
Unsupervised domain adaptation for automated knee osteoarthritis phenotype classification
Introduction
Osteoarthritis (OA) is a common degenerative disease. Ageing populations worldwide contribute to the increasing demand for OA diagnosis, staging and grading (1). Kellgren and Lawrence proposed a grading system with radiography, known as the Kellgren-Lawrence (K-L) grade (2), accessing patients’ knee joint space, osteophytes, sclerosis, and bone ends deformity (3). Despite its wide usage, K-L grade based on radiography provides a limited assessment of soft tissues like cartilage and meniscus in knee joints. Recently, several MRI-based grading systems were introduced. MRI-based knee OA grading systems like MRI Osteoarthritis Knee Score (MOAKS) (4) and Whole-Organ Magnetic Resonance Imaging Score (WORMS) (5) measure knee compartments in fine detail.
Manual grading of OA is time-consuming. Methods to perform automatic OA grading based on deep learning techniques have been reported. With radiographic data, previous work shows superior performance in classifying K-L grades by deep learning. Tiulpin and Saarakkala (6) demonstrated a convolutional neural network (CNN)-based multi-task classifier to classify K-L grade and Osteoarthritis Research Society International (OARSI) grades (7) on the radiographic data from Osteoarthritis Initiative (OAI) (8) and Multi-Center Osteoarthritis Study (MOST) (9) datasets. The authors reported an accuracy of 66.68% and 63.58% on K-L and OARSI grade classification tasks, respectively. Similarly, Zhang et al. (10) reported a K-L grade classification accuracy of 74.81% with a knee joint localisation before classification. A recent approach (11) synthesises radiographs with a Generative Adversarial Network (GAN) and mixes it with real images for training. This generative augmentation approach achieved the best testing accuracy of 75.76% when mixing the real images with 200% synthesised images. Han et al. (12) implemented an OA risk prediction pipeline with follow-up radiographic images synthesised from baseline, showing the diagnostic OA information is hidden inside the latent features. Their approaches achieved prediction accuracies of 84.8%, 19.7%, 44.9%, 64.0% and 59.8% for K-L grade 0, 1, 2, 3, and 4, respectively.
Compared to radiography, MRI provides more information for OA diagnostics, enables imaging of soft tissues such as cartilage and meniscus (13), and is more suitable for conducting phenotyping studies for clinical trials (14). Several works explored OA grading from MRI, with a focus on the tissues that cannot be imaged by radiograph. Astuto et al. classified knee abnormalities on cartilage, bone marrow edema, meniscus, and anterior cruciate ligament (ACL) with a two-step approach on 3D turbo spin echo (TSE)/fast spin echo (FSE) MRI. The author collected 1,435 knee MRIs to train the deep learning model and get the area under the receiver operating characteristic curve (AUROC) ranging from 0.83 to 0.93 (15). Tuya E and colleagues classified patellofemoral OA with a CNN on the axial radiograph, reporting an AUROC of 0.91 (16). With advanced deep learning techniques like semi-supervised learning, Hou et al. present a knee MRI cartilage grading without fully labelled data (17). However, none of the abovementioned literature proposed an automated OA grading for full MRI grading systems like MOAKS or WORMS. These fine-grained MRI grading scores provide elaborate quantitative descriptions of the knee, but they also bring a heavy workload to the radiologists and challenges to developing automatic grading systems.
Beyond grading systems, knee OA phenotyping is an emerging research topic that provides a new model for understanding OA (18). A knee OA phenotype is a single or a group of characteristics connected to OA outcomes, such as body mass index (BMI), external mechanical hurt, and knee structural changes (18). Bruyère et al. argues that OA is a combination of phenotypes rather than a single disease (19), considering the various factors that can trigger OA or contribute to the OA symptoms are phenotypes (20). Roemer and colleagues (21) define five phenotypes that describe knee structures under MRI: (I) inflammatory phenotype, defined by synovitis and/or joint effusion; (II) cartilage/meniscus phenotype, defined by meniscus pathology and reflects the cartilage loss; (III) subchondral bone, characterised by large bone marrow lesion (BML); (IV) atrophic; and (V) hypertrophic phenotype, defined by osteophytes. During the development, the authors employed a case-control study within the OAI dataset (8) dataset, pre-defined phenotypes with MOAKS, and evaluated the alignment between MOAKS and the simplified scoring system. Roemer et al. also conducted investigations of this set of phenotypes on the OAI dataset and found that knee OA progression is linked to both cartilage/meniscus (20) and subchondral bone (22) phenotypes. On the other hand, Namiri et al. (23) built classifiers with CNN on the OAI dataset for the phenotypes mentioned above. The classifiers take 2D sagittal and coronal MRI as input and are trained on a subset of OAI. This approach achieved a satisfactory performance on the test set, and the author predicted the phenotypes for the entire OAI dataset. With the phenotypes, the authors showed increased odds of developing OA on the knees with subchondral bone and hypertrophic phenotype in 4 years. He also reported there were increased odds of total knee replacement in 8 years for knees with inflammatory, subchondral bone, and hypertrophic phenotypes (23). The comprehensive set of knee phenotypes proposed by Roemer et al. (21) was verified clinically and succinctly and clearly defined, which makes it easy to be adapted to modern deep learning systems.
Domain shift problem is common in medical imaging due to the variations in the sampling population, image acquisition hardware, software, and imaging protocols. In certain scenarios, a model trained on one dataset may fail on another dataset because the training and inference domains are shifted. To address the domain shift problem, transfer learning and domain adaptation techniques have been extensively explored. Transfer learning, often referred to as pre-train/fine-tune, takes advantage of the large training data (source data) to create a model with strong generalisability (pre-train), allowing downstream users to fine-tune the model with a limited amount of labelled local data (target data) which has domain shift from the data used in pre-train (24). On the other hand, unsupervised domain adaptation (UDA) is a technique that enables CNN trained on a source domain to be adapted to a target domain without ground truth labels from the target data (24). UDA is one of the primary methods for transferring information from more extensive and more generalised datasets to smaller datasets. The UDA process enables downstream research on target data with zero labels (25). In medical image analysis, UDA allows researchers to take advantage of high-cost medical image datasets to expedite local research without additional labelling costs. UDA approaches align the source and target domains using various methods, such as domain translation (26), statistical matching and adversarial learning (27). UDA has been widely studied in medical image analysis, giving excellent outcomes in cross-modality, cross-organ, and cross-task applications. Dou et al. (28) adapt cardiac CT to MRI in a segmentation context; a similar CT-to-MRI segmentation by UDA was examined by Yang et al. at liver (29). Panfilov et al. (30) take UDA and other techniques to improve the robustness of knee segmentation. Gao et al. (31) apply UDA to decode brain states from functional MRI, in which UDA helps overcome various individual differences. Given the volume and complexity of UDA and transfer learning as a whole, Jiang et al. (24) and Guan and Liu (32) have published comprehensive reviews of UDA and its application in medical image analysis.
In this study, we proposed a novel approach for automatic OA phenotype classification by applying UDA based on the Adversarial Discriminative Domain Adaptation (ADDA) (27) framework. We used a CNN trained on a publicly available dataset for automated OA phenotype classification on a small dataset (n=50) from our hospital (Prince of Wales Hospital, Sha Tin, New Territories, Hong Kong SAR, China). We implemented a systematic UDA approach for automatic OA phenotype classification. In this study, we (I) proposed a UDA approach for automatic and objective MRI-based OA phenotype classification to address the challenges of collecting a large labelled dataset associated with supervised techniques; (II) compared the proposed method with two classifiers trained without UDA, demonstrating the improved phenotype classification performance of our UDA approach and (III) explored the application of UDA for OA phenotype classification tasks using datasets from multiple MRI vendors, acquisition protocols and research institutes. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-704/rc).
Methods
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The human study conducted at Prince of Wales Hospital (Sha Tin, New Territories, Hong Kong SAR, China) was approved by the Institutional Review Board, and informed consent was obtained from all patients and volunteers.
Data and MRI acquisition
We conducted a retrospective study to demonstrate the application of UDA for MRI-based OA phenotype classification. Table 1 summarises the demographics and data distribution of the source and the target data.
Table 1
| Characteristic | Source dataset | Target dataset (n=50) | |
|---|---|---|---|
| Cartilage/meniscus (n=318) | Subchondral bone (n=960) | ||
| Age (years) | 63.52±8.85 | 62.70±8.94 | 61.94±11.57 |
| BMI (kg/m2) | 28.90±4.88 | 28.85±4.75 | – |
| Gender | |||
| Male | 153 (48.11) | 386 (40.21) | 15 (30.00) |
| Female | 165 (51.89) | 574 (59.79) | 35 (70.00) |
| Knee | |||
| Left knee | 127 (39.94) | 390 (40.63) | 21 (42.00) |
| Right knee | 191 (60.06) | 570 (59.38) | 29 (58.00) |
| Cartilage/meniscus patients | 106 (33.33) | – | 8 (8.00) |
| Subchondral bone patients | – | 320 (33.33) | 5 (10.00) |
Data are presented as the mean ± standard deviation or n (%). The BMI for the target dataset is missing. BMI, body mass index.
The source dataset was a subset of the OAI dataset (8), including knee subjects randomly selected from the baseline and 4-year follow-up studies for the cartilage/meniscus and subchondral bone phenotype, respectively. Each data sample has a 3D MRI acquired with the double-echo steady-state sequence (DESS) (8) and corresponding MOAKS scores. The OAI dataset contains the MOAKS scores that were graded by experienced radiologists in previous studies (33-36). We combined the MOAKS sub-grades from these projects and selected subjects with knee MR images and the required MOAKS sub-grades to form the source dataset.
The target dataset contained knee MRI scans of 50 subjects collected at Prince of Wales Hospital (Sha Tin, New Territories, Hong Kong SAR, China) in 2020 and 2021 (37). Forty patients with radiographic OA and ten healthy controls received knee MRI exams. The average age of the participants in the target dataset was 61.94 years. Our target datasets were collected using three two-dimensional (2D) MRI sequences and a 3D TSE/FSE sequence (VISTA™) on a Philips Achieva TX 3.0T scanner (Philips Healthcare, Best, Netherlands). The 2D scans were used for manual MOAKS grading, and the 3D VISTATM scans were used for automated OA phenotype classification using the UDA pipeline. The detailed MRI protocol is reported in Table 2.
Table 2
| Sequence | 3D PD TSE (VISTA™) | PD TSE SAG | T2 SPAIR TSE COR | PD SPAIR TSE AX |
|---|---|---|---|---|
| Plane | Sagittal | Sagittal | Coronal | Axial |
| Fat suppression | SPAIR | None | SPAIR | SPAIR |
| No. of slices | 150 | 25 | 25 | 25 |
| Field of view (mm3) | 160×160×120 | 162×160×82 | 160×160×82 | 150×150×82 |
| TE/TR (ms/ms) | 26/900 | 30/3,451 | 62/5,429 | 30/5,864 |
| X-resolution (mm) | 0.71 | 0.4 | 0.227 | 0.293 |
| Y-resolution (mm) | 0.71 | 0.546 | 0.227 | 0.293 |
| Scan time (min: s) | 05:51 | 03:24 | 03:43 | 03:37 |
| Usage | Deep learning | MOAKS | MOAKS | MOAKS |
The 3D PD TSE (VISTA™) sequence was used for automatic phenotype classification. The remaining three sequences were prepared for MOAKS grading. 3D, three-dimensional; PD, proton density; TSE, turbo spin echo; SAG, sagittal; SPAIR, Spectral Attenuated Inversion Recovery; COR, coronal; AX, axial; TE, echo time; TR, repetition time; MOAKS, MRI Osteoarthritis Knee Score.
MOAKS grades and phenotype labels
Roemer et al. (21) developed the five knee OA phenotypes (inflammatory, cartilage/meniscus, subchondral bone, atrophic and hypertrophic) and introduced a protocol to convert MOAKS grades to knee OA phenotypes using the MRI collected from the OAI dataset. The phenotypes studied in this work were defined as follows. All subregions and grades were defined by the MOAKS grading system (4). (I) Cartilage/meniscus phenotype presents when at least one medial or lateral meniscus subregion was graded with one of complex tear, partial meniscal maceration or complete maceration, while any type of tear was identified on at least one of other subregions. Additionally, at least one of the cartilage grades, 2.1, 2.2, 3.2 or 3.3, should appear in the same knee subject. (II) Subchondral bone phenotype presents when at least one grade-3 BML was recorded from ten tibiofemoral joint (TFJ) subregions plus a grade-2 or grade-3 BML in additional two of ten TFJ subregions.
To quickly obtain the phenotype labels, we implemented a Python script to read the MOAKS grades from the source and target datasets and then convert MOAKS grades to binary phenotype labels following the definitions above. The MOAKS grades of the source dataset were obtained from the OAI, and the MOAKS grades for the target dataset were independently graded by two musculoskeletal radiologists following the grading protocol used by the OAI. Both radiologists had more than 6 years of experience. The MOAKS grades prepared by the two radiologists had an excellent intraclass correlation coefficient of 0.999 (P<0.01). We provided the data preparation workflow in Figure 1.
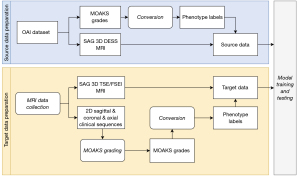
UDA pipeline
In this work, we utilise a UDA pipeline. We hypothesised that the UDA pipeline could learn from the source dataset and improve phenotype classification performance on the target dataset without involving the target phenotype labels.
Figure 2 illustrates the proposed UDA pipeline adapted from the ADDA (27) framework. ADDA minimises the domain shift between the source and target datasets in a discriminative manner. Besides pre-processing, the ADDA framework involves three steps, pre-training, adversarial adaptation, and testing. In this work, the pre-training step prepared an encoder and a classifier from source data by a supervised training process; during adversarial adaptation, we froze the pre-trained source encoder to generate feature representations for source samples and copy the same pre-trained encoder to generate feature representations for target samples (denoted as target encoder). This target encoder was initialised by the pre-trained weight from the source encoder and followed by a domain discriminator. Two neural networks were jointly optimised by a domain adversarial loss (38). The domain discriminator was designed to distinguish where the feature representation comes from (source or target) and improve the output feature from the target encoder. We finished the training for domain adaptation when the domain discriminator could not distinguish the feature origins. The last step was concatenating the target encoder with the classification head and evaluating this target classifier with the ground truth labels.
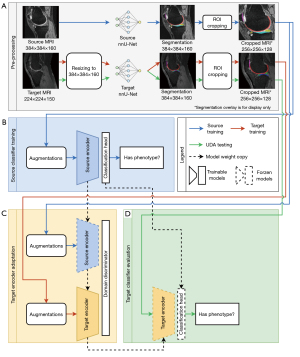
MR image pre-processing
As mentioned in the previous section, the phenotypes we adopt in this work are at the knee compartments at TFJ and patellofemoral joint (PFJ). Thus, we consider a volume that covers the TFJ and PFJ as a region of interest (ROI). We introduced an automatic cropping module as an MRI pre-processing, forcing the classification model to learn from the ROI to enhance training efficiency. Before cropping, the target MR images were first resized to 160×384×384 to align with the source MR images (Figure 2A). The cropping module was navigated by segmentation masks of femoral, tibial, patellar cartilages and meniscus. A bounding box was initially estimated by merging these segmentation masks, then we added offsets to the bounding box in the superior, inferior, and anterior directions to cover subchondral bone areas. The cropping module can automatically adjust the position of the ROI box with a fixed size (128×256×256) to cover the TFJ and PFJ. It also involves an automatic scaling scheme to guarantee full coverage of the knee joint in the cropped image. We used nnU-Net (39) to perform this segmentation for its state-of-the-art performance according to the comprehensive experiment reported by the authors. We also notice that the current deep learning segmentation methods have a close performance in terms of knee MRI segmentation from a benchmark on the OAI dataset (40). Thus, we trained two 2D nnU-Net models with the official implementation and default settings to provide segmentation masks. The source and target segmentation models were trained using 3D MR scans from the OAI-iMorphics dataset (141 for training, 35 for testing) and the labelled target dataset (17 for training, 8 for testing), respectively.
Source encoder training
Figure 2B shows the 3D DenseNet121 (41) encoder and classification head trained on the source dataset for each phenotype, giving the source classifier Cs. The augmentation scheme involved adding random Gaussian noise, scaling the intensity by a randomly selected factor from 0.8 to 1.2, rotating by a randomly chosen degree from −10 to 10 and scaling the image size by a randomly chosen factor from 1 to 1.1. Each augment item had a probability of 50%. We trained the classifiers with a batch size of 2, focal loss (42) with a gamma of 1, an Adam (43) optimiser with a learning rate of 10−6 and a weight decay of 10−3. The training process stopped when the validation loss increased by three times, and the model with the best area under the precision-recall curve on the validation set was selected.
Target encoder adaptation
As shown in Figure 2C, the domain adaptation process involved a source encoder with frozen weights, a target encoder initialised with the weights of the source encoder, and a randomly initialised domain discriminator. We adopted the hyperparameters from the study of Jiang et al. (24), in which the target encoder and domain discriminator was optimised by a domain adversarial loss (38) and a binary cross entropy objective function, respectively. A stochastic gradient descent (44) optimiser was set with an initial learning rate of 0.001, a momentum of 0.9 and a weight decay of 10−3 at a batch size of 2. The learning rate decays according to the following formula:
Where an+1 and an are the learning rates at the next epoch and the current epoch, respectively; n is the current epoch count and γ and λ are hyperparameters set at 0.0003 and 0.75, respectively. The same image augmentations used for the source encoder training were applied to both the source and target MR images. During the target encoder adaptation, we continuously fed samples from the source dataset while looping 49 target training samples for each epoch. Each training was run for 50 epochs, and the models from the last epoch were selected.
Target classifier evaluation
We attached the trained classification head from the source domain to the adapted target encoder, forming the target classifier CT-UDA (Figure 2D). Ablation studies were conducted by comparing three models, CT-UDA, Cs and CT for OA phenotype classification. Cs was the pre-trained source classifier. In the ablation studies, we directly inference Cs with the target data. Besides, the target classifier CT was trained with the target data only from scratch. CT was trained by the same hyperparameter as Cs. By comparing the classification performance of these three models, we want to evaluate the benefits of UDA in OA phenotype classification when the target dataset is relatively small.
Statistical analysis
For each phenotype, the source dataset was split into training (70%), validation (10%) and test (20%) sets to train the source classifier Cs. The target classifiers CT-UDA and CT were trained by a leave-one-out strategy that used 49 samples for training and 1 sample for testing. Dice similarity coefficient (DSC) scores were used to evaluate the segmentation models. AUROC, sensitivity, specificity and accuracy were used to assess the source and target classification models. When reporting AUROC scores, their 95% confidence intervals (CIs) and P value derived from DeLong et al. (45) are included. For all statistical tests, we set a two-sided significant level as P<0.05.
Implementation
The proposed system and scripts were implemented and tested using Python 3.10, PyTorch (46) 1.10, MONAI 0.8.1, PyTorch Lightning 1.6.3 and Transfer Learning Library (24) 0.2 (UDA only). An NVIDIA (Santa Clara, CA, USA) RTX A6000 graphics processing unit was used to run the deep learning experiments. We conducted the sample size analysis with MedCalc for Windows, version 20.1 (MedCalc Software, Ostend, Belgium), and other statistical analysis with SPSS for Windows, version 27 (IBM Crop, Armonk, NY, USA).
Results
Data collection
In this study, we have collected two datasets. The demographic for the datasets is available in Table 1.
Source data collection
The source data was collected from the OAI dataset. We first selected the participants with the MOAKS graded (33-36), then included the samples from the baseline and 48-month follow-up studies. Within this subgroup, we keep the MOAKS reading from the earliest reading project for those samples with multiple MOAKS readings. After we converted the MOAKS subgrades to phenotypes, we created a balanced dataset by randomly selecting negative-classified samples to keep the number of negative samples twice that of the positive samples. Figure 3 provides a flow chart for this procedure.
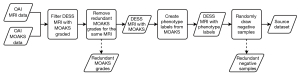
Target data collection
The target data was collected at Prince of Wales Hospital (Sha Tin, New Territories, Hong Kong SAR, China) (37). Forty patients and ten age-matched healthy controls were recruited following these criteria, (I) age equal to or greater than 18 years, (II) healthy and active knee joint (for healthy controls), (III) an initial OA status according to the classification standard from American College of Rheumatology (47), (IV) knee pain persists more than two months, and (V) presented evidence of radiographic OA. Then we excluded the participants have the following limitations, (I) restricted to MRI scans, (II) had a psychiatric disorder, (III) had claustrophobic, (IV) had inflammatory arthritis, (V) ongoing pregnancy and lactation, (VI) has a serious disease like advance cancer, (VII) has metal product in the knee, (VIII) has major blood system disease, (IX) has severe deformities of the lower limbs. For this study, we report that 50 samples are sufficient for a target AUROC of 0.9 when we set the type I error rate as 0.05 and the type II error rate as 0.2.
MR image pre-processing
We observed both segmentation models (nnU-Net) performed well. The DSC scores for segmented knee compartments ranged from 0.81 to 0.92 (detail available in Table 3). For the performance of the segmentation on the source dataset, it is comparable to the ones in previous work (40). We consider these scores to be satisfactory because the purpose of these predicted segmentation masks was to locate the PFJ and TFJ rather than accurate segmentation of tissues.
Table 3
| Knee compartment | Femoral cartilage | Lateral meniscus | Lateral tibial cartilage | Medial meniscus | Medial tibial cartilage | Patellar cartilage |
|---|---|---|---|---|---|---|
| Source nnU-Net | 0.92±0.02 | 0.91±0.02 | 0.92±0.02 | 0.87±0.05 | 0.90±0.03 | 0.87±0.08 |
| Target nnU-Net | 0.85±0.02 | 0.85±0.08 | 0.81±0.05 | 0.85±0.12 | 0.81±0.06 | 0.81±0.10 |
Numbers are the mean ± standard deviation from the leave-out test set.
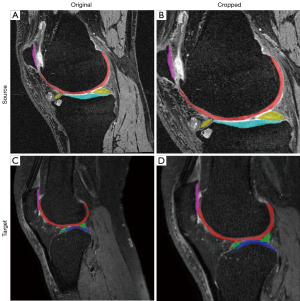
We use these models to generate segmentation for the knee samples. With the segmentations, we locate the knee joints and crop the ROI automatically. An example of automatic ROI selection is shown in Figure 4. The ROI selected the TFJ and PFJ and deleted the outer area.
Performance of source classifiers
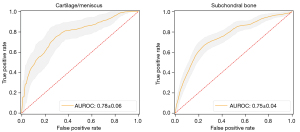
The source classifier Cs was evaluated on the hold-out test set (20% randomly selected samples) separately on each phenotype by bootstrapping 100 times. Figure 5 shows the receiver operating characteristics (ROC) curves for the classifiers. For the cartilage/meniscus and subchondral bone phenotype classifications, the means ± standard deviations of AUROC scores were 0.78±0.06 and 0.75±0.04, the sensitivities were 0.44±0.11 and 0.64±0.07, the specificities were 0.92±0.04 and 0.76±0.04 and the accuracies were 0.76±0.05 and 0.72±0.03, respectively.
Performance of target classifiers
The performance of target classifiers CT-UDA and Cs was improved compared with CT for both phenotypes. For the cartilage/meniscus phenotype, the UDA-trained classifier CT-UDA further outperformed the other classifiers (Cs and CT) in all parameters (AUROC, sensitivity, specificity and accuracy). Detailed performance is shown in Table 4. For the subchondral bone phenotype, we observed a similar performance among the UDA-trained classifier CT-UDA and the classifier Cstrained solely with the source dataset, but both classifiers outperformed CT, which was trained solely on the target dataset. Detailed performance is available in Table 5.
Table 4
| Classifier | AUROC | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|
| Cs | 0.76 (0.53–0.98), 0.02 | 50 (2/4) | 73.91 (34/46) | 72 (36/50) |
| CT | 0.52 (0.13–0.90), 0.93 | 25 (1/4) | 73.91 (34/46) | 70 (35/50) |
| CT-UDA(ours) | 0.90 (0.79–1.02), <0.01 | 75 (3/4) | 78.26 (36/46) | 78 (39/50) |
The AUROC column is AUROC value (95% CI), and P statistics. AUROC, area under the receiver operating characteristic curve; UDA, unsupervised domain adaptation; CI, confidence interval.
Table 5
| Classifier | AUROC | Sensitivity (%) | Specificity (%) | Accuracy (%) |
|---|---|---|---|---|
| Cs | 0.76 (0.55–0.98), 0.02 | 60 (3/5) | 66.67 (30/45) | 66 (33/50) |
| CT | 0.53 (0.33–0.73), 0.78 | 60 (3/5) | 42.22 (19/45) | 44 (22/50) |
| CT-UDA(ours) | 0.75 (0.56–0.94), 0.01 | 60 (3/5) | 64.44 (29/45) | 64 (32/50) |
The AUROC column is AUROC value (95% CI), and P statistics. AUROC, area under the receiver operating characteristic curve; UDA, unsupervised domain adaptation; CI, confidence interval.
Discussion
In this study, we proposed a UDA application for MRI-based OA phenotype classification. The proposed method adapted the CNN encoder trained on the large, publicly available OAI dataset to a smaller, locally collected target dataset. The performance of the target classifier trained with UDA, as evaluated by the AUROC, sensitivity, specificity, and accuracy, were improved compared with the classifier trained without UDA. The target classifier trained with UDA successfully captured the crucial information from the source dataset for OA phenotype classification and adapted it to the target dataset without target data labels, despite the differences in MRI sequences, scanner vendors, acquisition parameters and patient groups between the two datasets.
UDA is widely used in various medical imaging tasks, including segmentation (28,30,48) and diagnostics (49), and has been successfully applied for the analysis of images of multiple organs, including the heart (28), brain (31), breast (50), liver (48) and knee (30). UDA applications in medical imaging address the issue of insufficient data in the target dataset by taking the feature from the source to the target. To train an excellent deep-learning model for OA phenotype classification requires a large amount of data, which can be challenging for individual hospitals. We propose a method that leverages the publicly available OAI dataset to learn a deep representation that can facilitate downstream research on diagnostics and prognostics in local patient groups using deep learning. Our method has the potential to address the challenges of limited data and high labelling costs that often hamper such research.
We develop an automatic OA phenotype classification system based on clinically validated phenotypes and standard TSE/FSE MRI acquisition. The phenotypes were proposed by Roemer et al. (21), who conducted a case-control association analysis of phenotypes with multiple clinical assessments from 475 knee subjects from the Foundation for National Institutes of Health OA Biomarkers Consortium cohort, a subset of the OAI dataset (22). The analysis revealed a relationship between the subchondral bone phenotype and the risk of radiographic OA progression. In a longitudinal study of the entire OAI dataset by Namiri et al. (23), correlations between all phenotypes and concurrent structural OA were discovered. Note the phenotypes were designed for quick MRI knee structural screening for treatment. Our proposed method may speed up the screening process.
TSE/FSE sequences are widely available on MRI scanners and are regularly used in clinical routines and research. Astuto et al. (15) presented a knee OA staging system with multiple binary classifications from 1,786 3D TSE/FSE MR images, yielding AUROC values ranging from 0.83 to 0.93 for all knee compartments. Namiri et al. (51) performed severity staging on ACL injuries using a 3D TSE/FSE MR image dataset containing knee images from 1,243 subjects, yielding 89% and 92% accuracy with 3D and 2D CNNs, respectively. Although previous studies have generated promising results using supervised training with large high-quality datasets, those datasets are not publicly available. Meanwhile, the 3D MRI in publicly available datasets like OAI are collected by using sequence 3D DESS instead of 3D TSE/FSE. By using a publicly available and high-quality OAI dataset and adapting the trained features to a small 3D TSE/FSE dataset, our method provides OA grading based on common clinical TSE/FSE sequences.
Although UDA improved the classification performance compared with the non-UDA models, we were limited by small sample sizes and an imbalanced data distribution. The small sample size of the target dataset (n=50) constrained the statistical power and deep-learning performance. We also observed that the UDA classifier did not gain improvement compared with the direct inference on the source classifier in subchondral bone phenotype. This is likely due to the intricate characteristics associated with the subchondral bone phenotype, which hinders the model training. Additionally, the sample size and spatial geometry imbalance of our dataset affected the classification power. The patient groups had much smaller sample sizes than the control groups, and the geometry size of the image features that defined the phenotypes was small compared with the geometry size of the 3D knee volume. The small sample size and geometry imbalance may have led to the poor performance of the CNN. We attempted to overcome the sample size inequality by using focal loss (42) and the geometry imbalance by using ROI cropping. Future investigations to address the data imbalance issue are needed. For example, zero-shot learning (52,53), which learns from samples of the majority classes to improve the minor-class classification, may be useful in this application. The pathology detection approach proposed by Desai et al. (54) can potentially address the geometry imbalance issue by converting the phenotype classification to phenotype-related feature detection tasks.
Our work is limited by the structural OA phenotypes based on MRI. Roemer et al. (20) acknowledged that the MRI-based structural phenotypes always overlap where multiple phenotypes appear together. This property challenged the deep learning models to distinguish clear classification boundaries, especially with limited data size. In this work, we only studied two phenotypes due to limited phenotypes manifested in our small in-house dataset. Further work is needed to extend the proposed method to more OA phenotypes.
In conclusion, we report a UDA approach for automatic phenotype classification of knee OA in 3D TSE/FSE MR images. The proposed UDA implementation transfers OA phenotype classification information from the publicly available large OAI dataset to a small in-house dataset. The phenotype classification performance was significantly improved with UDA compared to without UDA. This is beneficial for downstream clinical research at individual hospitals, which often have insufficient training data. As UDA is transferable to various medical imaging tasks, this work may provide a useful reference for further research across institutions, imaging devices, acquisitions, and patient groups.
Acknowledgments
We would like to acknowledge Cherry Cheuk Nam Cheng and Ben Chi Yin Choi for assistance in patient recruitment and MRI exams, as well as Zongyou Cai for advising statistical analysis.
Funding: This study was supported by a grant from
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-704/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-704/coif). WC reports that this work was supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X). WC is a co-founder and a shareholder of Illuminatio Medical Technology Limited. JFG serves as an unpaid editorial board member of Quantitative Imaging in Medicine and Surgery. The other authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cui A, Li H, Wang D, Zhong J, Chen Y, Lu H. Global, regional prevalence, incidence and risk factors of knee osteoarthritis in population-based studies. EClinicalMedicine 2020;29-30:100587. [Crossref] [PubMed]
- Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis 1957;16:494-502. [Crossref] [PubMed]
- Kohn MD, Sassoon AA, Fernando ND. Classifications in Brief: Kellgren-Lawrence Classification of Osteoarthritis. Clin Orthop Relat Res 2016;474:1886-93. [Crossref] [PubMed]
- Hunter DJ, Guermazi A, Lo GH, Grainger AJ, Conaghan PG, Boudreau RM, Roemer FW. Evolution of semi-quantitative whole joint assessment of knee OA: MOAKS (MRI Osteoarthritis Knee Score). Osteoarthritis Cartilage 2011;19:990-1002. [Crossref] [PubMed]
- Peterfy CG, Guermazi A, Zaim S, Tirman PF, Miaux Y, White D, Kothari M, Lu Y, Fye K, Zhao S, Genant HK. Whole-Organ Magnetic Resonance Imaging Score (WORMS) of the knee in osteoarthritis. Osteoarthritis Cartilage 2004;12:177-90. [Crossref] [PubMed]
- Tiulpin A, Saarakkala S. Automatic Grading of Individual Knee Osteoarthritis Features in Plain Radiographs Using Deep Convolutional Neural Networks. Diagnostics (Basel) 2020.
- Altman RD, Gold GE. Atlas of individual radiographic features in osteoarthritis, revised. Osteoarthritis Cartilage 2007;15 Suppl A:A1-56.
- Peterfy CG, Schneider E, Nevitt M. The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 2008;16:1433-41. [Crossref] [PubMed]
- Segal NA, Nevitt MC, Gross KD, Hietpas J, Glass NA, Lewis CE, Torner JC. The Multicenter Osteoarthritis Study: opportunities for rehabilitation research. PM R 2013;5:647-54. [Crossref] [PubMed]
- Zhang B, Tan J, Cho K, Chang G, Deniz CM. Attention-based CNN for KL Grade Classification: Data from the Osteoarthritis Initiative. In: 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI); 2020:731-5.
- Prezja F, Paloneva J, Pölönen I, Niinimäki E, Äyrämö S. DeepFake knee osteoarthritis X-rays from generative adversarial neural networks deceive medical experts and offer augmentation potential to automatic classification. Sci Rep 2022;12:18573. [Crossref] [PubMed]
- Han T, Kather JN, Pedersoli F, Zimmermann M, Keil S, Schulze-Hagen M, Terwoelbeck M, Isfort P, Haarburger C, Kiessling F, Kuhl C, Schulz V, Nebelung S, Truhn D. Image prediction of disease progression for osteoarthritis by style-based manifold extrapolation. Nat Mach Intell 2022;4:1029-39.
- Roemer FW, Guermazi A, Demehri S, Wirth W, Kijowski R. Imaging in Osteoarthritis. Osteoarthritis Cartilage 2022;30:913-34. [Crossref] [PubMed]
- Roemer FW, Kwoh CK, Hayashi D, Felson DT, Guermazi A. The role of radiography and MRI for eligibility assessment in DMOAD trials of knee OA. Nat Rev Rheumatol 2018;14:372-80. [Crossref] [PubMed]
- Astuto B, Flament I. K Namiri N, Shah R, Bharadwaj U, M Link T, D Bucknor M, Pedoia V, Majumdar S. Automatic Deep Learning-assisted Detection and Grading of Abnormalities in Knee MRI Studies. Radiol Artif Intell 2021;3:e200165. [Crossref] [PubMed]
- E T. Wang C, Cui Y, Nai R, Zhang Y, Zhang X, Wang X. Automatic diagnosis and grading of patellofemoral osteoarthritis from the axial radiographic view: a deep learning-based approach. Acta Radiol 2023;64:658-65. [Crossref] [PubMed]
- Huo J, Ouyang X, Si L, Xuan K, Wang S, Yao W, Liu Y, Xu J, Qian D, Xue Z, Wang Q, Shen D, Zhang L. Automatic Grading Assessments for Knee MRI Cartilage Defects via Self-ensembling Semi-supervised Learning with Dual-Consistency. Med Image Anal 2022;80:102508. [Crossref] [PubMed]
- Dório M, Deveza LA. Phenotypes in Osteoarthritis: Why Do We Need Them and Where Are We At? Clin Geriatr Med 2022;38:273-86. [Crossref] [PubMed]
- Bruyère O, Cooper C, Arden N, Branco J, Brandi ML, Herrero-Beaumont G, Berenbaum F, Dennison E, Devogelaer JP, Hochberg M, Kanis J, Laslop A, McAlindon T, Reiter S, Richette P, Rizzoli R, Reginster JY. Can we identify patients with high risk of osteoarthritis progression who will respond to treatment? A focus on epidemiology and phenotype of osteoarthritis. Drugs Aging 2015;32:179-87. [Crossref] [PubMed]
- Roemer FW, Jarraya M, Collins JE, Kwoh CK, Hayashi D, Hunter DJ, Guermazi A. Structural phenotypes of knee osteoarthritis: potential clinical and research relevance. Skeletal Radiol 2023;52:2021-30. [Crossref] [PubMed]
- Roemer FW, Collins J, Kwoh CK, Hannon MJ, Neogi T, Felson DT, Hunter DJ, Lynch JA, Guermazi A. MRI-based screening for structural definition of eligibility in clinical DMOAD trials: Rapid OsteoArthritis MRI Eligibility Score (ROAMES). Osteoarthritis Cartilage 2020;28:71-81. [Crossref] [PubMed]
- Roemer FW, Collins JE, Neogi T, Crema MD, Guermazi A. Association of knee OA structural phenotypes to risk for progression: a secondary analysis from the Foundation for National Institutes of Health Osteoarthritis Biomarkers study (FNIH). Osteoarthritis Cartilage 2020;28:1220-8. [Crossref] [PubMed]
- Namiri NK, Lee J, Astuto B, Liu F, Shah R, Majumdar S, Pedoia V. Deep learning for large scale MRI-based morphological phenotyping of osteoarthritis. Sci Rep 2021;11:10915. [Crossref] [PubMed]
- Jiang J, Shu Y, Wang J, Long M. Transferability in Deep Learning: A Survey. ArXiv220105867 Cs 2022. [cited 2022 Apr 27]. doi:
10.48550 /arXiv.2201.05867. - Wilson G, Cook DJ. A Survey of Unsupervised Deep Domain Adaptation. ACM Trans Intell Syst Technol 2020;11:1-46. [Crossref] [PubMed]
- Zhu JY, Park T, Isola P, Efros AA. Unpaired Image-to-Image Translation Using Cycle-Consistent Adversarial Networks. In: 2017 IEEE International Conference on Computer Vision (ICCV); 2017:2242-51.
- Tzeng E, Hoffman J, Saenko K, Darrell T. Adversarial Discriminative Domain Adaptation. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:2962-71.
- Dou Q, Ouyang C, Chen C, Chen H, Heng PA. Unsupervised Cross-Modality Domain Adaptation of ConvNets for Biomedical Image Segmentations with Adversarial Loss. In: Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence; 2018:691-7. [cited 2022 Sep 26]. doi:
10.24963/ijcai.2018/96 . - Yang J, Dvornek NC, Zhang F, Chapiro J, Lin M, Duncan JS. Unsupervised Domain Adaptation via Disentangled Representations: Application to Cross-Modality Liver Segmentation. Med Image Comput Comput Assist Interv 2019;11765:255-63.
- Panfilov E, Tiulpin A, Klein S, Nieminen MT, Saarakkala S. Improving Robustness of Deep Learning Based Knee MRI Segmentation: Mixup and Adversarial Domain Adaptation. In: 2019 IEEE/CVF International Conference on Computer Vision Workshop (ICCVW); 2019:450-9.
- Gao Y, Zhang Y, Cao Z, Guo X, Zhang J. Decoding Brain States From fMRI Signals by Using Unsupervised Domain Adaptation. IEEE J Biomed Health Inform 2020;24:1677-85. [Crossref] [PubMed]
- Guan H, Liu M. Domain Adaptation for Medical Image Analysis: A Survey. IEEE Trans Biomed Eng 2022;69:1173-85. [Crossref] [PubMed]
- Sharma L, Hochberg M, Nevitt M, Guermazi A, Roemer F, Crema MD, Eaton C, Jackson R, Kwoh K, Cauley J, Almagor O, Chmiel JS. Knee tissue lesions and prediction of incident knee osteoarthritis over 7 years in a cohort of persons at higher risk. Osteoarthritis Cartilage 2017;25:1068-75. [Crossref] [PubMed]
- Wise BL, Niu J, Guermazi A, Liu F, Heilmeier U, Ku E, Lynch JA, Zhang Y, Felson DT, Kwoh CK, Lane NE. Magnetic resonance imaging lesions are more severe and cartilage T2 relaxation time measurements are higher in isolated lateral compartment radiographic knee osteoarthritis than in isolated medial compartment disease - data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 2017;25:85-93. [Crossref] [PubMed]
- Roemer FW, Kwoh CK, Hannon MJ, Hunter DJ, Eckstein F, Grago J, Boudreau RM, Englund M, Guermazi A. Partial meniscectomy is associated with increased risk of incident radiographic osteoarthritis and worsening cartilage damage in the following year. Eur Radiol 2017;27:404-13. [Crossref] [PubMed]
- Kraus VB, Collins JE, Hargrove D, Losina E, Nevitt M, Katz JN, Wang SX, Sandell LJ, Hoffmann SC, Hunter DJ. OA Biomarkers Consortium. Predictive validity of biochemical biomarkers in knee osteoarthritis: data from the FNIH OA Biomarkers Consortium. Ann Rheum Dis 2017;76:186-95. [Crossref] [PubMed]
- Zhao S, Cahill DG, Li S, Xiao F, Blu T, Griffith JF, Chen W. Denoising of three-dimensional fast spin echo magnetic resonance images of knee joints using spatial-variant noise-relevant residual learning of convolution neural network. Comput Biol Med 2022;151:106295. [Crossref] [PubMed]
- Ganin Y, Ustinova E, Ajakan H, Germain P, Larochelle H, Laviolette F, March M, Lempitsky V. Domain-Adversarial Training of Neural Networks. J Mach Learn Res 2016;17:1-35.
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Desai AD, Caliva F, Iriondo C, Mortazi A, Jambawalikar S, Bagci U, Perslev M, Igel C, Dam EB, Gaj S, Yang M, Li X, Deniz CM, Juras V, Regatte R, Gold GE, Hargreaves BA, Pedoia V, Chaudhari AS. The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset. Radiol Artif Intell 2021;3:e200078. [Crossref] [PubMed]
- Huang G, Liu Z, Van Der Maaten L, Weinberger KQ. Densely Connected Convolutional Networks. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:2261-9.
- Lin TY, Goyal P, Girshick R, He K, Dollar P. Focal Loss for Dense Object Detection. IEEE Trans Pattern Anal Mach Intell 2020;42:318-27. [Crossref] [PubMed]
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. ArXiv14126980 Cs 2017. [cited 2021 Mar 22]. doi:
10.48550/arXiv.1412.6980 . - Bottou L, Curtis FE, Nocedal J. Optimization Methods for Large-Scale Machine Learning. SIAM Rev 2018;60:223-311.
- DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837-45.
- Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In: Advances in Neural Information Processing Systems 2019:8026-37. [cited 2021 Jan 15]. Available online: https://papers.nips.cc/paper/2019/hash/bdbca288fee7f92f2bfa9f7012727740-Abstract.html
- Peat G, Thomas E, Duncan R, Wood L, Hay E, Croft P. Clinical classification criteria for knee osteoarthritis: performance in the general population and primary care. Ann Rheum Dis 2006;65:1363-7. [Crossref] [PubMed]
- Hong J, Yu SCH, Chen W. Unsupervised domain adaptation for cross-modality liver segmentation via joint adversarial learning and self-learning. Appl Soft Comput 2022;121:108729.
- Zhang Y, Wei Y, Wu Q, Zhao P, Niu S, Huang J, Tan M. Collaborative Unsupervised Domain Adaptation for Medical Image Diagnosis. IEEE Trans Image Process. 2020;29:7834-44.
- Hesse LS, Kuling G, Veta M, Martel AL. Intensity Augmentation to Improve Generalizability of Breast Segmentation Across Different MRI Scan Protocols. IEEE Trans Biomed Eng 2021;68:759-70. [Crossref] [PubMed]
- Namiri NK, Flament I, Astuto B, Shah R, Tibrewala R, Caliva F, Link TM, Pedoia V, Majumdar S. Deep Learning for Hierarchical Severity Staging of Anterior Cruciate Ligament Injuries from MRI. Radiol Artif Intell 2020;2:e190207. [Crossref] [PubMed]
- Bian C, Yuan C, Ma K, Yu S, Wei D, Zheng Y. Domain Adaptation Meets Zero-Shot Learning: An Annotation-Efficient Approach to Multi-Modality Medical Image Segmentation. IEEE Trans Med Imaging 2022;41:1043-56. [Crossref] [PubMed]
- Korkmaz Y, Dar SUH, Yurt M, Ozbey M, Cukur T. Unsupervised MRI Reconstruction via Zero-Shot Learned Adversarial Transformers. IEEE Trans Med Imaging 2022;41:1747-63. [Crossref] [PubMed]
- Desai AD, Schmidt AM, Rubin EB, Sandino CM, Black MS, Mazzoli V, Stevens KJ, Boutin R, Ré C, Gold GE, Hargreaves BA, Chaudhari AS. SKM-TEA: A Dataset for Accelerated MRI Reconstruction with Dense Image Labels for Quantitative Clinical Evaluation. arXiv; 2022. [cited 2023 Aug 16]. doi:
10.48550/arXiv.2203.06823 .


