
Development and validation of a fully automated system using deep learning for opportunistic osteoporosis screening using low-dose computed tomography scans
Introduction
Osteoporosis is a systemic disease characterized by a reduction in bone mineral density (BMD) and a systemic loss of microarchitecture, which leads to fragility fractures (1). Osteoporosis has become a common disease in the elderly population, affecting more than 200 million people worldwide (2). As osteoporosis is asymptomatic in its early stages, early diagnosis plays a vital role in preventing osteoporotic fractures caused by this disease (3).
BMD measurement is one of the main bases for the diagnosis of osteoporosis. Dual-energy X-ray is considered a standard technique for measuring BMD (4). Quantitative computed tomography (QCT) is another clinically validated method for measuring BMD which can also provide structural information (5). QCT provides more accurate BMD measurements and unlike dual-energy X-ray absorptiometry (DXA), it is not susceptible to factors such as severe degenerative changes in the hip or spine, vascular calcification, oral contrast, and certain foods or dietary supplements containing high levels of heavy minerals and elements (6-8). However, QCT still has several limitations, such as the need for dedicated software and phantom, fewer well-trained technicians, and the frequent need for calibration. In addition, even with standard scan protocols that typically include L1 and L2, the radiation dose in QCT is significantly higher than in DXA (9).
Nowadays, low-dose computed tomography (LDCT) is recommended in emphysema detecting, lung cancer screening, hepatic steatosis evaluation, and so on (10-12). Based on this, chest combined with upper abdomen LDCT scan is frequently used in physical checkups. As several studies have supported the viability of low-radiation-dose QCT in BMD measurement (13,14), the images obtained in the LDCT scans covering the L1 and L2 spine can be used for the BMD measurement using the dedicated QCT software without additional cost and exposure (15).
According to numerous studies, BMD is calculated using computed tomography (CT) values of cancellous bone obtained from standard CT scans, with a correlation coefficient between 0.399 and 0.891 (16). However, in addition to the different internal vertebral body characteristics, their CT values also depend on scanner characteristics, X-ray tube voltage, and other factors (17). Therefore, traditional CT scans are only partially effective in the diagnosis of osteoporosis because CT results obtained under different scanner models require calibration.
The deep learning (DL) technique has been successfully used for the analysis of radiological images, such as diagnosis of diseases (18), image processing (19-21), and parameter analysis (22-24). Numerous studies have also applied DL in the area of BMD measurement. Yasaka et al. (25) developed a convolutional neural network (CNN) model to predict the BMD obtained from DXA using unenhanced abdominal CT images. Pan et al. (26) developed a 3-dimensional (3D) segmentation model and a CT value based 1-degree linear function to automatically measure BMD using low dose chest CT scans. Fang et al. (27) presented a system combined with 2-dimensional (2D) U-Net for segmentation and DenseNet-121 for BMD calculation using sagittal images from standard spinal or abdominal CT scans.
In this study, we developed and validated a DL system using several 3D networks to automatically locate and segment the cancellous bones of T12, L1, and L2 vertebral bodies and measure BMD for opportunistic osteoporosis screening using LDCT scans. This was the first study to use a 3D DL system instead of 2D models for BMD measurements in LDCT scans without the need for calibration function. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1438/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the First Affiliated Hospital of Xi’an Jiaotong University and the requirement for individual consent for this retrospective analysis was waived.
Study population
Data were collected from general population who underwent chest and upper abdominal LDCT examination in the Department of Radiology at the First Affiliated Hospital of Xi’an Jiaotong University between April 2018 and July 2021. All data were anonymized before use.
The inclusion criteria were as follows:
- having undergone LDCT examination of the chest combined and upper abdomen, with scan range from the apical lung to the lower edge of the liver;
- willingness to participate in this clinical study.
The exclusion criteria were as follows:
- images could not be used for QCT analysis;
- the presence of a metal or bone cement implant;
- the target vertebrae had not been completely covered in the CT examination;
- secondary osteoporosis, such as osteoporosis caused by renal failure, diabetes, and hyperparathyroidism;
- Schmorl’s nodes, severe scoliosis, and vascular calcification.
Finally, 500 individuals from the general population (mean age 66.03±9.71 years, range 39 to 90 years, mean height 170.82±6.42 cm, mean weight 71.71±10.32 kg) were enrolled in this study; 121 were diagnosed with osteoporosis and 186 were diagnosed with osteopenia. Due to the limited computational power, we were limited to the number of enrollments, but were certain that the data for the final 500 cases enrolled were completely random and unselected. There were no missing data in this study. According to the size of the dataset, the 500 participants were randomly divided into a training set, a validation set, and a test set at a ratio of 6:2:2 using a 4-fold cross validation method to obtain more accurate model results from training. Figure 1 shows the illustration of our dataset and the 4-fold cross validation method.
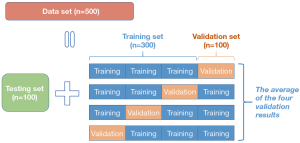
Image acquisition and reconstruction
All individuals underwent a plain CT scan on a 256-rows detector CT system (Revolution CT; GE, Waukesha, WI, USA) from the apical lung to the lower edge of the liver using the following scan parameters: 120 kVp, SmartmA (10–130 mA), gantry rotation time of 0.5 seconds per rotation, helical pitch of 0.992:1 to obtain a preset noise index of 16. The SmartmA is an automatic exposure control system that uses z-axis and angular tube current modulation to customize the dose to the size and shape of an individual. The range 10–130 mA was determined by the noise index set to 16. Noise index allows the user to select the amount of noise that will be present in the reconstructed images; before scanning, the user defines the desired image quality (noise index). A higher noise index is related with a lower milliampere-second, and as a result, a lower radiation dosage. The average volume CT dose index (CTDIvol) for the patient population in our study was low at 6.54±2.53 mGy.
Images were reconstructed with 512×512 reconstruction matrix and 1.25-mm slice thickness and interval using a standard reconstruction kernel.
QCT image post-processing
All CT images were post-processed by QCT Pro Model 4 (Mindways Software, Inc., Austin, TX, USA). The quality control analysis was performed by using a unified European Spine Phantom (ESP; No. 145, QRM GmbH, Möhrendorf, Germany). The central layer of the vertebral body was selected to calculate the average bone density value.
Although there are no consensus standards for diagnosing osteoporosis based on spine QCT measurements, some institutions provide a diagnostic category that approximately equates to the requirements of the World Health Organization (WHO) guidelines. Our diagnosis process was performed according to the guidelines introduced by the International Society for Clinical Densitometry (ISCD) and American College of Radiology (ACR): osteoporosis, if BMD values are below 80 mg/cm3; osteopenia, if BMD values are from 80 to 120 mg/cm3; and normal, if BMD values are over 120 mg/cm3 (28,29). In our study, diagnosis was made using the absolute lumbar QCT BMD values, averaged over the cancellous bone density of 2 lumbar vertebrae (commonly the L1 and L2 vertebrae, with T12 vertebra substituted if either L1 or L2 was non-measurable).
Development of the BMD measurement system
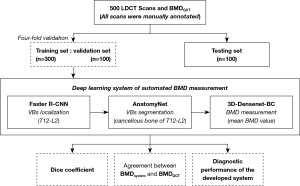
The development of this automated BMD measuring system consisted of the following 3 main stages. First, train a localization model to identify and locate the target vertebrae (T12–L2) in the region for coarse segmentation. Second, train a segmentation model to finely segment the cancellous bone of target vertebrae in the localized image. Finally, train a DenseNet based classification model for BMD measurement, the output of which is the mean BMD value. All models in this study are from open-source programs. Figure 2 shows the overall process of development and validation of this system.
Vertebra localization
Image preprocessing
This model needs to convert axial CT images into maximum intensity projection (MIP) images, which can be used to better show the location of target vertebrae (T12, L1, L2).
The image preprocessing consisted of the following 3 steps. First, select the maximum density projection map formed by the rays perpendicular to the coronal plane direction, where the horizontal coordinates correspond to the coordinates of the X-axis in the original CT data and the vertical coordinates correspond to the coordinates of the Z-axis. Second, use the labeling tool to label the 3 vertebrae in the generated maximum density projection map as a whole. The label was determined by a radiologist after appropriate widening from the edges of the annotated data. Third, divide data into a training set, a validation set, and a test set.
Development of a localization model
The localization model was mainly constructed to obtain the upper and lower coordinates of the target vertebrae in the Z-axis direction in the original CT data. The object detection algorithm chosen was the faster region-based convolutional neural network (R-CNN), which mainly consists of Fast R-CNN (Facebook AI Research) and Region Proposal Network (RPN) (30). RPN mainly provides region proposal information, whereas Fast R-CNN is mainly responsible for extracting input image feature-maps, receiving input image features from RPN input, and unifying the map size and outputting the class of the target object as well as the position information.
Vertebra segmentation
Image preprocessing
All 500 LDCT scans were manually annotated by a radiologist with 3 years of experience in CT imaging for the contours of target vertebrae, who was blinded to the diagnosis, to develop the segmentation model. Another radiologist with 5 years of experience performed a visual examination before inputting segmentation data into the model. The location of the 3 vertebrae in the cropped CT data were obtained by the localizing model, of which the X-, Y-, and Z-axes ranged from (180 to 330), (200 to 420), and (ZL-5 to ZU+5), respectively, where the ZL and ZU roots were based on the upper and lower positions of the three vertebral bodies in the Z-axis direction provided by the localizing model. All cropped data were processed using a window level of 0 Hounsfield units (HU) and window width of 400 HU. All CT values in the data were scaled to (0, 1) using the min-max normalization method. The size of the input segmentation model was set to 150×220×120. If the size of the cropped data was smaller than this set size, then the input condition was met by up- and down-complementing by 0.
Development of segmentation model
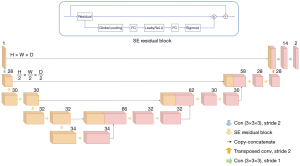
The segmentation model chosen was the 3D AnatomyNet model (Department of Computer Science, University of California, Irvine, CA, USA) (31), a variant of U-Net as can be seen from the structure picture. The purpose of using only 1 down-sampling layer in the encoder part was to occupy less GPU memory and improve the ability to segment small regions; secondly, the Squeeze-and-Excitation residual block was used instead of the previous convolution operation as the building block of AnatomyNet, which is equivalent to introducing an attention mechanism between channels, so that the interdependencies between channels can be adaptively modelled and calibrated, and therefore better able to extract effective features and increase the representational power of the network. The structure of AnatomyNet is shown in Figure 3.
BMD calculation
In this study, a 3D DenseNet model (Cornell University, Tsinghua University, and Facebook AI Research) for the calculation of averaged BMD value was constructed. The uniformly sized images obtained from the segmentation model were randomly divided into a training set, a validation set, and a test set in a ratio of 6:2:2 and used as the dataset for the input of BMD calculation model.
DenseNet-BC uses dense blocks that connect the entire layers. Each layer obtains inputs from previous layers and passes via its own feature maps to preserve the accuracy of classifier.
As shown in Figure 4, this detection model was a 121-layer 3D-DenseNet-BC with the following structure of the network in order: initial convolutional layer, max-pooling layer, dense block 1, transition layer 1, dense block 2, transition layer 2, dense block 3, transition layer 3, dense block 4, global average pooling layer, and fully connected layer.
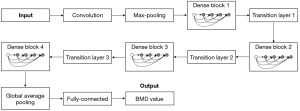
The Initial convolution layer consists of a convolution kernel of step stride 2, padding 3 and size 7×7×7, followed by a batch normalization layer and a rectified linear unit (ReLU) activation function. The maximum pooling layer consisted of a pooling kernel with a step stride of 2, a padding of 1, and a size of 3×3×3. The numbers of bottleneck layers for dense block 1 to dense block 4 were 6, 12, 24, and 16, respectively. The overall network structure of each bottleneck layer was in the following order: BN-ReLU-Conv(1×1×1)-BN. The overall network structure of each bottleneck layer was as follows: BN-ReLU-Conv(1×1×1)-BN-ReLU-Conv(3×3×3). Each transition layer connects 2 adjacent dense blocks. Each transition layer contains in turn a BN layer, a ReLU layer, a convolution layer, and an average pooling layer. A compression factor was added to the transition layer, which was set to 0.5 in the described method. In the convolution layer, the step size was 1 and the kernel size was 1×1×1. In the pooling layer, the step size was 2 and the kernel size was 2×2×2. Global average pooling was performed after dense block 4. The output feature map size was specified as 1×1×1. More specific structure and definition of models in this system are shown in Appendix 1.
Statistical analysis
The software SPSS 22.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. The Dice coefficient, which effectively quantifies the spatial overlap between segmentation and ground truth, was used to evaluate segmentation performance. It is calculated by taking twice the number of elements that are common to both the segmentation set and ground truth set and dividing it by the sum of the number of elements in each set. The normality of all continuous variables was verified. To compare the BMD values predicted by the proposed system with the ground truth obtained by QCT, linear regression and Bland–Altman (BA) analyses were performed. To evaluate the diagnostic performance of the proposed system for predicting osteoporosis and abnormal BMD (osteoporosis and osteopenia), the area under the curve (AUC) from receiver operating characteristic (ROC) curve analysis and the sensitivity, specificity, positive and negative predictive values were calculated. All analyses were 2-tailed and P<0.05 was considered statistically significant.
The training process was run on a Xeon E5-2664 v4 Gold (Intel, Santa Clara, CA, USA) CPU 3.20 GHz with NVIDIA (Santa Clara, CA, USA) Tesla P100 GPU.
Results
Vertebra localization
Manual inspection was performed to check if the targeted vertebrae were correctly localized by Faster R-CNN. We also used Recall and Precision to measure the probability of target retrieval and the probability of correct target classification. The results showed that the overall positioning effect for 3 vertebral bodies was very good, with both recall and accuracy reaching 100%, indicating that 3 vertebral cancellous bones could be found on each image.
Vertebra segmentation
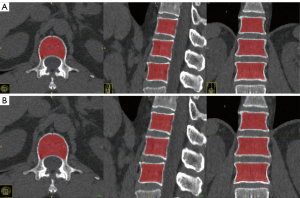
The proposed method achieved a mean Dice coefficient of 0.95 for vertebrae segmentation on the testing dataset, with Dice coefficients greater than 0.90 accounting for 96.6%. The average Dice coefficient for the validation set was 0.95, with 95% having a Dice coefficient greater than 0.9. In the training set, the average Dice coefficient for the segmentation evaluation metrics was 0.952, with 96.9% having a Dice coefficient greater than 0.9. Figure 5 shows an example of the segmentation results.
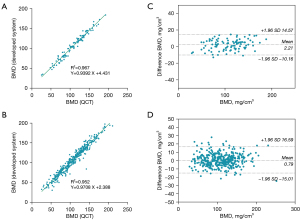
The performance of the proposed system in the measurement of mean BMD
The performance of the proposed system in the measurement of mean BMD is shown in Figure 6. Linear regression analysis showed a strong correlation between the proposed system and QCT for the measurement of mean BMD, with a correlation coefficient (R2) of 0.967 in the testing set and 0.952 in the validation set. The slopes of linear regressions using the proposed system and QCT were also close to 1, showing that the proposed system could measure vertebral BMD accurately. Additionally, BA analysis showed good agreement between the proposed system and QCT for mean BMD measurement. Compared with the results of QCT, BMD measurement by the proposed system generated mean bias of 2.21 and 0.79 mg/cm3 in the testing and validation set, respectively. The 95% limits of agreement were (−10.16, 14.57) mg/cm3 in the testing set and (−15.01, 16.59) mg/cm3 in the validation set.
Diagnostic performance of the proposed system for osteoporosis and osteopenia diagnosis
The diagnostic performance of the proposed system for detecting osteoporosis and abnormal BMD is shown in Table 1. The AUC of the proposed system for detecting osteoporosis was 0.987 in the validation set and 0.984 in the testing set. The AUC for distinguishing abnormal BMD (osteopenia and osteoporosis) from normal bone mass was 0.984 in the validation set and 0.993 in the testing set. For detection of osteoporosis, the proposed system achieved 98.99% in sensitivity and 92.36% in specificity in the validation set, and 95.45% in sensitivity and 91.03% in specificity in the testing set. The sensitivity and specificity of the proposed system for distinguishing abnormal BMD from normal bone mass were 95.98% and 92.72% in the validation set and 96.55% and 100.00% in the testing set, respectively.
Table 1
| Diagnosis | AUC (95% CI) | Sensitivity (n/N) | Specificity (n/N) | PPV (n/N) | NPV (n/N) |
|---|---|---|---|---|---|
| Validation set | |||||
| Osteoporosis | 0.987 (0.980–0.995) | 98.99% (98/99) | 92.36% (278/301) | 80.99% (98/121) | 99.64% (278/279) |
| Abnormal BMD (osteopenia and osteoporosis) | 0.984 (0.974–0.994) | 95.98% (239/249) | 92.72% (140/151) | 95.60% (239/250) | 93.33% (140/150) |
| Testing set | |||||
| Osteoporosis | 0.984 (0.965–1) | 95.45% (21/22) | 91.03% (71/78) | 75.00% (21/28) | 98.60% (71/72) |
| Abnormal BMD (osteopenia and osteoporosis) | 0.993 (0.982–1) | 96.55% (56/58) | 100.00% (42/42) | 100.00% (56/56) | 95.45% (42/44) |
QCT, quantitative computer tomography; AUC, area under the curve; CI, confidence interval; PPV, positive predictive value; NPV, negative predictive value; BMD, bone mineral density.
Discussion
In this study, we developed a 3D deep neural network model to automate BMD measurement for detecting osteoporosis and osteopenia using chest and abdomen LDCT scans. To our knowledge, this was the first study to automate BMD measurement in LDCT scans using a 3D DL system.
Vertebrae localization and segmentation in LDCT scans are the basis of automating BMD measurement. In this study, we used a Fast-RCNN network trained by MIP images to localize the target vertebral bodies. Besides, we employed 3D-AnatomyNet to conduct segmentation of the cancellous bone of the target vertebrae (T12–L2). The average Dice coefficients of segmentation on the testing and validation sets were both 0.95, demonstrating that the automated segmentation was highly associated with manual annotation. Additionally, over 95% cases had a Dice coefficient greater than 0.9.
We selected QCT as a reference standard for BMD measurement instead of DXA of the lumbar spine because QCT might give a more accurate evaluation of the performance of our proposed system. Although DXA is the most commonly used technology for diagnosing osteoporosis and performing serial BMD measurements, it is susceptible to abdominal aortic calcification and spinal degeneration. QCT is also a reliable indicator for measuring spinal BMD. It is a 3D metric that only measures the density of the cancellous bone of the vertebra. Unlike DXA, its measurement area does not contain cortical bone. However, QCT still requires frequent calibration, dedicated software, and complex post-processing work. Therefore, the application of this proposed 3D DL system may help to simplify the procedure of BMD measurement, thus making osteoporosis screening more easily accessible.
Previous studies (25-27) have mainly used 2D networks or linear function to calculate BMD values, meaning that the results are not representative of all anatomical information within cancellous bone or are affected by the instability of CT values. The methods used in our system are based on several leading 3D DL models, which use a classification model instead of converting CT values to BMD values, allowing the system to be independent of the linear relationship between CT values and BMD values in clinical use, thus achieving reliable results without regular calibration. At the same time, we segmented all cancellous parts of the vertebral body, thus avoiding the problems caused by manual selection of regions of interest (ROIs). Following the ISCD and ACR’s recommendations, the averaged L1 and L2 BMD (with T12 vertebra substituted if either L1 or L2 is non-measurable) was used as the diagnosis metric of our proposed system. Our results demonstrated that the BMD values measured by our system had a strong statistically significant correlation with the values acquired by QCT. The correlation coefficient (R2) between our system and QCT reached 0.967 in the testing set. For diagnostic performance, the testing set of our proposed system achieved 95.45% in sensitivity and 91.03% in specificity for diagnosing osteoporosis, and 96.55% in sensitivity and 100.00% in specificity for distinguishing abnormal bone mass (osteopenia and osteoporosis) from normal bone mass using the images in LDCT scans. Compared to the results of a previous study (26), our system had a better sensitivity as well as a higher AUC for the diagnosis of osteoporosis. In addition, the computational time to process a case in this model was only about 8.46 seconds on our server, making it possible to use it in a clinical situation. To reduce the potential risk associated with the radiation dose received by the patient, the LDCT data used in our study had a mean effective CT dose of 4.61±1.98 mSv and, for comparison, the normal dose for a routine application is approximately 9 mSv for the same scan range.
This paper still has several limitations. Firstly, our model was developed and evaluated using data from a single institution and a single scanner. As a result, more studies using data from several other institutions are needed to demonstrate the generalizability of the results, whereas adjustment of the training set with large amounts of data from different manufacturers’ sources at different institutions is needed to increase the reproducibility of this model. The segmentation data was annotated by 1 radiologist, and although a visual examination was conducted by another radiologist, it might still cause bias. The ROI mismatch between QCT and the developed system is also a limitation in this study, because this can cause make direct comparison of the results of the training model and QCT difficult.
In conclusion, the proposed 3D DL-based system enables automatic vertebrae segmentation and BMD calculation using LDCT images of the thorax and upper abdomen and provides high accuracy for diagnosing osteoporosis and osteopenia, meaning that this DL-based system extends the utility of LDCT scans beyond their intended routine clinical purpose and makes osteoporosis screening more accessible. This DL-based system extends the scope of purpose of LDCT scans beyond their intended routine clinical use and has potential to become a clinically available osteoporosis screening tool.
Conclusions
This study developed an automated DL-based system to measure BMD using LDCT scans for opportunistic osteoporosis screening with high accuracy.
Acknowledgments
Funding: This study received funding from the Key R&D Program of Shaanxi Province Universities and Colleges (No. 2020GXLH-Y-026).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-1438/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-1438/coif). JL is an employee of GE Healthcare, the manufacturer of the CT scanner used in this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of the First Affiliated Hospital of Xi’an Jiaotong University and the requirement for individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet 2011;377:1276-87. [Crossref] [PubMed]
- Aaseth J, Boivin G, Andersen O. Osteoporosis and trace elements--an overview. J Trace Elem Med Biol 2012;26:149-52. [Crossref] [PubMed]
- Kanis JA, Cooper C, Rizzoli R, Reginster JYScientific Advisory Board of the European Society for Clinical and Economic Aspects of Osteoporosis (ESCEO) and the Committees of Scientific Advisors and National Societies of the International Osteoporosis Foundation (IOF). Correction to: European guidance for the diagnosis and management of osteoporosis in postmenopausal women. Osteoporos Int 2020;31:801. [Crossref] [PubMed]
- Ott SM. Methods of determining bone mass. J Bone Miner Res 1991;6:S71-6; discussion S83-4. [Crossref] [PubMed]
- American College of Radiology. ACR practice parameter for performing and interpreting diagnostic computed tomography(ct). 2018; Available online: https://www.acr.org/-/media/ACR/Files/Practice-Parameters/QCT.pdf. Accessed October 13, 2020.
- Orwoll ES, Oviatt SK, Mann T. The impact of osteophytic and vascular calcifications on vertebral mineral density measurements in men. J Clin Endocrinol Metab 1990;70:1202-7. [Crossref] [PubMed]
- Liu G, Peacock M, Eilam O, Dorulla G, Braunstein E, Johnston CC. Effect of osteoarthritis in the lumbar spine and hip on bone mineral density and diagnosis of osteoporosis in elderly men and women. Osteoporos Int 1997;7:564-9. [Crossref] [PubMed]
- Yu EW, Thomas BJ, Brown JK, Finkelstein JS. Simulated increases in body fat and errors in bone mineral density measurements by DXA and QCT. J Bone Miner Res 2012;27:119-24. [Crossref] [PubMed]
- Engelke K, Adams JE, Armbrecht G, Augat P, Bogado CE, Bouxsein ML, Felsenberg D, Ito M, Prevrhal S, Hans DB, Lewiecki EM. Clinical use of quantitative computed tomography and peripheral quantitative computed tomography in the management of osteoporosis in adults: the 2007 ISCD Official Positions. J Clin Densitom 2008;11:123-62. [Crossref] [PubMed]
- de Torres JP, Bastarrika G, Wisnivesky JP, Alcaide AB, Campo A, Seijo LM, Pueyo JC, Villanueva A, Lozano MD, Montes U, Montuenga L, Zulueta JJ. Assessing the relationship between lung cancer risk and emphysema detected on low-dose CT of the chest. Chest 2007;132:1932-8. [Crossref] [PubMed]
- Diederich S, Wormanns D. Impact of low-dose CT on lung cancer screening. Lung Cancer 2004;45:S13-9. [Crossref] [PubMed]
- Boyce CJ, Pickhardt PJ, Kim DH, Taylor AJ, Winter TC, Bruce RJ, Lindstrom MJ, Hinshaw JL. Hepatic steatosis (fatty liver disease) in asymptomatic adults identified by unenhanced low-dose CT. AJR Am J Roentgenol 2010;194:623-8. [Crossref] [PubMed]
- Museyko O, Heinemann A, Krause M, Wulff B, Amling M, Püschel K, Glüer CC, Kalender W, Engelke K. A low-radiation exposure protocol for 3D QCT of the spine. Osteoporos Int 2014;25:983-92. [Crossref] [PubMed]
- Malekzadeh M, Abbasi-Rad S, Keyak JH, Nabil M, Asadi M, Mobini N, Naghdi P, Emadi H, Saligheh Rad H, Shiran MB. Liquid Calibration Phantoms in Ultra-Low-Dose QCT for the Assessment of Bone Mineral Density. J Clin Densitom 2020;23:108-16. [Crossref] [PubMed]
- Valentinitsch A, Trebeschi S, Kaesmacher J, Lorenz C, Löffler MT, Zimmer C, Baum T, Kirschke JS. Opportunistic osteoporosis screening in multi-detector CT images via local classification of textures. Osteoporos Int 2019;30:1275-85. [Crossref] [PubMed]
- Gausden EB, Nwachukwu BU, Schreiber JJ, Lorich DG, Lane JM. Opportunistic Use of CT Imaging for Osteoporosis Screening and Bone Density Assessment: A Qualitative Systematic Review. J Bone Joint Surg Am 2017;99:1580-90. [Crossref] [PubMed]
- Li F, Li D, Zhang Y. Influence of tube voltage on CT attenuation, radiation dose, and image quality: phantom study. Chinese Journal of Radiology 2013;458-461: [Crossref]
- Li J, Zhou Y, Wang P, Zhao H, Wang X, Tang N, Luan K. Deep transfer learning based on magnetic resonance imaging can improve the diagnosis of lymph node metastasis in patients with rectal cancer. Quant Imaging Med Surg 2021;11:2477-85. [Crossref] [PubMed]
- Hsieh J, Liu E, Nett B, Tang J, Thibault JB, Sahney S A new era of image reconstruction: TrueFidelity™. White Paper (JB68676XX), GE Healthcare 2019. Available online: https://www.gehealthcare.com.br/-/jssmedia/040dd213fa89463287155151fdb01922.pdf
- Cheng Y, Han Y, Li J, Fan G, Cao L, Li J, Jia X, Yang J, Guo J. Low-dose CT urography using deep learning image reconstruction: a prospective study for comparison with conventional CT urography. Br J Radiol 2021;94:20201291. [Crossref] [PubMed]
- Cao L, Liu X, Li J, Qu T, Chen L, Cheng Y, Hu J, Sun J, Guo J. A study of using a deep learning image reconstruction to improve the image quality of extremely low-dose contrast-enhanced abdominal CT for patients with hepatic lesions. Br J Radiol 2021;94:20201086. [Crossref] [PubMed]
- Yang H, Sun J, Yang L, Xu Z. A Unified Hyper-GAN Model for Unpaired Multi-contrast MR Image Translation. International Conference on Medical Image Computing and Computer-Assisted Intervention 2021. Springer, Cham: 2021:127-37.
10.1007/978-3-030-87199-4_12 10.1007/978-3-030-87199-4_12 - Zhao W, Huang X, Wang G, Guo J. PET/MR fusion texture analysis for the clinical outcome prediction in soft-tissue sarcoma. Cancer Imaging 2022;22:7. [Crossref] [PubMed]
- Liu L, Tang C, Li L, Chen P, Tan Y, Hu X, Chen K, Shang Y, Liu D, Liu H, Liu H, Nie F, Tian J, Zhao M, He W, Guo Y. Deep learning radiomics for focal liver lesions diagnosis on long-range contrast-enhanced ultrasound and clinical factors. Quant Imaging Med Surg 2022;12:3213-26. [Crossref] [PubMed]
- Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol 2020;30:3549-57. [Crossref] [PubMed]
- Pan Y, Shi D, Wang H, Chen T, Cui D, Cheng X, Lu Y. Automatic opportunistic osteoporosis screening using low-dose chest computed tomography scans obtained for lung cancer screening. Eur Radiol 2020;30:4107-16. [Crossref] [PubMed]
- Fang Y, Li W, Chen X, Chen K, Kang H, Yu P, Zhang R, Liao J, Hong G, Li S. Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. Eur Radiol 2021;31:1831-42. [Crossref] [PubMed]
- Zysset P, Qin L, Lang T, Khosla S, Leslie WD, Shepherd JA, Schousboe JT, Engelke K. Clinical Use of Quantitative Computed Tomography-Based Finite Element Analysis of the Hip and Spine in the Management of Osteoporosis in Adults: the 2015 ISCD Official Positions-Part II. J Clin Densitom 2015;18:359-92. [Crossref] [PubMed]
- American College of Radiology 2018. ACR–SPR–SSR practice parameter for the performance of musculoskeletal quantitative computed tomography (QCT). American College of Radiology,Reston. Available online: https://www. acr. org/-/media/ACR/Files/Practice-Parameters/QCT. pdf
- Guo Q, Liu L, Zhang C, Xu W, Jing W. Muti-scale Feature Fusion Network Based on Feature Pyramid Model. Chinese Journal of Engineering Mathematics 2020;37:521-30.
- Zhu W, Huang Y, Zeng L, Chen X, Liu Y, Qian Z, Du N, Fan W, Xie X. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med Phys 2019;46:576-89. [Crossref] [PubMed]


