
Can machine learning models improve early detection of brain metastases using diffusion weighted imaging-based radiomics?
Introduction
Intracranial metastases form when cancerous cells spread from the primary cancer site through the blood and form new tumors within the brain, leading to significant disease burden and patient morbidity. Metastatic complications are responsible for approximately 90% of cancer-related morbidity (1), and up to 40% of cancer patients will experience at least one intracranial metastasis within their lifetime (2), with the majority of metastases originating from lung, breast, or melanoma cancers. Conventional treatment options include surgical resection, whole-brain radiation therapy, stereotactic radiosurgery (SRS), systemic therapy, or a combination of these approaches (3,4). Prior to treatment with SRS, high resolution magnetic resonance imaging (MRI) is required to properly localize the metastasis to achieve local control while sparing the surrounding healthy brain tissue. The standard protocol for imaging brain metastases is using gadolinium-enhanced T1 (Gd-T1)-weighted MRI.
Brain metastasis formation is accompanied by cancer cells invading the tissue’s parenchyma. Angiogenesis and tumor growth lead to microstructural changes within the brain. As a result, the diffusion of water molecules change over time as the metastasis develops. Diffusion weighted imaging (DWI) is an MRI technique that utilizes the kinetics of water molecules within the body to create contrast (5), allowing for the imaging of these microstructural changes that may otherwise be undetected on conventional Gd-T1. Furthermore, apparent diffusion coefficient (ADC) maps, generated from DWI, provide a quantitative image set, allowing data from multiple DWI sessions taken at various times to be quantitatively compared. Most DWI research on brain metastases, to date, has focused on analyzing only one imaging session, or one image set prior to treatment with one or a few sets post-treatment. Our institution treats over two hundred SRS patients per year, with approximately 20% of those patients requiring retreatment for metastatic recurrence. This provides us with a unique longitudinal dataset to not only examine ADC maps post-SRS, but also at multiple points prior to metastatic manifestation on Gd-T1.
Given the large amount of imaging data inherent in longitudinal studies, proper data management and methodology are vital. Machine learning (ML) works by taking empirical data as observational input, identifying complex relationships and patterns, and outputting intelligent decisions based on these observations (6). Supervised ML is often used in medical research (7) since oncologists can provide valuable knowledge as the ‘ground truth’ when training prediction models. Additionally, advancements in data-characterization algorithms and texture analysis such as radiomics have allowed researchers to take raw images and extract useful metrics that have proven useful in solving many problems such as differentiating different histologies (8) and predicting future treatment outcomes (9,10). Previous studies have suggested that ADC maps are an effective dataset for ML modeling (9,11), but are still under investigation due to a lack of standardization in ADC protocols and small sample sizes. Multiple supervised ML algorithms also exist and are readily available for training, but it is unclear which models will perform the best given an ADC radiomics dataset.
Currently, metastases are diagnosed visually on computed tomography (CT) or MRI. Visual manifestation may take a long time however, and more subtle microstructural changes are likely to occur before they appear on conventional imaging. This presents an opportunity for using ML in combination with other imaging modalities sensitive to microstructural changes such as DWI, to aid in earlier detection of metastatic growth. The goal of this study is to develop and compare several supervised ML classifiers that can differentiate brain metastases from normal healthy tissue using radiomic features. By analyzing longitudinal DWI taken pre- and post-SRS, we hypothesize that subtle changes in local diffusion metrics can occur during metastasis formation but prior to their detection on conventional T1 MRI. We also present which ML classifier is recommended for an ADC dataset based on performance metrics. Our results will demonstrate an application of ML in the cancer clinic for early detection of cranial metastases. We present our research in accordance with the TRIPOD reporting checklist (12) (available at https://qims.amegroups.com/article/view/10.21037/qims-23-441/rc).
Methods
Patient population
This retrospective study included patients who underwent multiple (at least three) SRS treatments at our institution between 2017 and 2022. Patients who received resection at any point in time were excluded from the cohort. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and was approved by the Health Research Ethics Board of Alberta Cancer Committee (Ethics ID HREBA.CC-20-0041). This study was retrospective and thus, informed consent was waived. Figure 1 summarizes the overall study methodology and workflow.

SRS and imaging
SRS treatment plans were created in Eclipse (Varian Medical Systems, Palo Alto, US) using multiple non-coplanar 10 MV flattening filter free beams. A dose of 20–22 Gy was prescribed to the planning target volume (tumor plus 1 mm) at the 80% isodose level in a single fraction according to standard institutional SRS protocol. As part of standard institutional protocol for SRS, Gd-T1 and DWI MR sequences were acquired pre- and post-treatment. All patients received pre-SRS imaging within one to two weeks prior to treatment. Follow-up MR imaging was conducted to assess treatment response and disease progression at systematic intervals: every 3 months for the first year, every 4 months for the second year, and biannually thereafter. Follow-ups were also carried out indefinitely until the patient’s death. All Gd-T1 and DWI were acquired on a GE or Siemens scanner (1.5 and 3T). Vendor software was used to generate ADC maps from DWI using b values of either 0, 1,000 s·mm−2, or 0, 500 and 1,000 s·mm−2. Metastatic contours were drawn by the radiation oncologist at the time of treatment planning. Contours were defined as visible lesions, or gross target volume, on Gd-T1. Image sets were acquired for study between 2017 and 2022 via our institution’s picture archiving and communication system (PACS) client.
Preprocessing
Pre-processing was performed on a custom-built desktop running Windows 11 (Microsoft, Redmond, US) and WSL2 Ubuntu 20.04 LTS (Canonical, London, UK). Digital Imaging and Communications in Medicine (DICOM) T1, DWI, as well as clinical tumor contours were anonymized using dicognito (v. 0.13.0) to remove personal identification tags (13). Images were then converted to the Neuroimaging Informatics Technology Initiative (NIFTI) format using dcm2niix (v. 1.0.20181125) (14,15). Metastases contours that were treated with SRS were also converted from initial DICOM structure files to binary NIFTI masks using dcmrtstruct2nii (v. 1.0.19) (16).
The FSL library (17) was used to register all imaging modalities and sessions. Firstly, brain extraction was performed on CT and Gd-T1 (18) (see Figure 2). Tissue segmentation was conducted on Gd-T1 to generate binary masks of white matter, grey matter, and cerebral spinal fluid (CSF) (19). Secondly, all CT and Gd-T1 images were linearly registered (6 degrees of freedom) to the earliest available CT image set (reference CT) using FSL’s Linear Image Registration Tool (FLIRT) (20,21). FSL’s epi_reg tool and white matter segmentation data were used to linearly register ADC maps to the same imaging session Gd-T1 via boundary-based registration (22). Finally, individual registrations were concatenated, and net CT-to-ADC transformation matrices were generated for all patients and imaging dates. Transformation matrices were also applied to the metastasis binary masks to transfer these contours from clinical CT onto ADC maps.

All ADC maps were normalized across imaging sessions using the central ventricle’s CSF mean ADC value. This was performed by using additional linear registrations between Gd-T1 and the standard ICBM Average Brain MNI152 data set (23). Central ventricle probability maps from the MNI152 atlas were extracted and applied to CSF segmentation data to create patient-specific CSF contours (24). Mean ADC values were calculated for each ADC map using these contours and applied to scale all ADC voxels accordingly.
Control group
Control contours were generated using existing clinical metastasis contours flipped contralaterally (25,26) (see Figure 3). Each CT image and metastasis contour are mirrored laterally and re-registered to the reference CT. This new contralateral binary mask was then transferred back to the original CT and then transformed onto the normalized ADC maps as a control. Each control was verified to ensure that it was generated in normal appearing healthy brain tissue devoid of any abnormalities, and it did not overlap with any other controls or metastases contours.

Radiomics and clinical features
The open-source package Pyradiomics (27) was used to calculate radiomic features using the normalized ADC maps and binary masks (clinical metastases and healthy brain tissue controls). Both normalized ADC maps and filtered images were used as input with default parameters during extraction. Default filters included wavelet, Laplacian of Gaussian, square, square root, logarithmic, and exponential filters applied to the normalized ADC maps. Anatomical location data was also extracted by using the MNI152 atlas (28) and FSL’s atlasquery. Additional demographic and clinical data such as patient age at time of ADC imaging, gender, original primary histology, and chemotherapy regimen were recorded and added to the dataset as features. Chemotherapy features were assigned to each metastasis based on the type of chemotherapy received (normal chemotherapy, immunochemotherapy, targeted chemotherapy, hormonal chemotherapy, or any combination of treatments). Chemotherapy data was encoded using one-hot encoding.
Feature engineering
MRI obtained from pre-SRS imaging, post-SRS follow up, and all subsequent treatments were temporally aligned to create a unique longitudinal image timeline showing the evolution of ADC maps for each patient. Metastases were then arranged in chronological order of detection to establish which metastases were viable for analysis by having at least three pre-SRS image sets (see Figure 4). Each radiomic feature was calculated for every time point available pre- and post-SRS. Additional features included slope and intercept from linear regression of pre-SRS radiomic values. Finally, two random noise features were introduced to the data set to remove unimportant features that ranked less than noise during training. Features that were missing numerical data were imputed with zero.

Statistical analysis and ML
Correlation analysis was performed and any features with >95% Pearson correlation were excluded. The dataset was then divided into training and validation sets using an 80/20 split with stratification and scaled using Scikit-Learn’s StandardScaler (29). Four ensemble binary classification ML algorithms were trained and analyzed in this study: Random Forest (RF) Classifier (30), Linear Support Vector Machine (SVM) (31), Adaptive Boost (ADA) (32), Extreme Gradient Boost (XGB) (33). Each classifier performed supervised learning using the training set and optimized towards accuracy. For RF, ADA, and XGB, features were ranked based on each classifier’s respective importance metric. Any features that ranked lower than the two random noise features were excluded from further training using that specific algorithm. For SVM, iterative construction was performed to establish feature importance (25). New SVM sub-models were created, trained, and scored by iteratively adding the most important features one at a time based on their original SVM feature importance rankings. The sub-model’s feature set with the highest accuracy was selected as the final feature set. Each ML model’s selected features were based on their overall predictive power, without any input about the actual outcomes to further minimize bias and data leakage during training. Grid search was used to tune hyperparameters for each algorithm using 10-fold cross validation (CV), optimizing towards balanced accuracy. Finally, a summed classifier was generated by integrating the four models into a soft-voting pipeline. Classifiers were evaluated based on area under the receiver operating characteristic curve (AUC), accuracy, recall, and precision, and presented with 95% confidence intervals (34) for both the training set and the internal validation set. Additional comparisons were also performed against models trained only on first-order statistics (excluding radiomic features). Performance differences were analyzed using a Mann-Whitney U statistical test and P values are presented.
Results
Sample characteristics
A total of 116 patients received SRS between 2017 and 2022. Of those patients, 789 intracranial metastases were examined, and 189 metastases were excluded due to the insufficient pre-SRS imaging data. Five hundred ninety-five control contours were generated using contralateral healthy tissue and used as the negative case for ML prediction. Each patient underwent a median of 6 DWI scans as part of their SRS treatment or follow-up imaging protocols. Median follow up time post-SRS was 95 days. A total of 790 Gd-T1 image sets, 790 ADC maps, and 275 clinical CT were retrieved and processed. Patient characteristics, primary histology, and chemotherapy regimen are summarized in Table 1.
Table 1
| Characteristics | Total cohort | Training cohort | Internal validation cohort |
|---|---|---|---|
| Demographic | |||
| Sex (male/female) | 43/73 | ||
| Median age at DWI scan (years) [range] | 61.8 [35–89] | ||
| Median No. of metastases [range] | 5 [2–22] | ||
| Primary histology (by patient), n [%] | |||
| Lung | 57 [49] | ||
| Breast | 21 [18] | ||
| Melanoma | 14 [12] | ||
| Bladder | 3 [3] | ||
| Colon | 4 [3] | ||
| Other | 17 [15] | ||
| Primary histology (by metastasis), n [%] | |||
| Lung | 253 [42] | 198 [41] | 55 [46] |
| Breast | 133 [22] | 113 [24] | 20 [17] |
| Melanoma | 117 [20] | 94 [20] | 23 [19] |
| Bladder | 17 [3] | 12 [3] | 5 [4] |
| Colon | 13 [2] | 10 [2] | 3 [3] |
| Other | 67 [11] | 53 [11] | 14 [12] |
| Systemic therapy (by metastasis), n [%] | |||
| Chemotherapy | 304 [51] | 249 [52] | 55 [46] |
| Immunotherapy | 352 [59] | 280 [58] | 72 [60] |
| Targeted | 169 [28] | 143 [30] | 26 [22] |
| Hormonal | 19 [3] | 15 [3] | 4 [3] |
DWI, diffusion weighted imaging.
Feature importance
Each of the four algorithms used reduced feature subsets to decrease the complexity of the algorithm while improving ML performance and generalization. XGB, ADA, and RF used reduced feature sets created by removing features found less important than the introduced noise variables. Following reduction, there were 325, 141, and 258 features used to train and test XGB, ADA, and RF, respectively. SVM feature importance was cutoff using iterative construction, resulting in 823 features for the final SVM-IC feature set. Top 5 ranking features for each algorithm are shown in Table 2. Most features used by all trained classifiers related to a change in ADC radiomics pre-SRS, the linear intercept, or from the previous imaging session (lag1). Additionally, the majority of top radiomic features relied on derived ADC maps (mainly wavelet and log filtering) and not on the raw ADC maps.
Table 2
| Feature ranking | Linear SVM | Random Forest | Adaptive Boost | Extreme Gradient Boost |
|---|---|---|---|---|
| 1 | change_log-sigma-5-mm-3D_gldm_DependenceEntropy | original_gldm_DependenceNonUniformityNormalized | intercept_wavelet-HLH_glszm_LargeAreaLowGrayLevelEmphasis | change_log-sigma-2-mm-3D_firstorder_Mean |
| 2 | log-sigma-3-mm-3D_gldm_DependenceVariance | log-sigma-1-mm-3D_firstorder_10Percentile | intercept_wavelet-HLH_glszm_SizeZoneNonUniformityNormalized | change_log-sigma-1-mm-3D_glcm_MCC |
| 3 | change_log-sigma-5-mm-3D_glcm_Imc1 | lag1_wavelet-LLH_firstorder_Median | change_log-sigma-4-mm-3D_firstorder_Maximum | intercept_wavelet-LLH_glcm_InverseVariance |
| 4 | intercept_log-sigma-3-mm-3D_glrlm_RunVariance | intercept_wavelet-HHH_glszm_GrayLevelNonUniformityNormalized | intercept_wavelet-LHH_glszm_SmallAreaLowGrayLevelEmphasis | change_wavelet-LHH_firstorder_Median |
| 5 | lag1_wavelet-HHH_firstorder_Mean | change_logarithm_glszm_SizeZoneNonUniformityNormalized | lag1_log-sigma-4-mm-3D_gldm_LargeDependenceLowGrayLevelEmphasis | wavelet-HHL_firstorder_Median |
SRS, stereotactic radiosurgery; Gd-T1, gadolinium-enhanced T1; SVM, Support Vector Machine; LLH, low-low-high pass filter; HHH, high-high-high pass filter; HLH, high-low-high pass filter; LHH, low-high-high pass filter; MCC, maximal correlation coefficient; HHL, high-high-low pass filter.
ML analysis
Four ML binary classification models were analyzed, and results are summarized in Table 3. All analyzed algorithms underwent hyperparameter optimization and were tuned to optimize classification accuracy. Training set AUC curves are shown in Figure 5A. XGB provided the strongest prediction accuracy (0.877±0.021, P<0.05) during cross-validation training. XGB’s AUC also performed strongly (0.949±0.013, P<0.05) when compared to the other analyzed algorithms. SVM performed similarly to XGB in accuracy (0.857±0.021, P<0.05) and AUC (0.906±0.020, P<0.05) during training with its corresponding feature subset (found during SVM-IC). RF and ADA performed slightly lower than SVM and XGB both in accuracy (RF: 0.823±0.024, ADA: 0.810±0.024, P<0.05) and AUC (RF: 0.901±0.018, ADA: 0.894±0.020, P<0.05). Algorithm rankings regarding recall are identical, with XGB exhibiting the highest recall (0.880±0.021). The soft voting classifier, comprising of a combination of the four algorithms, performed strongly in AUC (0.921±0.016) and classification accuracy (0.837±0.023). XGB and RF provided the best overall accuracy and recall on both dataset splits while maintaining relatively low training times (XGB: 1.415, RF: 1.092, SVM: 0.975, and ADA: 11.288 min).
Table 3
| Classification algorithm | Training set | Internal validation set | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | Accuracy | Recall | Precision | AUC | Accuracy | Recall | Precision | ||
| Linear Support Vector Machine | 0.906±0.020 | 0.857±0.021 | 0.857±0.021 | 0.857±0.021 | 0.766±0.059 | 0.711±0.056 | 0.711±0.056 | 0.711±0.056 | |
| Random Forest | 0.901±0.018 | 0.823±0.024 | 0.823±0.024 | 0.823±0.024 | 0.922±0.034 | 0.858±0.045 | 0.858±0.046 | 0.858±0.046 | |
| Adaptive Boost | 0.894±0.020 | 0.810±0.024 | 0.811±0.024 | 0.810±0.024 | 0.882±0.043 | 0.816±0.051 | 0.817±0.051 | 0.816±0.050 | |
| Extreme Gradient Boost | 0.949±0.013 | 0.877±0.021 | 0.880±0.021 | 0.878±0.021 | 0.910±0.037 | 0.833±0.047 | 0.833±0.046 | 0.833±0.046 | |
| Soft Voting Classifier | 0.921±0.016 | 0.837±0.023 | 0.837±0.023 | 0.837±0.023 | 0.890±0.040 | 0.820±0.046 | 0.822±0.046 | 0.820±0.046 | |
Data are shown as performance value ± 95% confidence interval. ML, machine learning; AUC, area under the receiver operating characteristic curve.
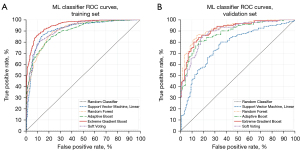
Looking at the internal validation test results, XGB again had higher accuracy in all calculated metrics (P<0.05) but was lower than RF, which performed the strongest. RF validation results were found to be slightly higher than the training results in all metrics. ADA performed very similarly to its training set results. The soft voting classifier performance worse compared to its training set results but remained comparable to the top performing XGB and RF. SVM performed significantly worse during its validation set when compared to its training set results, presenting the largest generalization gap seen from the ML algorithms. Validation set AUC curves are shown in Figure 5B.
The performance metrics of our radiomics-based models were also superior to the same models trained on first-order statistics only. Both XGB balanced accuracy (0.752±0.027, P<0.05) and AUC (0.843±0.023, P<0.05) dropped in performance significantly when compared to using radiomics data. Similar decreases were also found in RF balanced accuracy (0.753±0.026, P<0.05) and AUC (0.833±0.024, P<0.05).
Discussion
The goal of this study was to develop a DWI-based feature set and ML pipeline that can differentiate between metastatic and healthy brain tissue prior to their manifestation on standard T1 imaging. We trained multiple ML classifiers on longitudinal ADC maps taken pre-SRS. The radiomic and clinical data set, feature engineering methodology, and pipeline presented demonstrated that our ML models can identify metastases with an accuracy of 87.7% on the training set, and 85.8% on an unseen test set. This is in line with a previous systematic review reporting ML classification results between 84–93% (35). RF and XGB classifiers were the top performers in all ML metrics analyzed. While ADA performed comparably, training times were an order of magnitude longer (11.3 minutes for ADA versus <2 minutes for all other models). SVM displayed comparable training metrics but did not generalize well. The main features that contributed to our model accuracy included the change in ADC-based radiomic values months before the actual SRS treatment, suggesting that ML can be used to detect subtle diffusion changes prior to visual confirmation of metastases on conventional Gd-T1 MRI.
Changing ADC values have been shown to relate to increasingly restricted diffusion and increased hyper-cellularity or cell density of cancerous tissue (36,37). This is due to decreased mobility of water molecules in the extracellular space (38,39). Our previous work (40) built the foundation for this current study by showing that significant changes in first-order ADC metrics within metastatic regions occur months prior to their manifestation on Gd-T1. This supports other previous studies that have also suggested first-order statistics of diffusion imaging such as mean, median, and percentiles, can be valuable for studying brain metastases (41-44). The multiple differing conclusions regarding which ADC metric is best to use however implies that the exact relationship between metastasis formation and diffusion is unclear. First order statistics may fail to capture the structural complexity of malignant tissue, such as local voxel patterns and heterogeneity. In this study our processing pipeline and models accounted for this variation by allowing all these first-order metrics, and additional higher-order statistics, to play a role in the final calculation model. The significant difference in performance that we saw between the first-order only dataset and our radiomics dataset supports our methodology. The results suggest that changes in diffusion metrics are a key indicator for future metastatic occurrence given the large number of ‘change’ metrics that ranked highly in future importance across the ML models.
The main areas of interest for ML in radiology are image segmentation, registration, and computer-aided detection and diagnosis. ML has greatly risen in popularity, with modeling used to process the vast amount of imaging data taken in clinical practice (45). The idea of differentiating brain tissue using ML-aided detection was explored in our study, and has been investigated previously using other characterizations, such as between metastases and glioblastoma (8). Another example is a recent study demonstrating ML models achieving greater specificity compared to neuroradiologists when distinguishing radionecrosis from non-necrotic tissue post-SRS (9). While other ML studies often use modalities such as CT, T1, and even DWI as input data to their models, our methodology uniquely utilized extensive longitudinal ADC maps for radiomic data analysis. This allowed us to augment our feature set with the change in diffusion metrics over time, as opposed to single instance radiomic measurements. Finally, particular attention was given to improve ADC stability (36,46) and generalization of the trained models through normalization of ADC maps and multipoint regression analysis of radiomic features.
We acknowledge several limitations with the study. Firstly, the imaging dataset used was limited to a single institution and may generalize better if multi-institutional ADC maps from a heterogenous dataset were integrated into the ML model input. Secondly, the patients chosen in this study all underwent SRS to treat cranial metastases, with controls generated from those same patients due to a lack of longitudinal DWI data from healthy patients. The lack of an external validation group may restrict the results to SRS patients and limit our models’ usage in other patient demographics. Lastly, there may be more clinical features that are missing from our feature set. The subgroup analysis within different therapies and primaries is an interesting topic but was not pursued in this study due to the large sample size required for each group, instead we tried to incorporate as many relevant patient and therapy features that may potentially impact ADC, e.g., chemotherapy & immunotherapy into the analysis.
Results of this study indicate that longitudinal DWI-based ML models can be used as a tool for early brain metastases detection. Future work will explore extending this model for generalized search across the brain for high-risk patients. Integration of such a tool would require minimal additional resources as DWI is commonly included in standard imaging protocols for SRS patients. Exporting the calculated ADC maps and going through our proposed processing pipeline, whether in a standalone application or integrated into existing treatment planning software, can be done in parallel to current standards and would serve as an additional source of information for the physician. If the model identifies areas at risk of developing metastases due to diffusion changes, closer surveillance with shorter interval imaging can be considered. Conversely, if no high-risk areas are noted, less frequent imaging follow-up may be appropriate, allowing departmental resources to be deployed according to patients’ underlying risk of recurrence.
Conclusions
Longitudinal diffusion-based ML models were trained to accurately differentiate intracranial metastatic tissue from healthy brain tissue. Given the complexity of diffusion changes within the brain, ADC-based radiomics provided the necessary data for training the model, with XGBoost and RF classifiers providing the best predictive power. The main features that contributed to our model accuracy primarily were changes in diffusion metrics months before the actual SRS treatment of the metastasis and detection on Gd-T1. Although ML in the cancer clinic is still being investigated, our results and methodology using longitudinal diffusion-based radiomics opens the door for the proactive screening and early detection of future cranial metastases using artificial intelligence. Future work will focus on clinical integration of the model as a diagnostic aid.
Acknowledgments
This work was presented during the European Society for Therapeutic Radiology and Oncology (ESTRO) 2022 Conference in Copenhagen, Denmark: Joseph M, Ekaterina T, Nicolas P. PO-1762: Early detection of brain metastases using diffusion weighted imaging radiomics and machine learning. ESTRO Congress 2022 [Internet]. 2022 May. Available from: https://www.estro.org/Congresses/ESTRO-2022/661/radiomics-modellingandstatisticalmethods/11369/earlydetectionofbrainmetastasesusingdiffusionweigh
Funding: This work was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-441/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-441/coif). ET and NP have received a grant from the Brain Tumour Foundation of Canada to support this work. SF works in an education and advisory position with Sanofi. JM received funding from Alberta Innovates: Data-Enabled Innovation grant and the Brain Tumour Foundation of Canada. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Health Research Ethics Board of Alberta Cancer Committee (Ethics ID HREBA.CC-20-0041), and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Brabletz T, Lyden D, Steeg PS, Werb Z. Roadblocks to translational advances on metastasis research. Nat Med 2013;19:1104-9. [Crossref] [PubMed]
- Wong J, Hird A, Kirou-Mauro A, Napolskikh J, Chow E. Quality of life in brain metastases radiation trials: a literature review. Curr Oncol 2008;15:25-45.
- Jenkinson MD, Haylock B, Shenoy A, Husband D, Javadpour M. Management of cerebral metastasis: evidence-based approach for surgery, stereotactic radiosurgery and radiotherapy. Eur J Cancer 2011;47:649-55. [Crossref] [PubMed]
- Samson K. Benefits of Stereotactic Brain Radiosurgery Demonstrated in Phase III Trial. Oncology Times 2016;38:32-42.
- Baliyan V, Das CJ, Sharma R, Gupta AK. Diffusion weighted imaging: Technique and applications. World J Radiol 2016;8:785-98. [Crossref] [PubMed]
- Bishop CM. Pattern recognition and machine learning. New York: Springer; 2006.
- Hastie T. The elements of statistical learning data mining, inference, and prediction. New York: Springer New York; 2009:757.
- Qian Z, Li Y, Wang Y, Li L, Li R, Wang K, Li S, Tang K, Zhang C, Fan X, Chen B, Li W. Differentiation of glioblastoma from solitary brain metastases using radiomic machine-learning classifiers. Cancer Lett 2019;451:128-35. [Crossref] [PubMed]
- Peng L, Parekh V, Huang P, Lin DD, Sheikh K, Baker B, et al. Distinguishing True Progression From Radionecrosis After Stereotactic Radiation Therapy for Brain Metastases With Machine Learning and Radiomics. Int J Radiat Oncol Biol Phys 2018;102:1236-43. [Crossref] [PubMed]
- Karami E, Soliman H, Ruschin M, Sahgal A, Myrehaug S, Tseng CL, Czarnota GJ, Jabehdar-Maralani P, Chugh B, Lau A, Stanisz GJ, Sadeghi-Naini A. Quantitative MRI Biomarkers of Stereotactic Radiotherapy Outcome in Brain Metastasis. Sci Rep 2019;9:19830. [Crossref] [PubMed]
- Vidić I, Egnell L, Jerome NP, Teruel JR, Sjøbakk TE, Østlie A, Fjøsne HE, Bathen TF, Goa PE. Support vector machine for breast cancer classification using diffusion-weighted MRI histogram features: Preliminary study. J Magn Reson Imaging 2018;47:1205-16. [Crossref] [PubMed]
- Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med 2015;13:1.
- Conrad B. blairconrad/dicognito [Internet]. 2022 [accessed 2022 Apr 20]. Available online: https://github.com/blairconrad/dicognito
- Li X, Morgan PS, Ashburner J, Smith J, Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods 2016;264:47-56. [Crossref] [PubMed]
- rordenlab/dcm2niix [Internet]. Chris Rorden’s Lab; 2022 [accessed 2022 Apr 20]. Available online: https://github.com/rordenlab/dcm2niix
- Phil T. Sikerdebaard/dcmrtstruct2nii: v1.0.19 [Internet]. Zenodo; 2020 [accessed 2022 Apr 20]. Available online: https://zenodo.org/record/4037865
- Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 2004;23:S208-19. [Crossref] [PubMed]
- Smith SM. Fast robust automated brain extraction. Hum Brain Mapp 2002;17:143-55. [Crossref] [PubMed]
- Zhang Y, Brady M, Smith S. Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 2001;20:45-57. [Crossref] [PubMed]
- Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal 2001;5:143-56. [Crossref] [PubMed]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 2002;17:825-41. [Crossref] [PubMed]
- Greve DN, Fischl B. Accurate and robust brain image alignment using boundary-based registration. Neuroimage 2009;48:63-72. [Crossref] [PubMed]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 2001;356:1293-322. [Crossref] [PubMed]
- Terada Y, Toda H, Okumura R, Ikeda N, Yuba Y, Katayama T, Iwasaki K. Reticular Appearance on Gadolinium-enhanced T1- and Diffusion-weighted MRI, and Low Apparent Diffusion Coefficient Values in Microcystic Meningioma Cysts. Clin Neuroradiol 2018;28:109-15. [Crossref] [PubMed]
- Oladosu O, Liu WQ, Pike BG, Koch M, Metz LM, Zhang Y. Advanced Analysis of Diffusion Tensor Imaging Along With Machine Learning Provides New Sensitive Measures of Tissue Pathology and Intra-Lesion Activity in Multiple Sclerosis. Front Neurosci 2021;15:634063. [Crossref] [PubMed]
- Horváth A, Perlaki G, Tóth A, Orsi G, Nagy S, Dóczi T, Horváth Z, Bogner P. Increased diffusion in the normal appearing white matter of brain tumor patients: is this just tumor infiltration? J Neurooncol 2016;127:83-90. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3-D model-based neuroanatomical segmentation. Human Brain Mapping 1995;3:190-208.
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 2011;12:2825-30.
- Breiman L. Random Forests. Machine Learning 2001;45:5-32. [Crossref] [PubMed]
- Cortes C, Vapnik V. Support-vector networks. Machine Learning 1995;20:273-97.
- Schapire RE. Explaining adaboost. In: Empirical Inference. Berlin Heidelberg: Springer; 2013:37-52.
- Chen T, Guestrin C. XGBoost: A Scalable Tree Boosting System. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining 2016;785-94.
- Efron B. Bootstrap Methods: Another Look at the Jackknife. The Annals of Statistics 1979;7:1-26.
- Cho SJ, Sunwoo L, Baik SH, Bae YJ, Choi BS, Kim JH. Brain metastasis detection using machine learning: a systematic review and meta-analysis. Neuro Oncol 2021;23:214-25. [Crossref] [PubMed]
- Peerlings J, Woodruff HC, Winfield JM, Ibrahim A, Van Beers BE, Heerschap A, Jackson A, Wildberger JE, Mottaghy FM, DeSouza NM, Lambin P. Stability of radiomics features in apparent diffusion coefficient maps from a multi-centre test-retest trial. Sci Rep 2019;9:4800. [Crossref] [PubMed]
- Matsushima N, Maeda M, Takamura M, Takeda K. Apparent diffusion coefficients of benign and malignant salivary gland tumors. Comparison to histopathological findings. J Neuroradiol 2007;34:183-9. [Crossref] [PubMed]
- Lee EK, Lee EJ, Kim MS, Park HJ, Park NH, Park S 2nd, Lee YS. Intracranial metastases: spectrum of MR imaging findings. Acta Radiol 2012;53:1173-85. [Crossref] [PubMed]
- Berghoff AS, Spanberger T, Ilhan-Mutlu A, Magerle M, Hutterer M, Woehrer A, Hackl M, Widhalm G, Dieckmann K, Marosi C, Birner P, Prayer D, Preusser M. Preoperative diffusion-weighted imaging of single brain metastases correlates with patient survival times. PLoS One 2013;8:e55464. [Crossref] [PubMed]
- Madamesila J, Ploquin N, Faruqi S, Tchistiakova E. Investigating diffusion patterns of brain metastases pre- and post-stereotactic radiosurgery: a feasibility study. Biomed Phys Eng Express 2021; [Crossref]
- Wang S, Summers RM. Machine learning and radiology. Med Image Anal 2012;16:933-51. [Crossref] [PubMed]
- Bozdağ M, Er A, Çinkooğlu A. Histogram Analysis of ADC Maps for Differentiating Brain Metastases From Different Histological Types of Lung Cancers. Can Assoc Radiol J 2021;72:271-8. [Crossref] [PubMed]
- Lee SL, Ravi A, Morton G, Loblaw A, Tseng CL, Haider M, Murgic J, Nicolae A, Semple M, Chung HT. Changes in ADC and T2-weighted MRI-derived radiomic features in patients treated with focal salvage HDR prostate brachytherapy for local recurrence after previous external-beam radiotherapy. Brachytherapy 2019;18:567-73. [Crossref] [PubMed]
- Weiss E, Ford JC, Olsen KM, Karki K, Saraiya S, Groves R, Hugo GD. Apparent diffusion coefficient (ADC) change on repeated diffusion-weighted magnetic resonance imaging during radiochemotherapy for non-small cell lung cancer: A pilot study. Lung Cancer 2016;96:113-9. [Crossref] [PubMed]
- Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, Allison T, Arnaout O, Abbosh C, Dunn IF, Mak RH, Tamimi RM, Tempany CM, Swanton C, Hoffmann U, Schwartz LH, Gillies RJ, Huang RY, Aerts HJWL. Artificial intelligence in cancer imaging: Clinical challenges and applications. CA Cancer J Clin 2019;69:127-57. [Crossref] [PubMed]
- Merisaari H, Taimen P, Shiradkar R, Ettala O, Pesola M, Saunavaara J, Boström PJ, Madabhushi A, Aronen HJ, Jambor I. Repeatability of radiomics and machine learning for DWI: Short-term repeatability study of 112 patients with prostate cancer. Magn Reson Med 2020;83:2293-309. [Crossref] [PubMed]


