonary atelectasis, pulmonary embolism, pulmonary eosinophilic infiltrates, pulmonary vasculitis, or diffuse parenchymal lung disease; (II) a history of lung surgery or chest radiotherapy; (III) low-quality image; or (IV) absence of lung window thin-section CT images.
The final cohort consisted of 181 patients. Among them, 61 cases were diagnosed as lobar pneumonia, 60 cases as lobular pneumonia, and 60 cases as interstitial pneumonia. Figure 1 illustrates typical CT image slices for these three pneumonia types. The selected cases were divided into a training cohort and an independent test cohort. The training cohort consisted of 135 cases, with 45 cases each of lobar pneumonia, lobular pneumonia, and interstitial pneumonia. For training, a 5-fold cross-validation method was used, with each part of the training dataset being involved in both training and validation. The independent testing cohort comprised 46 cases, including 16 cases of lobar pneumonia, 15 cases of lobular pneumonia, and 15 cases of interstitial pneumonia.

Patient demographic statistics are summarized in Table 1. The flowchart for the participant selection is shown in Figure 2.
Table 1
| Characteristics | Training cohort | Testing cohort | P value |
|---|---|---|---|
| Number of patients | 135 | 46 | – |
| Age, mean ± SD, years | 43.7±19.44 | 49.8±17.89 | 0.062 |
| Sex, N (%) | 0.079 | ||
| Male | 59 (43.7) | 27 (58.7) | |
| Female | 76 (56.3) | 19 (41.3) |
SD, standard deviation.
CT image data acquisition
CT scans were obtained with equipment from different manufacturers using standard imaging protocols. The acquisition and reconstruction parameters of these scans are summarized in Table 2. CT images were reconstructed with a 512×512 matrix and a slice thickness of 1 mm.
Table 2
| CT scanners | Siemens SOMATOM Definition | Siemens SOMATOM go. Top | United Imaging uCT 790 |
|---|---|---|---|
| Scan number | 61 | 81 | 39 |
| Tube voltage, kVp | 110–120 | 120 | 120 |
| Tube current | Automatic mA modulation | Automatic mA modulation | Automatic mA modulation |
| Pitch | 1.2 | 1.0 | 1.1875 |
| Detector configuration, mm | 64×0.6 | 64×0.6 | 80×0.5 |
| Resolution | 512×512 | 512×512 | 512×512 |
| Section thickness, mm | 1 | 1 | 1 |
CT, computed tomography.
AI model
CT image annotation
Firstly, three junior radiologists (W.G., with 7 years; M. L., with 5 years; and Q.Z., with 4 years of chest image diagnostic experience) independently segmented different cases according to the three types of pneumonia patterns shown in Figure 1 within the dataset using the ITK-SNAP software (ITK-SNAP, University of Pennsylvania, PA, USA). Then, a specialist (X.W., with 20 years of chest CT diagnostic experience) manually reviewed and modified the segmentations in ITK-SNAP software. These manual segmentations were used as the ground truth to optimize and evaluate the quality of the automatic segmentation model. The experienced radiologists’ segmentation outcomes served as the gold standard. Subsequently, two AI models were developed respectively based on annotations from the three junior radiologists and the specialist (a senior radiologist).
Network framework of deep learning algorithms
All cases used thin-layer image data (approximately 300–700 layers per patient). Initially, we acquired the CT data and used the threshold method to extract the lung parenchyma’s mask (Figure 3A,3B). We then utilized this mask’s boundary values to crop the original data and identify the 3D volume of interest (VOI), representing the specific region of the lungs under examination (Figure 3C). Once isolated, this data underwent resampling and normalization before being fed into a specialized neural network known as a 3D U-net (Figure 3D). This network was designed to process data in chunks, known as subvolumes, each with its own width, height, and depth. Similar to 2-dimensional (2D) methods, we input these subvolumes sequentially, later combining their results to achieve a comprehensive segmentation map. In this process, we classified three different types of pneumonia as distinct segmentation tasks, assigning each a unique label. The network then produced the CT scan’s segmentation results, incorporating the type of pneumonia into the segmentation results label (Figure 3E). Finally, we performed post-processing on these segmentation results, restoring them to their original image size (Figure 3F). This methodology assists in identifying the shape and type of lesion, which in turn aids physicians in their diagnostic process.

3D SE-U-Net network architecture
The 3D SE-U-Net network structure diagram is shown in Figure 4. The network structure consisted of three parts: encoder, decoder, and skip connection. The encoder consisted of four downsampling modules, each containing two 3×3×3 pixels convolution layers (Conv3D), each of which was immediately followed by an instance normalization (IN) layer, a leaky rectified linear unit (leaky ReLU), and a squeeze and excitation (SE) module. At the end of the downsampling module was a 2×2×2 pixels max pooling layers with a step size of 2 pixels. The decoder structure was similar to the encoder, except that the max pooling layer was replaced by a 2×2×2 pixels transposed convolution layer. Skip connection connected the feature maps before the max pooling layers at the same depth with the output feature maps of the transposed convolutional layer in the upsampling module.
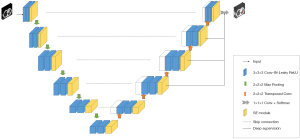
We added spatial and channel SE modules (23) to enable the model to adaptively learn the importance of each channel and position and adjust the channel and position contributions in the feature map according to the needs of the task. This attention mechanism helped the network to better focus on important feature channels and positions, thereby improving model performance.
In the upsampling of the 3D U-Net model mentioned above, except for the two bottommost layers, deep supervision (24) was added to each layer. By adding two additional auxiliary classifiers as network branches to supervise the backbone network, the shallow layer can be trained more fully to prevent the gradient disappearance and the convergence speed being too slow.
AI model training and prediction
During training, we used a 5-fold cross-validation method to divide the cohort into five equal parts, rotating as validation sets to identify the optimal hyperparameters and train the model. The independent testing cohort was not used for either training or internal validation. During training, data augmentation methods such as random rotation, random scaling, random elastic transformation, gamma correction, and mirroring were used to increase the amount of training data and improve the generalization ability of the model. The trained models were used for predicting target CT images, and the Dice scores were respectively calculated both in the validation and the test dataset.
Statistical analysis
The data were subjected to normality and homoscedasticity tests via a Q-Q (quantile-quantile) plot and Levene’s test, respectively. For variables following a normal distribution, an independent sample t-test was deployed. The chi-square test was employed for inter-group comparisons of categorical variables. The diagnostic efficacy of the AI models for pneumonia was evaluated using a receiver operating characteristic (ROC) curve. The test dataset was used to compare diagnostic performance based on sensitivity, specificity, and accuracy, thus facilitating classification into lobar, lobular, or interstitial pneumonia. Additionally, the area under the curve (AUC) for each model was computed to assess overall classification performance. The software SPSS 25.0 (IBM Corp., Armonk, NY, USA) was used for conducting demographic statistics. The open-source statistical software Python version 3.6.5 (Python Software Foundation, Wilmington, DE, USA) was deployed for the analysis and evaluation of the AI model and its diagnostic performance. P values of less than 0.05 (2-sided) were deemed statistically significant.
Results
Segmentation of lung infection region
We used the original nnU-Net as a benchmark to compare the accuracy of single-task segmentation and multi-task segmentation, as well as the accuracy of our model compared to the benchmark for multi-task segmentation. Single-task segmentation refers to the use of lobar pneumonia, lobular pneumonia, or interstitial pneumonia alone for segmentation tasks. Examples of the infection region segmentation for lobar, lobular, and interstitial pneumonia patients in CT are shown in Figure 5. The segmentation performance contrast is depicted in Table 3.

Table 3
| Model | Validation Dice (95% CI) | Test Dice (95% CI) |
|---|---|---|
| Original nnU-Net | 0.841 (0.769–0.913) | 0.670 (0.575–0.766) |
| Original nnU-Net + classify | 0.655 (0.562–0.748) | 0.683 (0.564–0.803) |
| Our model | 0.743 (0.657–0.826) | 0.723 (0.602–0.845) |
| Lobar only | 0.855 (0.786–0.924) | 0.840 (0.764–0.917) |
| Lobular only | 0.654 (0.560–0.747) | 0.638 (0.358–0.918) |
| Interstitial only | 0.730 (0.643–0.817) | 0.627 (0.468–0.785) |
| Average of three | 0.746 (0.661–0.831) | 0.701 (0.605–0.799) |
nnU-Net, no new U-Net; CI, confidence interval.
The original nnU-Net yielded a Dice coefficient of 0.841 [95% confidence interval (CI): 0.769–0.913] in the validation set for single-task segmentation, yet it declined significantly to a Dice coefficient of 0.670 (95% CI: 0.575–0.766) in the test set. When adopting a multi-task learning approach and introducing a classification task, the Dice coefficient in the validation set fell to 0.655 (95% CI: 0.562–0.748), whereas in the test set, it improved modestly to 0.683 (95% CI: 0.564–0.803). Our model achieved more substantial improvements, with a Dice coefficient of 0.743 (95% CI: 0.657–0.826) in the validation set and 0.723 (95% CI: 0.602–0.845) in the test set, which was significantly superior to the original nnU-Net (P=0.033<0.05), leading to more accurate lesion segmentation. It also surpassed the average Dice coefficients of the three models, thereby corroborating the effectiveness of multi-task learning.
Model performance
The diagnostic performance of the model is shown in Table 4. The ROC curve analysis results are shown in Figure 6. Our model achieved an AUC of 0.989 (95% CI: 0.969–1.000), accuracy of 0.927 (95% CI: 0.876–0.978), precision of 0.889 (95% CI: 0.827–0.951), sensitivity of 0.889 (95% CI: 0.827–0.951), and specificity of 0.946 (95% CI: 0.902–0.990) in the internal test queue. Additionally, although the diagnostic performance for lobar pneumonia decreased slightly in comparison to specialized single-task models, it was significantly improved for both lobular and interstitial pneumonia.
Table 4
| Model | Accuracy (95% CI) | Precision (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) |
|---|---|---|---|---|
| The original nnU-Net | ||||
| Lobar | 0.934 (0.885–0.982) | 0.842 (0.771–0.913) | 1.000 (1.000–1.000) | 0.900 (0.841–0.959) |
| Lobular | 0.869 (0.802–0.935) | 0.846 (0.775–0.917) | 0.733 (0.646–0.820) | 0.935 (0.887–0.983) |
| Interstitial | 0.891 (0.830–0.952) | 0.857 (0.788–0.926) | 0.800 (0.722–0.878) | 0.935 (0.887–0.983) |
| Average | 0.899 (0.839–0.958) | 0.848 (0.778–0.918) | 0.844 (0.773–0.915) | 0.923 (0.871–0.975) |
| Our model | ||||
| Lobar | 0.978 (0.949–1.000) | 0.941 (0.895–0.987) | 1.000 (1.000–1.000) | 0.966 (0.930–1.000) |
| Lobular | 0.913 (0.858–0.968) | 0.866 (0.799–0.932) | 0.866 (0.799–0.932) | 0.935 (0.887–0.983) |
| Interstitial | 0.891 (0.830–0.952) | 0.857 (0.788–0.926) | 0.800 (0.722–0.878) | 0.935 (0.887–0.983) |
| Average | 0.927 (0.876–0.978) | 0.889 (0.827–0.951) | 0.889 (0.827–0.951) | 0.946 (0.902–0.990) |
nnU-Net, no new U-Net; CI, confidence interval.

Influence of senior doctors on model performance enhancement
Our model was initially trained using data annotated by junior doctors. Subsequently, the training was continued on this pre-existing model using labels annotated by senior doctors. This 2-step training process led to an enhancement in the model’s performance. Specifically, the Dice coefficient of the model’s segmentation improved by 0.014, increasing from 0.709 (95% CI: 0.589–0.830) to 0.723 (95% CI: 0.602–0.845). Additionally, the AUC also witnessed an increase of 0.042, rising from 0.947 to 0.989. The ROC is displayed in Figure 7.

Discussion
In the present study, we have developed and rigorously evaluated an AI model with the primary aim of identifying pneumonia and further categorizing it into lobar pneumonia, lobular pneumonia, and interstitial pneumonia. The robust performance metrics exhibited by our model underscore its potential in assisting the identification and classification of pneumonia based on thin-section chest CT images. Such a tool, based on our findings, could be instrumental in providing valuable insights for a more precise diagnosis of pneumonia types. Additionally, this tool has the potential to optimize the clinical workflow, leading to expedited patient care and improved health outcomes.
CNNs are one of the representative algorithms for deep learning and have achieved very good results in medical image processing tasks. For many traditional tasks in medical imaging such as classification, segmentation, and detection, CNNs are one of the go-to choices due to their proven efficacy (25). This is consistent with our research goal of distinguishing pneumonia based on anatomical classification. Generative learning refers to algorithms or models that learn to generate new samples/data that resemble a given set of training samples. Gulakala et al. (26) developed a progressively growing generative adversarial network (PGGAN) for generative learning and obtained high resolution X-ray images to achieve rapid diagnosis of COVID-19 infections, which resulted in 40% lighter models as compared to the state-of-the-art models (ResNet and DenseNet) and a 5% increase in test accuracy. Accuracy in generative models is more subjective as refers to how well the model generates data that resembles real data, but they do not provide an “accuracy” in the same sense as classification tasks. If used for tasks such as data augmentation in medical imaging, the accuracy of the subsequent model (e.g., a classifier) would be a more relevant metric than the accuracy of the generative model itself (27). Our study chose the 3D U-Net model for segmentation and diagnosis of pneumonia based on its following advantages: 3D U-Net model preserves the 3D spatial context of volumetric data, making it more suitable for tasks where the relationship between volumetric regions is crucial; 3D U-Net has a symmetric contracting path (encoder) and expansive path (decoder), which allows high-resolution feature maps in the decoder. This aids in precise localization for segmentation tasks. 3D U-Net employs skip connections between the encoder and decoder parts, enabling the network to use features from multiple resolutions. This helps in capturing both global and local information. Given the right augmentations and the depth of the network, 3D U-Net can achieve good performance even with a limited amount of labeled data (28).
Segmentation of lung infection region
In this study, we developed a multi-stage AI system with a primary focus on extracting the region of lung infection, which is a key step in pneumonia diagnosis. The original nnU-Net displayed a significant drop in Dice coefficient from 0.841 (95% CI: 0.769–0.913) in the validation set to 0.670 (95% CI: 0.575–0.766) in the test set. This drop reveals the model’s limited generalizability and suggests potential overfitting during the training phase (13,29). However, by incorporating a multi-task learning approach and adding a classification task, we observed a complex dynamic between the model’s performance on the validation and test datasets. Although the Dice coefficient on the validation set decreased to 0.655 (95% CI: 0.562–0.748)—a trade-off in single-task performance—the model’s generalizability modestly improved, with the test set Dice coefficient rising to 0.683 (95% CI: 0.564–0.803) (30).
Our proposed model further improved these outcomes, achieving a Dice coefficient of 0.743 (95% CI: 0.657–0.826) in the validation set and 0.723 (95% CI: 0.602–0.845) in the test set, indicating superior performance in lesion segmentation. Machado et al. (31) utilized 2D Inf-Net for auto-segmentation of COVID-19 and other types of pneumonia using CT scans. The mean F1 score of the auto-segmentation algorithm was 0.72, similar to our results. Moreover, our model surpassed the average Dice coefficients of the three models, validating the efficiency of the multi-task learning approach. These findings not only underline the potential of multi-task learning in developing more robust and generalizable models but also illuminate the challenges of balancing multiple tasks. They underscore the need for optimization strategies that ensure that performance across all tasks aligns with the “do no harm” principle for responsible machine learning in healthcare (32). Future research will aim to refine these multi-task learning strategies to nurture models that maintain high levels of accuracy, precision, sensitivity, and specificity—all critical factors for reliable diagnostic applications.
Model performance
This study further assessed our model’s performance in distinguishing three distinct types: lobar, lobular, and interstitial pneumonia. The model exhibited remarkable diagnostic efficacy, reflecting an AUC of 0.989, accuracy of 0.927 (95% CI: 0.876–0.978), precision of 0.889 (95% CI: 0.827–0.951), sensitivity of 0.889 (95% CI: 0.827–0.951), and specificity of 0.946 (95% CI: 0.902–0.990) in the internal testing cohort. To the best of our knowledge, there currently exist no other datasets that focus on the anatomical classification of pneumonia in chest CT scans. Prior research has primarily focused on differentiating specific types of pneumonia. For instance, Zheng et al. (33) developed a deep learning model that leveraged CT images to differentiate between COVID-19, bacterial pneumonia, typical viral pneumonia, and healthy controls. This model attained an overall accuracy of 0.94 and an AUC of 0.96. Similarly, Li et al. (9) employed a ResNet50 model to differentiate COVID-19 from non-pneumonia or community-acquired pneumonia, achieving per-scan sensitivity and specificity rates of 90% and 96%, respectively.
The diagnostic performance of our model for lobar pneumonia showed a slight decline compared to specialized single-task models. This observation aligns with existing literature suggesting that task-specific models can sometimes surpass generalized models. However, this marginal decrease does not significantly impact the model’s overall diagnostic capacity, given its impressive general performance metrics. Importantly, our model showed a significant improvement in diagnosing both lobar and interstitial pneumonia. This finding is encouraging, considering the inherent complexity and overlapping symptoms of these conditions that make traditional diagnostic methods challenging (34). Despite its multi-task orientation, our model effectively identifies nuanced features associated with lobar and interstitial pneumonia, thereby enhancing its diagnostic potential for these conditions.
Influence of senior doctors on model performance enhancement
Our results illuminate the influence of senior doctors’ involvement in model training, underlining the value of their expertise in the iterative process of machine learning model development. We implemented a 2-step training process, initially incorporating junior doctors before integrating annotations from senior doctors. This process led to an improvement in model performance, signifying the critical role of domain expertise in developing effective machine learning models for healthcare. The 2-step training approach, which integrates insights from both junior and senior doctors, offers a potential pathway to refine model performance. Future research could explore the possible benefits of extending this approach, such as integrating multidisciplinary expertise or using a tiered annotation strategy. Nevertheless, we must balance these potential improvements against the additional time and resources required for senior clinicians’ multiple annotation rounds.
Limitations
This study recognizes several limitations. Firstly, the small sample size from a single institution potentially limits the findings’ broad applicability and generalizability. We recommend that future studies increase the sample size and include external validation to improve the model’s versatility across diverse clinical settings. Secondly, the model’s improved performance, partly due to the inclusion of senior doctors’ annotations, introduces variability. Differences in expertise between less experienced and seasoned clinicians could lead to unevenness within the training dataset, potentially affecting the proposed model’s overall transferability. Thirdly, although the AI model we developed demonstrates a superior ability to identify distinct types of pneumonia, it is important to remember that CT scan findings should be interpreted alongside the patient’s clinical history, physical examination results, and laboratory data. Although the correlation between pathogens and pneumonia’s anatomical categorization is useful, overlap exists, indicating that different pathogens might sometimes produce identical CT patterns. Future studies could benefit from using AI-identified CT features and types as labels and combining these with clinical data for comprehensive model construction. This integrated approach may enable more precise pathogenic diagnosis of pneumonia, ultimately providing more effective clinical support.
Conclusions
Our study presents a robust multi-task learning model with substantial promise in enhancing the segmentation and classification of pneumonia in medical imaging. Our findings indicate that the model possesses the capability to accurately detect pneumonia lesions and classify them according to their anatomical type. Notably, the model’s performance was influenced positively by the involvement of senior doctors in the iterative training process. This multi-level expert involvement in the model’s development emphasizes the importance of domain expertise in machine learning for healthcare. The successful classification of various pneumonia types could prove invaluable for clinicians, potentially facilitating more accurate diagnoses and informing tailored treatment decisions.
Acknowledgments
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-945/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-945/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Ethics Board of the Third Hospital of Peking University (No. M2022854) and the requirement for informed consent for this retrospective study was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mackenzie G. The definition and classification of pneumonia. Pneumonia (Nathan) 2016;8:14. [Crossref] [PubMed]
- Cilloniz C, Liapikou A, Torres A. Advances in molecular diagnostic tests for pneumonia. Curr Opin Pulm Med 2020;26:241-8. [Crossref] [PubMed]
- McAllister DA, Liu L, Shi T, Chu Y, Reed C, Burrows J, Adeloye D, Rudan I, Black RE, Campbell H, Nair H. Global, regional, and national estimates of pneumonia morbidity and mortality in children younger than 5 years between 2000 and 2015: a systematic analysis. Lancet Glob Health 2019;7:e47-57. [Crossref] [PubMed]
- GBD 2019 LRI Collaborators. Age-sex differences in the global burden of lower respiratory infections and risk factors, 1990-2019: results from the Global Burden of Disease Study 2019. Lancet Infect Dis 2022;22:1626-47. [Crossref] [PubMed]
- Garin N, Marti C, Scheffler M, Stirnemann J, Prendki V. Computed tomography scan contribution to the diagnosis of community-acquired pneumonia. Curr Opin Pulm Med 2019;25:242-8. [Crossref] [PubMed]
- Sharma S, Maycher B, Eschun G. Radiological imaging in pneumonia: recent innovations. Curr Opin Pulm Med 2007;13:159-69. [Crossref] [PubMed]
- Khanday NY, Sofi SA. Deep insight: Convolutional neural network and its applications for COVID-19 prognosis. Biomed Signal Process Control 2021;69:102814. [Crossref] [PubMed]
- Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, Lungren MP, Ng AY. Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv preprint arXiv:1711.05225 2017.
- Li L, Qin L, Xu Z, Yin Y, Wang X, Kong B, Bai J, Lu Y, Fang Z, Song Q, Cao K, Liu D, Wang G, Xu Q, Fang X, Zhang S, Xia J, Xia J. Using Artificial Intelligence to Detect COVID-19 and Community-acquired Pneumonia Based on Pulmonary CT: Evaluation of the Diagnostic Accuracy. Radiology 2020;296:E65-71. [Crossref] [PubMed]
- Cao Y, Xu Z, Feng J, Jin C, Han X, Wu H, Shi H. Longitudinal Assessment of COVID-19 Using a Deep Learning-based Quantitative CT Pipeline: Illustration of Two Cases. Radiol Cardiothorac Imaging 2020;2:e200082. [Crossref] [PubMed]
- Huang L, Han R, Ai T, Yu P, Kang H, Tao Q, Xia L. Serial Quantitative Chest CT Assessment of COVID-19: A Deep Learning Approach. Radiol Cardiothorac Imaging 2020;2:e200075. [Crossref] [PubMed]
- Ronneberger O, Fischer P, Brox T. U-Net: Convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer; 2015.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Gonzalez C, Gotkowski K, Bucher A, Fischbach R, Kaltenborn I, Mukhopadhyay A. Detecting when pre-trained nnu-net models fail silently for covid-19 lung lesion segmentation. In: de Bruijne M, Cattin PC, Cotin S,Padoy N, Speidel S, Zheng Y, Essert C. editors. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2021. Cham: Springer; 2021.
- Wong PK, Yan T, Wang H, Chan IN, Wang J, Li Y, Ren H, Wong CH. Automatic detection of multiple types of pneumonia: Open dataset and a multi-scale attention network. Biomed Signal Process Control 2022;73:103415. [Crossref] [PubMed]
- Wang G, Liu X, Li C, Xu Z, Ruan J, Zhu H, Meng T, Li K, Huang N, Zhang S. A Noise-Robust Framework for Automatic Segmentation of COVID-19 Pneumonia Lesions From CT Images. IEEE Trans Med Imaging 2020;39:2653-63. [Crossref] [PubMed]
- Chen W, Han X, Wang J, Cao Y, Jia X, Zheng Y, Zhou J, Zeng W, Wang L, Shi H, Feng J. Deep diagnostic agent forest (DDAF): A deep learning pathogen recognition system for pneumonia based on CT. Comput Biol Med 2022;141:105143. [Crossref] [PubMed]
- Cook AE, Garrana SH, Martínez-Jiménez S, Rosado-de-Christenson ML. Imaging Patterns of Pneumonia. Semin Roentgenol 2022;57:18-29. [Crossref] [PubMed]
- Washington L, O'Sullivan-Murphy B, Christensen JD, McAdams HP. Radiographic Imaging of Community-Acquired Pneumonia: A Case-Based Review. Radiol Clin North Am 2022;60:371-81. [Crossref] [PubMed]
- Franquet T. Imaging of Community-acquired Pneumonia. J Thorac Imaging 2018;33:282-94. [Crossref] [PubMed]
- Koo HJ, Lim S, Choe J, Choi SH, Sung H, Do KH. Radiographic and CT Features of Viral Pneumonia. Radiographics 2018;38:719-39. [Crossref] [PubMed]
- Hani C, Trieu NH, Saab I, Dangeard S, Bennani S, Chassagnon G, Revel MP. COVID-19 pneumonia: A review of typical CT findings and differential diagnosis. Diagn Interv Imaging 2020;101:263-8. [Crossref] [PubMed]
- Hu J, Shen L, Sun G. Squeeze-and-excitation networks. Proc. IEEE Conf. Comput. Vis. Pattern Recognit 2018:7132-41.
- Lee C Y, Xie S, Gallagher P, Zhang Z, Tu Z. Deeply-supervised nets. Proc. Artif. Intell. Statist., 2015: 562-70.
- Liang S, Liu H, Gu Y, Guo X, Li H, Li L, Wu Z, Liu M, Tao L. Fast automated detection of COVID-19 from medical images using convolutional neural networks. Commun Biol 2021;4:35. [Crossref] [PubMed]
- Gulakala R, Markert B, Stoffel M. Rapid diagnosis of Covid-19 infections by a progressively growing GAN and CNN optimisation. Comput Methods Programs Biomed 2023;229:107262. [Crossref] [PubMed]
- Bowles C, Chen L, Guerrero R, Bentley P, Gunn R, Hammers A, Dickie DA, Hernández MV, Wardlaw J, Rueckert D. Gan augmentation: Augmenting training data using generative adversarial networks. arXiv preprint arXiv:1810.10863 2018.
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D u-net: learning dense volumetric segmentation from sparse annotation. arXiv preprint arXiv:1606.06650 2018.
- Ching T, Himmelstein DS, Beaulieu-Jones BK, Kalinin AA, Do BT, Way GP, et al. Opportunities and obstacles for deep learning in biology and medicine. J R Soc Interface 2018;15:20170387. [Crossref] [PubMed]
- Ruder S. An overview of multi-task learning in deep neural networks. arXiv preprint arXiv:1706.05098 2017.
- Machado MAD, Silva RRE, Namias M, Lessa AS, Neves MCLC, Silva CTA, Oliveira DM, Reina TR, Lira AAB, Almeida LM, Zanchettin C, Netto EM. Multi-center Integrating Radiomics, Structured Reports, and Machine Learning Algorithms for Assisted Classification of COVID-19 in Lung Computed Tomography. J Med Biol Eng 2023;43:156-62. [Crossref] [PubMed]
- Wiens J, Saria S, Sendak M, Ghassemi M, Liu VX, Doshi-Velez F, Jung K, Heller K, Kale D, Saeed M, Ossorio PN, Thadaney-Israni S, Goldenberg A. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019;25:1337-40. [Crossref] [PubMed]
- Zheng F, Li L, Zhang X, Song Y, Huang Z, Chong Y, Chen Z, Zhu H, Wu J, Chen W, Lu Y, Yang Y, Zha Y, Zhao H, Shen J. Accurately Discriminating COVID-19 from Viral and Bacterial Pneumonia According to CT Images Via Deep Learning. Interdiscip Sci 2021;13:273-85. [Crossref] [PubMed]
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]



