
Assessment of ultrashort echo time (UTE) T2* mapping at 3T for the whole knee: repeatability, the effects of fat suppression, and knee position
Introduction
Ligaments, tendons, and menisci are connective tissues in the knee which impact the symptoms and outcomes of knee disorders such as osteoarthritis (OA) (1-3). The menisci are vital for proper knee function and protection of the articular cartilage, and ligaments and tendons contribute to joint stability (4,5). Previous studies reported that elevated T2*, T2 and/or T1ρ relaxation times are often associated with meniscal damage/tears, anterior cruciate ligament (ACL) injury, and knee OA (6-10). Ligaments, tendons, and menisci have high concentrations of organized collagen fibers, a lower concentration of proteoglycan and less water (11) compared to other tissues, resulting in short T2* relaxation times. Short T2* relaxation times cause tissues to have little to no detectable signal with conventional magnetic resonance (MR) techniques, but imaging sequences with ultrashort echo times (UTE) have been used to quantify T2* relaxation time in ligaments, tendons, and menisci (6,12-18). Therefore, UTE-type sequences are attractive candidates for detecting signal from tissues with short or ultrashort T2* relaxation times (13). Quantitatively assessing tissues with ultrashort T2* relaxation times offers the potential to identify imaging biomarkers that are associated with early knee OA (6).
Variation in UTE based T2* relaxation time introduced by clinically relevant imaging parameters and patient-related conditions require evaluation to determine the reliability and sensitivity needed to distinguish diseased from control tissues. Previous work has evaluated the reproducibility of UTE-T2* quantifications for articular cartilage (19), patellar tendon (PT) (20), and compared T2* to UTE-T2* relaxation times quantified in the meniscus (16). However, these studies have not evaluated the effect that some other clinically relevant parameters have on T2* quantifications, such as fat suppression and patient positioning. Tissues with short T2* relaxation times may be partially saturated by the fat suppression pulse, leading to a lower signal intensity at UTEs, and overestimation of the tissue’s T2* relaxation time (12,21). UTE scans can be performed with and without fat suppression, and an understanding of the effect that fat suppression has on T2* relaxation time quantifications would help comparisons between studies in the literature. Patient-related conditions, such as the positioning of the knee with respect to the magnetic field, may also have an effect on T2* quantifications. Strong dipolar interactions in collagen-rich tissues are subject to the magic angle effect, which can contribute to considerable overestimation of T2* relaxation times (22-24).
The goals of this study were to evaluate (I) the repeatability of T2* relaxation time measured by a UTE research application sequence, and evaluate (II) the effect of fat suppression and (III) knee position on T2* relaxation time quantification. The repeatability of UTE based T2* relaxation time mapping was evaluated against a conventional gradient echo (GRE) sequence using scan-rescan images from standard gadolinium (Gd) phantoms and in vivo subjects. Effects of fat suppression on quantitative results were evaluated in phantoms and an ex-vivo pig knee specimen. The effects of knee position were evaluated using UTE-T2* and conventional GRE-T2* knee tissue relaxation times using in-vivo imaging of human subjects.
Methods
Pulse sequence
We utilized a dual-echo, three-dimensional non-selective, radial center-out UTE research application sequence on a clinical 3T scanner throughout this study. The 3D radial UTE research application sequence consisted of a short non-selective rectangular radio frequency (RF) pulse followed by a 20 µs transmit/receive switch time and a 100% asymmetric (half spokes) FID acquisition starting at the k-space center. To achieve an ultrashort TE, data acquisition started during ramp-up of the readout gradient. K-space was sampled using a 3D radial center-out trajectory with a koosh ball like distribution of spokes (Figure 1).

This 3D radial UTE prototype sequence can provide either single-echo or dual-echo acquisition. After the RF excitation pulse, the first echo was acquired from the k-space center to the surface of a sphere (inside-out readout) with UTE. A second echo can be acquired with either inside-out or outside-in trajectory corresponding to the polarity (monopolar or bipolar) of the second readout gradient. All dual-echo UTE scans were performed with monopolar readout; therefore, an additional rewinder gradient was used after the first read out gradient (Figure 1). Image reconstruction was based on a conventional non-uniform fast Fourier transform (NUFFT) by Cartesian gridding using a Kaiser-Bessel window function. To reduce trajectory deviations due to gradient non-linearity, axis-specific gradient delay correction was used to during reconstruction.
Phantom, ex-vivo and in-vivo image acquisition and processing
Phantom, ex-vivo and in-vivo experiments were carried out for this study. All data were acquired without pre-scan normalization, which is a correction method that impacts image intensity uniformity. Acquiring images without pre-scan normalization avoids variations in signal intensity scaling between acquisitions and facilitates interleaving of multiple acquisitions with different echo times (TE). Manual frequency, transmit adjustment, and 3D shimming were performed before each test with a manually selected shim volume. Scans were performed on a 3T scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) which as has a maximum gradient amplitude of 80 mT/m, and a slew rate of 200 T/m/s. All phantom and in-vivo experiments used a knee coil (1Tx/15Rx, QED), and the ex-vivo experiments used a flexible coil (4Rx). Multi-echo data were aligned with increasing TEs and voxel-wise T2* relaxation time was determined by using mono-exponential least square fitting.
Phantom experiment
Two Gd phantoms with 15 and 30 mM concentrations were prepared and three sequences were utilized: (I) single-echo UTE, (II) dual-echo UTE, and (III) multi-echo GRE (25,26) (Table 1). To ensure that single- and dual-echo UTE sequences yielded similar signal for common TEs, “signal ratio” maps were generated, which divided echo images from the single-echo sequence by the corresponding echo-image from the dual-echo sequence. The mean signal ratio was calculated across a representative slice approximately perpendicular to the phantom’s length. Regions of interest (ROI) for each phantom were generated from a representative axial, sagittal, and coronal slice with an automatic regional grow algorithm that was initialized with a manually defined seed point within each slice. Mean and standard deviation (SD) of T2* relaxation times within each ROI were calculated.
Table 1
| Study type | Sequence | |||
|---|---|---|---|---|
| Single-echo UTE/DESS | Dual-echo UTE | Multi-echo GRE | ||
| Gd phantom | Single-echo UTE | |||
| Scan time (min:s) | 8:17 (per echo time) | 8:17 (per acquisition for two echoes) | 13:39 | |
| FOV (mm3) | 130×130×130 | 130×130×130 | 130×130×128 | |
| Matrix size (freq × phase × slice) | 240×240×240 | 240×240×240 | 256×256×128 | |
| Resolution (mm2) | 0.542×0.542 | 0.542×0.542 | 0.508×0.508 | |
| Slice thickness (mm) | 0.542 | 0.542 | 1.000 | |
| TR (ms) | 24.9 | 24.9 | 25 | |
| TE (ms) | 0.06, 2, 4.92, 7.5, 8.62, 10, 19.5 | 0.06/7.5, 2/8.62, 4.92/10, 7.5/19.5 | 4.92, 8.61, 12.80, 20.00 | |
| Flip angle (degrees) | 6 | 6 | 6 | |
| BW (Hz/pixel) | 465 | 465 | 455 | |
| Radial views | 20,000 | 20,000 | – | |
| Fat suppression | Off | Off/on (spokes: 5, 10, 15, and 20) | Off | |
| Ex-vivo/in-vivo | DESS | |||
| Scan time (min:s) | 5:50 | 10:09 per acquisition × 2 | 9:13 | |
| FOV (mm3) | 140×140×112 | 160×160×160 | 160×160×120 | |
| Matrix size (freq × phase × slice) | 384×384×120 (ex-vivo); 384×384×160 (in-vivo) |
240×240×240 | 256×256×120 | |
| Resolution (mm2) | 0.365×0.365 | 0.667×0.667 | 0.625×0.625 | |
| Slice thickness (mm) | 0.700 | 0.667 | 1.000 | |
| TR (ms) | 17.55 | 12.9 | 29 | |
| TE (ms) | 6.02 | 0.06/4.92, 2/8 | 2.48, 6.86, 11.24, 15.62, 20, 24.38 | |
| Flip angle (degrees) | 25 | 6 | 12 | |
| BW (Hz/pixel) | 185 | 465 | 500 | |
| Radial views | – | 45,000 | – | |
| Fat suppression | On | Ex-vivo: off/on (spokes: 5, 10, 15, and 20); in-vivo: on, 20 spokes |
On | |
Gd, gadolinium; UTE, ultra-short echo time; DESS, dual echo steady state; GRE, gradient echo; FOV, field of view; TR, repetition time; TE, echo time; BW, bandwidth.
The phantoms were also used to evaluate the effect of fat suppression on UTE-T2* relaxation time quantification. Dual-echo UTE acquisitions without fat suppression and with different number of radial spokes (5, 10, 15, and 20) per fat suppression pre-pulse were acquired in both Gd phantoms. All fat suppressed acquisitions used a spectrally selective fat saturation pulse.
Ex-vivo experiment
The ex-vivo experiment utilized one secondary use pig hind limb. The hind limb was disarticulated at the hip, leaving the knee joint capsule and surrounding soft tissue undisturbed, then stored at −20 ℃ until it was thawed and brought to room temperature over a 36-hour period prior to imaging. The ex-vivo sample remained unfixed throughout the experiment. The ex-vivo limb was used to evaluate the effects of fat suppression on UTE-T2* relaxation time quantification. Similar to the phantom experiment, all acquisitions utilized a dual-echo UTE sequence without fat suppression and with different number of radial spokes (5, 10, 15, and 20) per fat suppression pre-pulse.
In addition to the UTE sequence, dual-echo steady state (DESS) and multi-echo GRE sequences (Table 1) were acquired. The DESS images were used to segment anatomical structures. Three adjacent representative sagittal slices near the mid-substance of the ACL, posterior cruciate ligament (PCL), PT and the medial and lateral meniscus (MEN) were manually segmented. The DESS image was rigidly registered to the first echo of the UTE and GRE images. The estimated registration parameters from the DESS to the first UTE echo were applied to the four tissue segmentations to align the corresponding ROI in the UTE and GRE images (27). Mean and SD UTE-T2* and GRE-T2* relaxation time within each ROI for both Gd phantoms and ex-vivo knee tissue were calculated for the UTE and GRE images, respectively.
In-vivo experiment
Five healthy volunteers (2 males and 3 females) took part in the in-vivo study. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Institutional Review Board of the Cleveland Clinic, written informed consent was obtained from the participants prior to human subject experiments. Subjects did not have reported knee pain, a history of knee injury, or inflammatory disease. Each volunteer was imaged with their knee in an extended position then imaged again with their knee flexed within the knee coil to assess the effects of knee position on T2* relaxation time. For each knee position, dual-echo UTE, multi-echo GRE, and DESS images were collected (Table 1).
To evaluate T2* repeatability, subjects were rescanned using the UTE (n=4) and GRE (n=3) sequences. Rescan images were collected within the same session as the initial images but after subjects had exited and re-entered the scanner. For each knee position, all UTE and GRE image data were rigidly registered to the first echo of the GRE image (17). Representative slices of four tissues (PT, ACL, PCL, MEN) were manually segmented on the GRE images in both the extended and flexed knee position (8 segmentations total for each volunteer). For each tissue, three sagittal slices were segmented near the midsection of the ligaments and tendon, and the body of the MEN. To ensure consistent ROIs between scan and rescan images, the rescan image data were also rigidly registered to the first echo of the original scan’s GRE image. Each tissue’s segmentation was manually inspected in the rescanned images, and if the mask was misaligned, then new segmentations were generated using the registered rescan image. Mean and SD of the T2* relaxation time was calculated in each ROI. To avoid fitting to echoes with low signal to noise ratio (SNR), the T2* relaxation time for each tissue was recalculated with the longest echo excluded if the average mean T2* value across subjects (either in extended or flexed positions) was less than half the longest echoes’ TE (Table 2).
Table 2
| Sequence | ACL | PCL | PT | MEN |
|---|---|---|---|---|
| UTE (ms) | None | None | 8.00 | None |
| GRE (ms) | None | 24.38 | 15.62, 20.00, 24.38 | 20.00, 24.38 |
ACL, anterior cruciate ligament; PCL, posterior cruciate ligament; PT, patellar tendon; MEN, meniscus; UTE, ultra-short echo time; GRE, gradient echo.
The knee position in extended and flexed positions was quantified using the DESS images. A semi-automated approach was utilized to define ROIs for the femur and tibia bones in the extended and flexed knee positions, and surfaces were generated from the segmentations. An automated approach used the bone surfaces to define the fixed femoral and tibial coordinate systems (28), and the coordinate systems were used to determine the joint kinematics for each position (29).
Statistical analysis
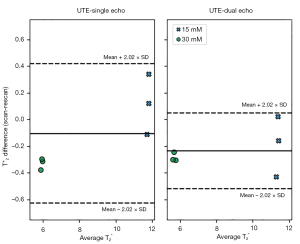
The phantom and in-vivo studies were used to evaluate the repeatability of the UTE-T2* relaxation time. The percent difference between each initial and repeated scan for the phantom and in-vivo images was calculated to evaluate the repeatability of mean T2* relaxation time for the UTE and GRE sequences (Table 3). The percent difference was calculated for each subject’s ROI, then averaged across all subjects. Additionally, a Bland-Altman analysis was used to evaluate the scan-rescan repeatability of T2* relaxation time, where the limits of agreement were determined using the SD of the scan-rescan difference from the 95% confidence intervals of the t-distribution. The percent difference and intraclass correlation coefficient (ICC) between imaging sequences was calculated to evaluate the agreement between T2* relaxation time measures. All of the percent differences were calculated as the absolute difference between two values divided by the mean of the two values, all multiplied by 100. ICC was calculated using R Statistical Software (V4.1.3; R Core Team) using the irr package [V0.84.1; Gamer et al. (30)] with a two-way random model and type of absolute agreement. An ICC value above 0.90 indicates excellent agreement while values below 0.50 indicates poor repeatability (31). Additionally, the root mean square deviation (RMSD) in mean T2* relaxation times between initial and repeated scans across subjects (Table 3) was calculated.
Table 3
| Study | Comparison | Statistical measure |
|---|---|---|
| Phantom | Scan vs. rescan | % Diff, ICC, Bland-Altman |
| UTE vs. GRE | ICC | |
| Fat suppression | CV, ICC | |
| Ex vivo | Fat suppression | CV, ICC |
| In vivo | Scan vs. rescan | % Diff, ICC, RMSD, Bland-Altman |
| UTE vs. GRE | % Diff | |
| Extension vs. flexion | % Diff, ICC, RMSD |
% Diff, percent difference; ICC, intraclass correlation coefficient; CV, coefficient of variation; RMSD, root mean square deviation; UTE, ultra-short echo time; GRE, gradient echo.
The effect of fat suppression on T2* relaxation time quantification was evaluated using the phantom and ex-vivo images. The amount of variation introduced by fat suppression was evaluated by calculating the coefficient of variation (CV) across all levels of fat suppression (number of radial spokes per fat saturation pulse) for each ROI in the phantom and ex-vivo images. The ICC across non-fat suppressed and all fat suppressed images was calculated to evaluate the agreement between mean T2* relaxation times (Table 3).
The reproducibility of T2* relaxation time quantification for the UTE and GRE sequences was evaluated using the in vivo images in the extended and flexed knee position. The percent difference in mean T2* values between knee positions was calculated for each ROI and averaged across subjects. Additionally, the RMSD for mean T2* relaxation times between extended and flexed knee positions across subjects was calculated (Table 3). The agreement between calculated T2* relaxation times using data acquired in the extended and flexed position was also evaluated by calculating ICC for each sequence.
Results
Images of both Gd phantoms had homogeneous signal distribution with no observable artifacts (Figure 2A,2B). The minimum mean coefficient of determination (R2) (±SD) of the monoexponential fit across both phantoms and across the single and dual echo UTE and GRE sequences was 0.98 (±0.06). Within the phantom’s ROI, the mean signal ratio between the single- and dual-echo acquisitions showed values above 1.00, and increased as the TE increased (Figure 2C,2D). T2* relaxation time maps calculated from the single and dual-echo UTE sequences showed similar variations, where the average SD across slices was 0.66 and 0.64 ms, respectively.

The average mean T2* relaxation times across the imaged planes for the single-echo UTE, dual-echo UTE and the multi-echo GRE images was 11.85, 11.25, 11.31 ms for the 15 mM Gd phantom, and 5.76, 5.52, and 5.59 ms for the 30 mM Gd phantom, respectively (Table 4). All of the sequence’s percent difference in mean T2* relaxation time between scan-rescan images was less than 8%, and the Bland-Altman analysis showed a mean difference less than 0.25 ms (Figure 3). The inter-sequence percent difference values were less than 3.5%, and inter-sequence ICC showed excellent agreement between estimated T2* relaxation times (Table 4).
Table 4
| Sequence | 15 mM Gd phantom | 30 mM Gd phantom | |||||
|---|---|---|---|---|---|---|---|
| Axial | Coronal | Sagittal | Axial | Coronal | Sagittal | ||
| Single-echo UTE, ms | |||||||
| First scan | 11.88±0.58 | 11.98±0.90 | 11.67±1.41 | 5.83±0.13 | 5.78±0.33 | 5.69±0.53 | |
| Second scan | 11.76±0.61 | 11.64±1.00 | 11.79±1.29 | 6.14±0.28 | 6.07±0.36 | 6.06±0.57 | |
| % Diff | 1.04 | 2.88 | 0.96 | 5.21 | 4.98 | 6.44 | |
| Dual-echo UTE, ms | |||||||
| First scan | 11.32±0.47 | 11.37±0.69 | 11.06±1.34 | 5.59±0.12 | 5.53±0.33 | 5.44±0.52 | |
| Second scan | 11.48±0.62 | 11.35±0.99 | 11.49±1.29 | 5.90±0.33 | 5.77±0.37 | 5.74±0.64 | |
| % Diff | 1.38 | 0.20 | 3.81 | 5.33 | 4.33 | 5.34 | |
| Multi-echo GRE, ms | |||||||
| First scan | 11.35±1.20 | 11.33±1.31 | 11.23±1.52 | 5.65±0.52 | 5.58±0.65 | 5.55±0.76 | |
| Second scan | 11.90±0.80 | 11.84±1.00 | 11.68±1.32 | 6.01±1.01 | 6.04±1.19 | 5.98±1.02 | |
| % Diff | 4.79 | 4.35 | 3.96 | 6.27 | 7.97 | 7.46 | |
| Inter-sequence | |||||||
| CV | 2.75% | 3.12% | 2.82% | 2.20% | 2.35% | 2.22% | |
| ICC | 0.993 (0.893–0.999) | ||||||
The inter-sequence CV and ICC values were calculating across the single-, dual-echo UTE, and the GRE T2* values. Data are presented as the mean ± standard deviation, % Diff, CV and the ICC with 95% CIs. Gd, gadolinium; UTE, ultra-short echo time; GRE, gradient echo; CV, coefficient of variation; ICC, intraclass correlation coefficient; % Diff, percent difference; CI, confidence interval.
The ex-vivo UTE images showed that the ACL and PT had rapidly decaying signals between 0.06 and 8.00 ms (Figure 4). The minimum mean R2 value of the monoexponential fit across all of the ex vivo images was 0.98 and 0.93 for the UTE and GRE sequences, respectively. Mean UTE-T2* relaxation time showed little variation between scans with fat suppression and scans with different numbers of radial spokes after fat suppression (Table 5). The CV between scans was less than 5% for both phantoms and knee tissues, however, the tissues showed the shortest and longest UTE-T2* relaxation time in the scan without fat suppression and with 5 spokes after fat suppression, respectively (Table 5). The largest difference in mean UTE-T2* relaxation time between scans with and without fat suppression was less than 1.5 ms. The mean UTE-T2* relaxation times calculated from the ex-vivo images were shorter than the GRE-T2* relaxation times for each evaluated tissue, with the largest difference being nearly 8 ms (Table 5).
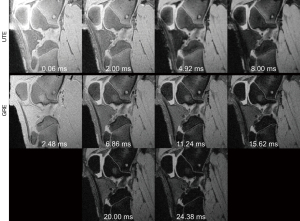
Table 5
| Sequence | 15 mM Gd | 30 mM Gd | ACL | PCL | PT | MEN | |
|---|---|---|---|---|---|---|---|
| GRE-T2* (ms) | 16.14±6.58 | 21.54±8.49 | 9.00±2.69 | 11.65±4.92 | |||
| UTE-T2* (ms) | |||||||
| None | 11.31±0.20 | 5.59±0.013 | 9.80±3.15 | 12.69±2.76 | 4.18±0.56 | 8.12±1.90 | |
| Spokes # 5 | 11.31±0.19 | 5.76±0.029 | 10.94±3.80 | 14.11±2.98 | 4.51±0.95 | 8.30±2.06 | |
| Spokes # 10 | 11.22±0.18 | 5.70±0.032 | 10.69±3.59 | 13.91±3.06 | 4.33±0.91 | 8.27±1.96 | |
| Spokes # 15 | 11.16±0.20 | 5.68±0.033 | 10.64±3.63 | 13.67±2.88 | 4.23±0.85 | 8.28±1.99 | |
| Spokes # 20 | 11.14±0.18 | 5.65±0.031 | 10.44±3.73 | 13.61±3.14 | 4.20±0.85 | 8.19±2.05 | |
| Overall CV (UTE-T2*) | 0.72% | 1.10% | 4.10% | 4.01% | 3.17% | 0.91% | |
| Fat suppressed CV (UTE-T2*) | 0.68% | 0.82% | 1.93% | 1.66% | 3.24% | 0.58% | |
| Overall ICC (UTE-T2*) | 0.993 (0.975–0.999) | ||||||
Overall CV and ICC values were calculated across all of the images. The fat suppressed CV values were calculated across only the fat suppressed images. Data are presented as mean ± standard deviation, CV, and the ICC with 95% confidence intervals. GRE, gradient echo; UTE, ultra-short echo time; CV, coefficient of variation; ICC, intraclass correlation coefficient; Gd, gadolinium; ACL, anterior cruciate ligament; PCL, posterior cruciate ligament; PT, patellar tendon; MEN, meniscus.
The in-vivo UTE images had shorter mean T2* relaxation times compared to the corresponding tissues in the GRE images (Figure 5, Table 6). The average mean R2 (±SD) of the monoexponential fit across subjects was greater than 0.95 (±0.02) for all tissues for both UTE and GRE sequences. For repeated scans, the UTE and GRE sequences had percent differences and RMSD less than 8% and 0.6 ms, respectively (Table 7). Similarly, the Bland-Altman analysis showed a mean difference in T2* relaxation time between scan and rescan images less than 0.2 ms for both UTE and GRE sequences (Figure 6). Additionally, the UTE and GRE sequences had scan/rescan ICC values greater than 0.98 (Table 7).

Table 6
| Sequence | ACL | PCL | PT | MEN |
|---|---|---|---|---|
| UTE-T2* (ms) | ||||
| Extended knee | 11.91±1.64 | 7.52±0.68 | 2.87±0.66 | 7.44±0.30 |
| Flexed knee | 8.48±1.10 | 9.03±0.69 | 6.06±2.69 | 7.64±0.63 |
| ICC | 0.511 (0.089–0.775) | |||
| GRE-T2* (ms) | ||||
| Extended knee | 20.09±2.17 | 10.82±1.13 | 7.22±1.78 | 8.87±0.39 |
| Flexed knee | 15.04±2.12 | 12.04±0.88 | 6.17±1.01 | 8.88±0.77 |
| ICC | 0.769 (0.501–0.902) | |||
| UTE vs. GRE (% Diff) | ||||
| Extended knee | 51.45±6.05 | 35.84±6.76 | 83.05±31.15 | 17.46±6.27 |
| Flexed knee | 55.73±3.92 | 28.55±2.78 | 25.70±18.98 | 14.91±3.72 |
Data are presented as the mean ± standard deviation and the ICC with the 95% confidence interval. UTE, ultra-short echo time; GRE, gradient echo; % Diff, percent difference; ICC, intraclass correlation coefficient; ACL, anterior cruciate ligament; PCL, posterior cruciate ligament; PT, patellar tendon; MEN, meniscus.
Table 7
| Sequence | ACL | PCL | PT | MEN |
|---|---|---|---|---|
| UTE-T2* | ||||
| Extend/flex (n=5) | ||||
| RMSD (ms) | 3.93 | 1.87 | 4.31 | 0.76 |
| % Diff | 33.17±17.66 | 18.28±12.96 | 63.23±38.65 | 8.26±5.33 |
| Scan/rescan (n=4) | ||||
| RMSD (ms) | 0.24 | 0.34 | 0.39 | 0.54 |
| % Diff | 1.89±1.65 | 3.26±2.30 | 7.29±7.09 | 6.67±3.05 |
| ICC | 0.985 (0.958–0.995) | |||
| GRE-T2* | ||||
| Extend/flex (n=5) | ||||
| RMSD (ms) | 5.66 | 1.64 | 2.22 | 0.46 |
| % Diff | 28.86±14.41 | 12.84±6.04 | 22.47±18.39 | 4.87±2.17 |
| Scan/rescan (n=3) | ||||
| RMSD (ms) | 0.22 | 0.26 | 0.32 | 0.48 |
| % Diff | 1.05±0.88 | 1.98±1.46 | 4.91±2.38 | 4.80±2.14 |
| ICC | 0.997 (0.988–0.999) | |||
Data are presented as the mean ± standard deviation and the ICC with the 95% confidence interval. RMSD, root mean square deviation; % Diff, percent difference; ICC, intraclass correlation coefficient; ACL, anterior cruciate ligament; PCL, posterior cruciate ligament; PT, patellar tendon; MEN, meniscus; UTE, ultra-short echo time; GRE, gradient echo.

The effect of knee position on the quantified T2* relaxation times varied between tissues. Between the extended and flexed positions, subjects showed a mean ± SD change in flexion angle of 4.6°±3.2°. Overall, there was poor agreement between T2* relaxation times determined from extended and flexed images, with ICC values of 0.51 and 0.77 for the UTE and GRE sequences, respectively (Table 6). The UTE and GRE sequences showed the PT and ACL as having the highest RMSD in T2* relaxation time between extended and flexed knee positions, respectively (Table 7). Both sequences showed the meniscus as having smallest change in T2* relaxation times between knee positions, with RMSD and percent difference values less than 1.0 ms and 10%, respectively (Table 7).
Discussion
In this work, we presented and evaluated a 3D radial UTE prototype sequence to quantify T2* relaxation time and tested its repeatability, the effect of fat suppression, and knee position for whole knee imaging against corresponding measures obtained from a more conventional GRE T2* sequence. Previous studies have used other UTE sequences to evaluate the repeatability and reproducibility of UTE-T2* relaxation time quantified for knee imaging (19,20,32), and evaluated the effects of fat suppression on UTE-T2* quantification (21,33). This current study utilizes a prototype sequence to quantify the variation in UTE-T2* relaxation time introduced by clinically relevant factors such as knee positioning, and the quantified variations can be used to help identify the sensitivity needed to distinguish diseased from control tissues. In phantom scans with different Gd concentrations, low percent difference values indicate that both single-echo and dual-echo UTE sequences have excellent repeatability of T2* relaxation times as well as high agreement with the conventional GRE sequence (25,26). The agreement in UTE-T2* relaxation time between dual-echo UTE images acquired with and without fat suppression for the phantom and ex-vivo scans indicates that UTE-T2* relaxation time is not sensitive to fat suppression. For in vivo human knees, both UTE and GRE T2* relaxation time was repeatable, but showed variations between extended and flexed positions, indicating that orientation will have an effect on T2* relaxation time quantifications.
The results from the phantoms show that quantification of the T2* relaxation time is repeatable between scanning sessions, although there was some variation between phantoms. The percent difference values for 30 mM phantom were higher than the 15 mM Gd values across sequences. One potential reason for the observed differences could be shorter T2* relaxation time and rapidly decaying signal in the 30 mM phantom (Figure 2) causing later echoes to be more affected by noise and introducing variation in T2* quantifications. The phantom images from the GRE sequence generally had higher percent difference for mean T2* relaxation time compared to the UTE sequence for both phantoms (Table 4), which may be because the GRE sequence has a relatively long TE for the first echo, leading to lower SNR in the first TE image compared with the UTE sequence (19). In general, both UTE and GRE T2* repeatability are sensitive to image SNR, and efforts should be made to increase SNR and minimize the noise-induced variations to provide better quantitative reproducibility.
Fat suppression and the number of radial spokes used after fat suppression had little effect on UTE-T2* relaxation times in phantoms and tissue, as indicated by the low CVs between results with and without fat suppression, which are further supported by the high ICC value (0.99) indicating excellent agreement (Table 5). It was observed that the ex vivo tissue’s UTE-T2* relaxation times with fat suppression were slightly longer compared to those without fat suppression (Table 5). One potential reason for this difference could be that fat suppression has little effect on the homogeneous phantom with T2* >5 ms (for both phantoms), but the ultra-short T2* (<5 ms) components in the tissues may be affected by fat suppression. Short T2* tissues have a broader spectral bandwidth which can overlap with the frequency band of the fat saturation pulse, and signals in the ultra-short T2* component tissues could therefore also be partially saturated, leading to a lower signal intensity at UTE. A relatively long duration fat suppression pulse with narrower bandwidth could reduce signal attenuation of short T2* tissues but would potentially decrease the quality of fat saturation (21).
In vivo UTE-T2* relaxation times of ligament, tendon, and menisci obtained from our sequence were generally consistent with the literature where similar sequences were implemented. The present ACL UTE-T2* relaxation time was 11.9 ms, which is similar to that reported from in vitro (11.3 ms) (18), but shorter than has been reported in vivo (~16 ms) (15,17). This current study’s meniscus UTE-T2* relaxation time (7.4 ms) was near the lower range of values previously reported for the anterior and posterior meniscus regions (8.5–11.7 ms) (6,16,17). The difference between our results and those previously reported could stem from regional differences within the menisci, where the current study evaluated the meniscal body (Figure 5) rather than the anterior or posterior meniscal horns. The reported PT UTE-T2* relaxation time (2.9 ms) fell within the range that has been reported for healthy subjects (2.0–5.7 ms) (14,17), and the current study’s PCL UTE-T2* relaxation time (7.5 ms) was similar to previously reported (8.8 ms) for in-vivo subjects (17), and shorter than has been reported for ex-vivo specimens (11.1 ms) (34).
T2* relaxation times quantified from the phantoms showed excellent agreement between UTE and GRE sequences (ICC =0.99, Table 4), however, the values quantified from knee tissues showed poor agreement between sequences, with average percent differences ranging between 15–85% (Table 6). One potential reason for this difference could be that knee tissues are characterized by short and long T2* components (35), and the UTE sequence detects short T2* components which are not well captured by the conventional GRE sequence (36). The additional signal from the ultrashort echo could result in different T2* relaxation time quantifications between UTE and GRE sequences. The phantoms may not show the same variation between sequences because their signal decay may be characterized by a single component, resulting in similar T2* relaxation times between UTE and GRE sequences. Implementing bi-exponential fitting in both UTE- and GRE-derived images may resolve these in vivo sequence-specific differences and could be explored in future work. Additionally, the difference in T2* relaxation time between the UTE and GRE sequences could be due to the ex-vivo and in-vivo UTE sequence parameters having a longest echo that is one third of the longest echo in the GRE sequence (Table 1). Adding longer echoes to the UTE sequence that are comparable to the GRE sequence would provide a more direct comparison, however it would also prohibitively increase the scan time.
The UTE and GRE sequences demonstrated repeatability in vivo, however, knee position introduced variability into T2* relaxation time quantifications for the evaluated ligaments and tendon. Both sequences showed ICC values greater than 0.98 (Table 7) between in vivo scan-rescan images, demonstrating excellent agreement. The meniscus had a consistently low RMSD between T2* values quantified from extended and flexed knee positions. Chu et al. (6) showed significant differences of 2.7 and 5.0 ms in meniscal UTE-T2* relaxation time between uninjured controls and preoperative ACL tear patients with uninjured and injured menisci, respectively. This current study showed that the RMSD in meniscus UTE-T2* relaxation for repeated scans and different knee positions was less than 1 ms (Table 7), which suggests that the evaluated UTE sequence would be sensitive enough to detect clinically relevant changes in meniscus T2* relaxation times. The same group (15) also showed a significant 2 ms change in mean ACL graft UTE-T2* relaxation time between 1 and 2 years post-operatively, which is less than this current study’s reported RMSD of 4 ms between knee positions, but markedly greater than the RMSD of 0.24 ms reported between repeated scans (Table 7). The RMSD in ACL UTE-T2* values between repeated scans and knee positions indicates that the applied UTE sequence can be sensitive enough to evaluate ACL graft maturation in vivo, however, the reproducibility of ACL UTE-T2* quantifications is impacted by knee position reproducibility.
The sensitivity of T2* relaxation time to knee position can originate from multiple sources. One explanation could be the magic angle effect, which causes increasing overestimation of T2* relaxation time as the orientation of the tissue’s organized collagen approaches 54° relative to the static B0 field (22-24). The anatomic orientation of the ACL, PCL and PT may make these tissues more susceptible to the magic angle effect in the evaluated knee positions, resulting in variation of T2* relaxation time quantification between extended and flexed positions. Conversely, the meniscus is likely less affected by the magic angle effect because of its anatomic orientation in the evaluated joint positions. Changes in ligament and tendon tensioning between knee positions could be another source of variation in mean T2* values between knee positions, where T2* relaxation time may change as ligament tensioning changes between extended and flexed positions (37-39).
There are several limitations for our study worth noting. First, the sample size for this study was small, and including additional subjects would enhance the robustness of our findings. Additionally, only two knee positions were considered when evaluating the effect that knee position has on T2* relaxation time quantifications. The results show that UTE and GRE T2* quantifications can change due to knee position, however, a more systematic approach with multiple knee positions would be needed to characterize the orientation dependence of T2* relaxation time. A more systematic approach could also evaluate changes in joint rotations aside from flexion, such as varus and internal tibial rotation to account for potential coupled kinematics. As suggested earlier, the use of a mono-exponential fitting function to determine T2* relaxation times could introduce bias to the tissue’s UTE- and GRE-T2* relaxation time calculations and implementing a bi-exponential fitting function as others have done (35,40) may reduce this sequence-specific bias. Further to this point, separating short and long T2* components could provide additional sensitivity to detect clinically meaningful tissue remodeling processes and is of interest in future work developing these sequences.
Conclusions
In summary, we showed that the 3D radial UTE sequence can be used to repeatably quantify T2* relaxation time for knee ligaments, tendons, and menisci. The meniscus showed little variation in UTE-T2* values between evaluated knee positions, suggesting that the presented UTE sequence can yield reproducible T2* quantifications. The scan-rescan repeatability of the ACL UTE-T2* values indicates that the presented sequence may be used to detect clinically relevant changes in mean T2* relaxation time, however, reproducibility of these values is impacted by knee position consistency between scans.
Acknowledgments
Funding: This study was partially supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-459/coif). Kecheng Liu is employed by Siemens Medical Solutions USA, Inc., Malvern. Xiaodong Zhong was an employee of Medical Solutions USA, Inc. in Los Angeles until June 30, 2023. Stefan Sommer is employed by Siemens Healthineers International AG, Switzerland. Jillian E. Beveridge and William Zaylor received partial salary support from NIH/NIAMS R00-AR069094, and William Zaylor also received salary support from NIH T32AR007505. Xiaojuan Li and Xiaoliang Zhang received partial salary support from the grant NIH/NIBIB U01EB023829, and the same grant partially supported the work presented in this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the Institutional Review Board of the Cleveland Clinic, written informed consent was obtained from the participants prior to human subject experiments.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hasegawa A, Otsuki S, Pauli C, Miyaki S, Patil S, Steklov N, Kinoshita M, Koziol J, D'Lima DD, Lotz MK. Anterior cruciate ligament changes in the human knee joint in aging and osteoarthritis. Arthritis Rheum 2012;64:696-704. [Crossref] [PubMed]
- Levy YD, Hasegawa A, Patil S, Koziol JA, Lotz MK, D'Lima DD. Histopathological changes in the human posterior cruciate ligament during aging and osteoarthritis: correlations with anterior cruciate ligament and cartilage changes. Ann Rheum Dis 2013;72:271-7. [Crossref] [PubMed]
- Naoum S, Mitseas P, Koutserimpas C, Kotsapas M, Raptis K, Gantsos AS. Correlation of Cruciate Ligament Histological Findings with Coronal Plane Deformities and Characteristics of Patients Undergoing Total Knee Arthroplasty. Maedica (Bucur) 2022;17:323-8. [Crossref] [PubMed]
- Thorpe CT, Screen HR. Tendon Structure and Composition. Adv Exp Med Biol 2016;920:3-10. [Crossref] [PubMed]
- Benjamin M, Milz S, Bydder GM. Magnetic resonance imaging of entheses. Part 2. Clin Radiol 2008;63:704-11. [Crossref] [PubMed]
- Chu CR, Williams AA, West RV, Qian Y, Fu FH, Do BH, Bruno S. Quantitative Magnetic Resonance Imaging UTE-T2* Mapping of Cartilage and Meniscus Healing After Anatomic Anterior Cruciate Ligament Reconstruction. Am J Sports Med 2014;42:1847-56. [Crossref] [PubMed]
- Wang A, Pedoia V, Su F, Abramson E, Kretzschmar M, Nardo L, Link TM, McCulloch CE, Jin C, Ma CB, Li X MR. T1ρ and T2 of meniscus after acute anterior cruciate ligament injuries. Osteoarthritis Cartilage 2016;24:631-9. [Crossref] [PubMed]
- Rauscher I, Stahl R, Cheng J, Li X, Huber MB, Luke A, Majumdar S, Link TM. Meniscal measurements of T1rho and T2 at MR imaging in healthy subjects and patients with osteoarthritis. Radiology 2008;249:591-600. [Crossref] [PubMed]
- Baum T, Joseph GB, Karampinos DC, Jungmann PM, Link TM, Bauer JS. Cartilage and meniscal T2 relaxation time as non-invasive biomarker for knee osteoarthritis and cartilage repair procedures. Osteoarthritis Cartilage 2013;21:1474-84. [Crossref] [PubMed]
- Su X, Zhang Y, Gao Q, Liang Z, Wan L, Zhang L, Tang G. Preliminary study on the assessment of early cartilage degeneration by quantitative ultrashort echo time magnetic resonance imaging in vivo. Quant Imaging Med Surg 2022;12:3803-12. [Crossref] [PubMed]
- Amiel D, Frank C, Harwood F, Fronek J, Akeson W. Tendons and ligaments: a morphological and biochemical comparison. J Orthop Res 1984;1:257-65. [Crossref] [PubMed]
- Robson MD, Gatehouse PD, Bydder M, Bydder GM. Magnetic resonance: an introduction to ultrashort TE (UTE) imaging. J Comput Assist Tomogr 2003;27:825-46. [Crossref] [PubMed]
- Chang EY, Du J, Chung CB. UTE imaging in the musculoskeletal system. J Magn Reson Imaging 2015;41:870-83. [Crossref] [PubMed]
- Kijowski R, Wilson JJ, Liu F. Bicomponent ultrashort echo time T2* analysis for assessment of patients with patellar tendinopathy. J Magn Reson Imaging 2017;46:1441-7. [Crossref] [PubMed]
- Chu CR, Williams AA. Quantitative MRI UTE-T2* and T2* Show Progressive and Continued Graft Maturation Over 2 Years in Human Patients After Anterior Cruciate Ligament Reconstruction. Orthop J Sports Med 2019;7:2325967119863056. [Crossref] [PubMed]
- Yi J, Lee YH, Song HT, Suh JS. Comparison of T2* mapping between regular echo time and ultrashort echo time with 3D cones at 3 tesla for knee meniscus. Medicine (Baltimore) 2018;97:e13443. [Crossref] [PubMed]
- Wu M, Zhao W, Wan L, Kakos L, Li L, Jerban S, Jang H, Chang EY, Du J, Ma YJ. Quantitative three-dimensional ultrashort echo time cones imaging of the knee joint with motion correction. NMR Biomed 2020;33:e4214. [Crossref] [PubMed]
- Jerban S, Hananouchi T, Ma Y, Namiranian B, Dorthe EW, Wong JH, Shojaeiadib N, Wu M, Du J, D'Lima D, Chung CB, Chang EY. Correlation between the elastic modulus of anterior cruciate ligament (ACL) and quantitative ultrashort echo time (UTE) magnetic resonance imaging. J Orthop Res 2022;40:2330-9. [Crossref] [PubMed]
- Williams A, Qian Y, Chu CR. UTE-T2* mapping of human articular cartilage in vivo: a repeatability assessment. Osteoarthritis Cartilage 2011;19:84-8. [Crossref] [PubMed]
- Agergaard AS, Malmgaard-Clausen NM, Svensson RB, Nybing JD, Boesen M, Kjaer M, Magnusson SP, Hansen P. UTE T2* mapping of tendinopathic patellar tendons: an MRI reproducibility study. Acta Radiol 2021;62:215-24. [Crossref] [PubMed]
- Carl M, Nazaran A, Bydder GM, Du J. Effects of fat saturation on short T2 quantification. Magn Reson Imaging 2017;43:6-9. [Crossref] [PubMed]
- Shao H, Pauli C, Li S, Ma Y, Tadros AS, Kavanaugh A, Chang EY, Tang G, Du J. Magic angle effect plays a major role in both T1rho and T2 relaxation in articular cartilage. Osteoarthritis Cartilage 2017;25:2022-30. [Crossref] [PubMed]
- Du J, Pak BC, Znamirowski R, Statum S, Takahashi A, Chung CB, Bydder GM. Magic angle effect in magnetic resonance imaging of the Achilles tendon and enthesis. Magn Reson Imaging 2009;27:557-64. [Crossref] [PubMed]
- Wu M, Ma Y, Wan L, Jerban S, Jang H, Chang EY, Du J. Magic angle effect on adiabatic T(1ρ) imaging of the Achilles tendon using 3D ultrashort echo time cones trajectory. NMR Biomed 2020;33:e4322. [Crossref] [PubMed]
- Beveridge JE, Machan JT, Walsh EG, Kiapour AM, Karamchedu NP, Chin KE, Proffen BL, Sieker JT, Murray MM, Fleming BC. Magnetic resonance measurements of tissue quantity and quality using T(2) * relaxometry predict temporal changes in the biomechanical properties of the healing ACL. J Orthop Res 2018;36:1701-9. [Crossref] [PubMed]
- Flannery SW, Murray MM, Badger GJ, Ecklund K, Kramer DE, Fleming BC, Kiapour AM. Early MRI-based quantitative outcomes are associated with a positive functional performance trajectory from 6 to 24 months post-ACL surgery. Knee Surg Sports Traumatol Arthrosc 2023;31:1690-8. [Crossref] [PubMed]
- Li X, Benjamin Ma C, Link TM, Castillo DD, Blumenkrantz G, Lozano J, Carballido-Gamio J, Ries M, Majumdar S. In vivo T(1rho) and T(2) mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage 2007;15:789-97. [Crossref] [PubMed]
- Peters JR, Zaylor W, Chokhandre SK, Klonowski EM, Erdemir A, Beveridge JE, Li X. An Automated Approach to Define Femoral and Tibial Coordinate Systems with Limited Diaphysis Length. In: Orthopaedic Research Society Annual Meeting. Dallas Texas; 2023.
- Grood ES, Suntay WJ. A joint coordinate system for the clinical description of three-dimensional motions: application to the knee. J Biomech Eng 1983;105:136-44. [Crossref] [PubMed]
- Gamer M, Lemon J, Fellows I, Singh P. irr: Various Coefficients of Interrater Reliability and Agreement [Internet]. 2019 [cited 2022 May 5]. Available from: https://CRAN.R-project.org/package=irr
- Koo TK, Li MY. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J Chiropr Med 2016;15:155-63. [Crossref] [PubMed]
- Zhu D, Wu W, Yu W, Hong G, Fang Y, Li S, Ma Y. Ultrashort echo time magnetization transfer imaging of knee cartilage and meniscus after long-distance running. Eur Radiol 2023;33:4842-54. [Crossref] [PubMed]
- Chen Y, Li L, Le N, Chang EY, Huang W, Ma YJ. On the fat saturation effect in quantitative ultrashort TE MR imaging. Magn Reson Med 2022;87:2388-97. [Crossref] [PubMed]
- Wilms LM, Radke KL, Latz D, Thiel TA, Frenken M, Kamp B, Filler TJ, Nagel AM, Müller-Lutz A, Abrar DB, Nebelung S. UTE-T2* versus conventional T2* mapping to assess posterior cruciate ligament ultrastructure and integrity-an in-situ study. Quant Imaging Med Surg 2022;12:4190-201. [Crossref] [PubMed]
- Liu J, Nazaran A, Ma Y, Chen H, Zhu Y, Du J, Li S, Zhou Q, Zhao Y. Single- and Bicomponent Analyses of T2* Relaxation in Knee Tendon and Ligament by Using 3D Ultrashort Echo Time Cones (UTE Cones) Magnetic Resonance Imaging. Biomed Res Int 2019;2019:8597423. [Crossref] [PubMed]
- Williams A, Qian Y, Bear D, Chu CR. Assessing degeneration of human articular cartilage with ultra-short echo time (UTE) T2* mapping. Osteoarthritis Cartilage 2010;18:539-46. [Crossref] [PubMed]
- Csapo R, Juras V, Heinzle B, Trattnig S, Fink C. Compositional MRI of the anterior cruciate ligament of professional alpine ski racers: preliminary report on seasonal changes and load sensitivity. Eur Radiol Exp 2020;4:64. [Crossref] [PubMed]
- Jerban S, Nazaran A, Cheng X, Carl M, Szeverenyi N, Du J, Chang EY. Ultrashort echo time T2(*) values decrease in tendons with application of static tensile loads. J Biomech 2017;61:160-7. [Crossref] [PubMed]
- Wilms LM, Radke KL, Abrar DB, Latz D, Schock J, Frenken M, Windolf J, Antoch G, Filler TJ, Nebelung S. Micro- and Macroscale Assessment of Posterior Cruciate Ligament Functionality Based on Advanced MRI Techniques. Diagnostics (Basel) 2021;11:1790. [Crossref] [PubMed]
- Chang EY, Du J, Statum S, Pauli C, Chung CB. Quantitative bi-component T2* analysis of histologically normal Achilles tendons. Muscles Ligaments Tendons J 2015;5:58-62.


