
An artificial intelligence-assisted diagnosis modeling software (AIMS) platform based on medical images and machine learning: a development and validation study
Introduction
Computer-assisted analysis of medical images plays a key role in a variety of applications, such as population screening, diagnosis, treatment delivery, therapeutic evaluation, and auxiliary prognosis, across a broad range of medical fields (1-3). Benefiting from the ever-growing number of medical images, data-driven methods, such as radiomics (4) and deep learning (5), can achieve excellent performance by extracting (or learning) discriminative features from a massive set of medical images, facilitating human-level computer-assisted diagnosis systems for some specific tasks (6-9).
Radiomics, a machine learning-based methodology for medical image quantitative analysis, was first proposed in 2012 (4). Aerts et al. (10) subsequently made a breakthrough in decoding tumor phenotype by using radiomics and revealed its powerful ability in radiation oncology. Radiomics extracts high-throughput quantitative image features based on the hypothesis that radiologic phenotypes may reflect genetic alterations in carcinogenesis and tumor biology. The discriminative features represent the morphologic and textural changes of lesions that are associated with disease processes, which are considered to be noninvasive biomarkers capable of predicting the biologic behavior of the tumor (11,12). Compared with the visual assessments of medical images, radiomics supplies quantitative, objective, and comprehensive biomarkers for noninvasive diagnosis and prognosis (13). However, the current radiomics architecture still confronts users with several challenges: (I) building optimal radiomics models is labor intensive for radiologists and researchers since the workflow of radiomics modeling includes tedious steps, including image preprocessing, lesion labeling, feature extraction, feature selection, classifier training, evaluation, and performance comparison. These interrelated steps contain many candidate algorithms and their combinations. (II) The radiomics features are human-defined, which leads the majority of the high throughput features potentially lacking discriminative power for a specific task. This means that the performance of the radiomics model may depend highly on feature selection. (III) Unreliable segmentation may have an adverse effect on feature extraction and even lead to incorrect predictions in the radiomics analysis (14). Generally, radiomics modeling requires expertise in machine learning and programming ability in building a customized and optimal model for a specific application.
Deep learning based on a convolutional neural network (CNN) is another well-known machine learning paradigm and is currently being used in a core role within computer-assisted analysis systems due to its ability to perform feature self-learning from medical image datasets. The CNN models contain deeply nested compositions of simple parameterized functions (principally consisting of convolutions, scalar nonlinearity, moment normalizations, and their linear combinations), which are optimized by minimizing a loss function (15). Deep learning has been widely applied in medical image analysis tasks, including segmentation, classification, detection, and registration in various anatomical sites (e.g., brain, heart, lung, breast, abdomen, and prostate) (16,17). It also has demonstrated excellent performance in complex medical diagnosis tasks (18). In contrast to the radiomics architecture, the CNN can be trained in an end-to-end manner, automatically learning the features associated with the pathological diagnosis from imaging data. Thus, a precise lesion boundary and human-defined feature extraction are not required. This means that the original images without segmentation masks can be input into a deep learning network, which can avoid the adverse effects of unreliable segmentation (15). Deep learning methods are widely employed in a variety of applications, such as pulmonary nodule detection (19), liver fibrosis staging (20), and the determination of breast cancer hormonal receptor status (21). Despite its proven value in medical image analysis, deep learning presents several challenges: (I) similar to the radiomics paradigm, deep learning also comprises many interconnected components in its workflow, including image preprocessing, network building, model training and optimization, evaluation, and performance comparison, which requires that radiologists and researchers have a high level of expertise in deep learning to build optimal models. (II) The feature representation is not directly interpretable, which means that deep learning architectures are conceptually similar to black boxes for researchers, resulting in an unknown association and correspondence between learned deep features and diagnosis results.
Generally speaking, it is challenging and tedious to build an application-specialized machine learning model (either with radiomics or deep learning) if radiologists or researchers do not have expertise in machine learning or lack programming skills. To solve this problem, it is highly necessary to develop accessible and user-friendly software that can provide a convenient workflow for radiomics and deep learning-based medical image analysis. We thus developed an artificial intelligence-assisted diagnosis modeling software (AIMS) platform to alleviate the abovementioned problems. Moreover, the functionality and effectiveness of AIMS was evaluated by 3 experiments. The main advantages of AIMS are as follows: (I) it comprises both radiomics- and CNN-based deep learning paradigms and their corresponding standardized workflows; (II) the standardized workflows and modular design allow users to rapidly configure/build, evaluate, and compare different models for specific applications; (III) the user-friendly graphical user interface (GUI) enables users to automatically analyze large numbers of medical images without programming or editing scripts; and (IV) it provides an image-processing toolkit (such as segmentation, registration, and morphological operations) for convenient handling of lesion-labeling tasks. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-23-20/rc).
Methods
Design goals
The AIMS platform is aimed at providing a user-friendly infrastructure for radiologists and researchers to thus lower the barrier to building customized machine learning-based computer-assisted diagnosis models for medical image analysis. The design and implementation of AIMS follows several core principles, which satisfies the following key requirements:
- Completeness: the developed software should supply multiple machine learning frameworks to cover the frequently used paradigms (radiomics- and CNN-based deep learning). It should also provide complete machine learning workflows for medical image analysis, including data preparing/preprocessing, lesion labeling, classifier training, evaluation, performance comparison, and statistical result visualization.
- Standardization: the software should provide standardized machine learning workflows for medical image analysis.
- Friendliness: the platform should be simple, user-friendly, and accessible to everyday users without expertise in machine learning or programming. Specifically, this means the developed software should be able to conveniently build machine learning models using GUI wizards without the need for any editing of scripts or configuration of files.
- Flexibility: the software should include a flexible image processing toolkit for complex applications, such as semiautomatic segmentation for pixel-wise lesion labeling, registration for multiseries (both multimodality and multiphase) image analysis, and morphological operation for peritumoral analysis.
Software characteristics
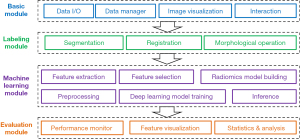
The first characteristic of the AIMS platform is that it is an all-in-one software platform for medical image analysis based on machine learning. We designed a loosely coupled and modular architecture in AIMS, which comprised several modules (Figure 1): (I) the basic module set mainly includes data inputs and outputs (I/Os), a data manager, image visualization, and human-computer interaction. (II) The labeling module set, mainly includes segmentation, registration, and morphological operation. (III) The machine learning module set mainly includes feature extraction, selection, and modeling in the radiomics workflow, and includes image preprocessing, model training, and inferencing in the deep learning workflow. In this way, AIMS facilitates complete and standardized pipelines for radiomics and deep learning paradigms, and users can switch between the radiomics workflow and deep learning workflow to conveniently build optimal machine learning models in their studies. (IV) The evaluation module set mainly includes performance monitoring, feature visualization, statistics generation, and analysis.
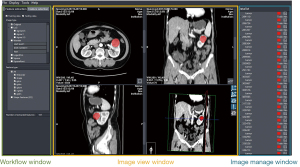
The second characteristic of AIMS is that it provides user-friendly GUIs (Figure 2) to guide users in building their own models. This includes the pipelines of image processing and machine learning modeling. Various data I/Os are also provided to cooperate with other domain-optimized software or toolkits. For example, AIMS could output a radiomics feature matrix as a Comma-Separated Values (CSV) file and internal statistical results as a binary MATLAB® (MAT) file. Moreover, AIMS could support the construction of a comprehensive classification model by concatenating radiomics features and other biomarkers (22), such as clinical features and immunohistochemical indicators.
The third characteristic of AIMS is that it provides various lesion-labeling tools to rapidly generate lesion masks. Lesion labeling is a common but time-consuming task in machine learning-based medical image analysis. In AIMS, some classical semiautomatic labeling methods and a UNet-based automatic segmentation (23) method are provided to rapidly create mask images. Semiautomatic labeling methods mainly include manual brush labeling, threshold segmentation, hole filling, and connected region extraction. In order to maximize the convenience of lesion labeling, registration is integrated with a map moving image and its mask with a fixed image (without a mask image) to create masks for multimodality or multiphase images, which is an ingenious and convenient way to create an initial mask for the fixed image. AIMS also provides morphological processing tools, which can be used to create peritumoral masks for peritumoral analysis in radiation oncology (24).
The comparison between AIMS and its competitive software is shown in Table 1.
Table 1
| Competing software | Free access | Segmentation | Registration | Morphological operation | Feature extraction | Radiomics workflow | Deep learning workflow |
|---|---|---|---|---|---|---|---|
| PyRadiomics (25) | √ | × | × | × | √ | × | × |
| SlicerRadiomicsa | √ | √ | √ | √ | √ | √ | × |
| Qualia Radiomicsb | √ | √ | √ | × | √ | √ | × |
| FeAture Explorer (26) | √ | × | × | × | √ | √ | × |
| CaPTk (27) | √ | √ | √ | × | √ | √ | × |
| FeTS (28) | √ | √ | × | × | × | × | × |
| GaNDLFc | √ | √ | × | × | × | × | √ |
| MITK (29) | √ | √ | √ | √ | × | × | × |
| MevisLabd | √ | √ | √ | √ | × | × | × |
| AIMS | √ | √ | √ | √ | √ | √ | √ |
a, an extension for 3D Slicer (https://github.com/AIM-Harvard/SlicerRadiomics), which encapsulates the PyRadiomics library to calculate a variety of radiomics features. b, this provides some contour tools and grow-cut segmentation for labeling small solid nodules and ground-glass shadow nodules (https://github.com/taznux/radiomics-tools). c, a generalizable application framework for segmentation, regression, and classification using PyTorch (https://github.com/mlcommons/GaNDLF). d, this is a modular framework for image processing research and development with a special focus on medical imaging. It includes modules for segmentation, registration, and volumetry, as well as quantitative morphological and functional analysis (https://www.mevislab.de/mevislab). AIMS, artificial intelligence-assisted diagnosis modeling software; CaPTk, Cancer Imaging Phenomics Toolkit; FeTS, federated tumor segmentation tool; GaNDLF, generally nuanced deep learning framework; MITK, Medical Imaging Interaction Toolkit.
AIMS is developed by using Qt1 for GUI, DCMTK2 for Digital Imaging and Communications in Medicine (DICOM) I/O, the Visualization Toolkit (VTK) (30) for image visualization, the Insight Toolkit (ITK) (30) for classical image processing (semiautomatic segmentation, registration, morphological operation), PyRadiomics (25) for image feature extraction, scikit-learn (31) in Python for radiomics modeling, and MONAI3 for deep learning-based CNN modeling. All extracted features in the radiomics pipeline are compliant with definitions of the Imaging Biomarker Standardization Initiative (IBSI) (32). AIMS is currently executable for the Microsoft Windows 10 (64 bit) operating system. Researchers can freely access this software by contacting the corresponding author (Yakang Dai) or by https://github.com/AIMSibet/AIMS. The recommended configuration of computer is listed in in Table S1 in Appendix 1.
Generally, AIMS provides both radiomics- and CNN-based deep learning to construct completeness and end-to-end pipelines. In each pipeline, it includes standardized machine learning workflows with GUI to satisfy the requirements of standardization and friendliness. AIM supplies various and flexible image processing tools for complex applications. More details of AIMS are described in the following subsections.
Software workflow
Users can build customized models by following a step-by-step GUI guide of AIMS. AIMS mainly contains 4 steps to build models (Figure 3): project configuration, lesion labeling/cropping, machine learning modeling (including radiomics and deep learning), and performance evaluation.

Project configuration
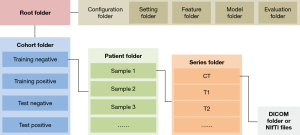
Image preparation is the first procedure in the workflow. AIMS accepts 3D or 2D images in both DICOM file and Neuroimaging Informatics Technology Initiative (NIfTI) file formats. In order to simplify the preparation of multimodality or multiphase images, we designed a “patient-series-image” document architecture referring to and simplifying the 4 hierarchical architectures in the DICOM standard. After image preparation, users can configure the training dataset and validation dataset in experimental and control cohorts. The file architecture is shown in Figure 4.
Lesion labeling/cropping
The second step of machine learning modeling for medical image analysis is creating lesion masks (or subimages in deep learning workflow). AIMS provides a convenient image-labeling toolkit to accelerate the time-consuming pixel-wise lesion labeling. Radiomics extracts high-throughput features from the regions of interest (ROIs), which are usually generated by manually delineating the outline of the entire lesion in all contiguous slices. In order to meet the challenge of pixel-wise labeling in medical images, AIMS supplies a convenient and flexible segmentation toolkit, which incorporates classical semiautomatic methods, morphology methods, registration-based labeling methods, and deep learning-based automatic labeling methods. As lesion masks are not necessary for the end-to-end training of CNN models, AIMS provides a cropping tool to crop subimages from a large source of images to reduce the input image size, which can greatly accelerate CNN training. Finally, all masks and their corresponding source images (or subimages) are automatically saved as NIfTI files.
Machine learning modeling
AIMS can implement either a radiomics workflow or a deep learning workflow according to the user’s configuration. In the workflow of radiomics analysis, AIMS offers a step-by-step GUI wizard (feature extraction, feature selection, model building, and performance analysis) to guide users in building radiomics models. In the workflow of deep learning, AIMS offers a straightforward GUI for users to configure the parameters of the CNN models.
Radiomics-based modeling
The fundamental assumption of radiomics is that the distinguishing texture features macroscopically reflect the irregularity of nuclear shape and arrangement. Radiomics extracts quantitative features from large-scale sets of medical images, quantitatively analyzes these representative features, and maps them to clinical conclusions for diagnosis and prediction (12). The workflow of radiomics analysis in AIMS follows the guidelines of the IBSI (33).
AIMS builds reproducible radiomics features (32), which are categorized into 7 feature types: (I) first-order features; (II) shape-based features; (III) gray-level co-occurrence matrix (GLCM) features; (IV) gray-level run length matrix (GLRLM) features; (V) gray-level size zone matrix (GLSZM) features; (VI) neighboring gray-tone difference matrix (NGTDM) features; and (VII) gray-level dependence matrix (GLDM) features. AIMS extracts features not only from the ROIs in the original images but also from images derived with image filters. The image filters include Laplacian of Gaussian, square, square root, logarithmic, exponential, gradient, local binary pattern, and wavelets. AIMS can combine the extracted features from multiple images or multiple regions (e.g., tumor mass and peritumor) if their names are input into the GUI wizard. AIMS would then save the extracted and selected features into separate CSV files. Moreover, researchers can combine more complex features by copying other biomarkers (e.g., clinical features, immunohistochemical indicators) into CSV files to build a more elaborate machine learning model.
The purpose of feature selection is to reduce the number of features and remove relevant features to prevent overfitting. Feature selection is critical for building a radiomics model with high-performance, repeatability, and interpretability. In this step, AIMS first normalizes each feature column to avoid the effect of different scales. It subsequently supplies 4 types of methods for selecting normalized features: t-test, correlation analysis, minimum redundancy maximum relevance (mRMR) (34), and sequential feature selection (35).
AIMS provides 7 machine learning algorithms for building radiomics models, which are nu-support vector classification (Nu-SVC), C-support vector classification (C-SVC), random forest, adaptive boosting (AdaBoost), extreme gradient boosting (XGBoost), least absolute shrinkage and selection operator (LASSO), and logistic regression based on scikit-learn. AIMS can automatically choose the appropriate hyperparameters through grid search, and researchers can refine these hyperparameters to obtain better performance. AIMS can further execute cross-validation on the training cohort and evaluate the prediction performance of optimized models on the independent test cohort. In order to intuitively analyze the role and relationship of selected features, AIMS automatically creates and saves feature heatmaps, which could be displayed in a statistical chart window of AIMS. Video 1 shows the operation of a radiomics workflow in AIMS.
Deep learning-based modeling
Deep learning-based modeling is an alternative paradigm for medical image analysis, which has been broadly applied in various applications due to its proven performance. Different from radiomics-based models that use handcrafted features, the deep learning-based classifier can be trained in an end-to-end manner to automatically perform high-level feature self-learning with little prior task-specific knowledge needed. Therefore, we integrated a deep learning-based workflow in AIMS for medical image analysis. Specifically, DenseNet-169 (36) was incorporated as the default classification network in AIMS. The cross-entropy loss function was used, and the Adam optimizer with a default learning rate of 10−4 was adopted. Similar to the deep learning-based segmentation in AIMS, a variety of hyperparameters of deep network can be set in classification tasks. In addition, DenseNet-121 and DenseNet-201 can be selected to adapt to different-scale classification tasks. Video 2 shows the operation of a deep learning workflow in AIMS.
Evaluation and visualization
AIMS offers various metrics to evaluate the classification performance of the constructed machine learning models. The metrics include confusion matrix (true positive, true negative, false positive, false negative), accuracy, sensitivity, specificity, precision, receiver operating characteristic (ROC), area under the curve (AUC) with 95% confidence interval (CI), geometric mean (G-mean), F1-score, and Matthews correlation coefficient (MCC). AIMS can output a study report with the following information: study and dataset description, configuration of the entire machine learning workflow, and experimental results. The final model can be saved if the performance satisfies the need of the task. This model can be implemented using AIMS to predict prospective data for radiation oncology.
Results
In order to verify functionality and effectiveness of AIMS, we performed various retrospective analysis tasks in radiation oncology: multiphase (multiphase contrast-enhanced) computed tomography (CT) analysis and multiregion (tumor mass region and peritumoral region) analysis for Fuhrman grading of clear cell renal cell carcinoma (ccRCC) and multimodality [biparametric magnetic resonance imaging (bpMRI)] analysis for Gleason grading of prostate cancer. The ccRCC Fuhrman grading was analyzed with the radiomics workflow, and the prostate cancer Gleason grading was performed with a deep learning workflow. In the experiments, AUC with 95% CI, accuracy, sensitivity, specificity, precision, MCC, and ROC were used to measure the performance of prediction models on the test cohorts. All image data were anonymous, and researchers were blinded to the clinical and histopathological reports of test cohorts.
This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and the experiment was approved by the institutional review board of The Second Affiliated Hospital of Soochow University and Suzhou Science & Technology Town Hospital. Individual consent for this retrospective analysis was waived.
Multiphase analysis for ccRCC Fuhrman grading
Some studies have shown that the performance of models using multiphase images is higher than that of those using single-phase images (37-39). In this experiment, we used AIMS to build radiomics models with features extracted from multiphase images for ccRCC Fuhrman grading.
We collected 247 samples with contrast-enhanced CT images, which were acquired from the Second Affiliated Hospital of Soochow University (Suzhou, China). The dataset included CT images, histopathology reports, and clinical data of patients who had undergone surgical resections of ccRCC between January 2009 and January 2019. A patient was included in this study if he/she underwent a preoperative contrast-enhanced computed tomography (CECT) with a 3-phase renal mass CT imaging protocol [corticomedullary phase (CP), nephrographic phase (NP), delay phase] had a histopathology report of ccRCC with a diagnosis with Fuhrman grades. Ultimately, 187 were finally enrolled after the following exclusion criteria were applied: (I) lack of Fuhrman grades in histopathology reports (n=36); (II) lack of CT images (n=4); (III) incomplete contrast-enhanced phases (n=17); (IV) incomplete lesion in CT images (n=2); and (V) suboptimal CT imaging quality (n=1). The patient characteristics are shown in Table S2 in Appendix 2. All samples included in the study were confirmed by pathology reports, and a simplified 2-tiered Fuhrman grade system (40) was used, which categorized samples into low grade and high grade. Consequently, 131 samples (low grade =95, high grade =36) were assigned into the training cohort, and 56 samples (low grade =40, high grade =16) were assigned into the test cohort. The lesion masks were created by 2 radiologists (with 25 years of experience) through delineating the outline of entire tumor in all contiguous slices. The radiologists only labeled lesions in the CP of contrast-enhanced CT images. The tumor masks of other phase images were created with an affine registration method integrated in AIMS (Figure 5).

The software extracted 107 quantitative 3D radiomics features from each lesion region in a single-phase image and then created a phase-combined feature set for each lesion. Each feature was normalized to achieve a zero mean and unit variance across the entire training and test cohorts to avoid the effect of different scales. In order to select discriminative features, a sophisticated feature selection procedure was performed as follows. First, low reproducibility features were removed if the variance of the normalized feature value was smaller than 10−3. The intraclass and interclass correlation coefficients (ICCs) of segmentation were computed to assess the inter- and intra-observer reproducibility, respectively. The features with an ICC lower than 0.75 were considered to have poor agreement and were therefore removed. Second, Pearson correlation analysis was performed to identify the distinctiveness of features and to remove the redundant features if their absolute correlations were higher than 0.1. The mRMR method was applied to identify the most important features for the criteria of both minimum redundancy and maximum relevance. Finally, only the top-20 most important features in each feature set from the training cohort were selected and input into a C-SVC (41) to build multiphase-feature combined models. The performance of the multiphase-feature combined models was assessed using the test cohort. We built 7 types of radiomics models to verify the availability and effectiveness of the multiphase features: (I) features extracted from the CP; (II) features extracted from the NP; (III) features extracted from the excretory phase (EP); (IV) features extracted from the corticomedullary and nephrographic phases (CNP); (V) features extracted from the corticomedullary and excretory phases (CEP); (VI) features extracted from the nephrographic and excretory phases (NEP); and (VII) features extracted from the corticomedullary, nephrographic, and excretory phases (CNEP).
The statistical performance of the 7 models on the test cohort is shown in Table 2 and Figure 6. The statistical results clearly show that the CNP model achieved the best overall performance among 7 models, which verified the effectiveness of multiphase analysis provided by AIMS. Our results also showed that the features extracted from the CP had the most discriminating power for grading the malignancy of ccRCC among the 3 phases of contrast-enhanced CT, which is consistent with a conclusion of another study (38). It is well known that ccRCC may enhance heterogeneously, with its peak enhancement occurring during CP, followed by a progressive washout (42).
Table 2
| Model | AUC (95% CI) | Acc | Sen | Spe | Pre | MCC |
|---|---|---|---|---|---|---|
| CP | 0.659 (0.508–0.810) | 0.377 | 0.610 | 0.695 | 0.333 | 0.242 |
| NP | 0.711 (0.562–0.859) | 0.626 | 0.701 | 0.767 | 0.652 | 0.219 |
| EP | 0.559 (0.402–0.716) | 0.539 | 0.605 | 0.881 | 0.586 | 0.162 |
| CNP | 0.776 (0.653–0.899) | 0.707 | 0.732 | 0.705 | 0.723 | 0.263 |
| CEP | 0.695 (0.553–0.838) | 0.388 | 0.600 | 0.695 | 0.431 | 0.202 |
| NEP | 0.658 (0.502–0.812) | 0.475 | 0.595 | 0.486 | 0.362 | 0.178 |
| CNEP | 0.633 (0.473–0.791) | 0.461 | 0.547 | 0.595 | 0.321 | 0.201 |
The data are presented as the mean value. AUC, area under the curve; CI, confidence interval; Acc, accuracy; Sen, sensitivity; Spe, specificity; Pre, precision; MCC, Matthews correlation coefficient; CP, corticomedullary phase; NP, nephrographic phase; EP, excretory phase; CNP, corticomedullary and nephrographic phases; CEP, corticomedullary and excretory phases; NEP, nephrographic and excretory phases; CNEP, corticomedullary, nephrographic, and excretory phases.

Multiregion analysis for ccRCC Fuhrman grading
Several previous studies have demonstrated that the peritumoral microenvironment accurately and comprehensively reflects the characteristics and the heterogeneity of tumors (43), which suggests that features from the peritumoral region can help to distinguish the malignancy grades of tumors. In this experiment, we used AIMS to build radiomics models with multiregion features for ccRCC Fuhrman grading.
We collected 267 CT images of CP images, histopathology reports, and the clinical data of patients who had undergone surgical resections for ccRCC Fuhrman grading. All data were downloaded from The Cancer Genome Atlas-Kidney Renal Clear Cell Carcinoma (TCGA-KIRC) dataset (44,45), and Fuhrman grades were confirmed by pathology reports. The images in TCGA-KIRC were obtained from 7 centers4 in the United States between November 2010 and May 2014, with devices from multiple manufactures (GE HealthCare, Siemens Healthineers, and Philips, New York, Pittsburgh, Rochester, University of North Carolina Chapel Hill, Bethesda, MD, Houston, TX, Buffalo, NY, USA). The inclusion criteria in this experiment were as follows: (I) pathology-confirmed ccRCC after surgery and (II) CT scans in the NP before surgery and radiotherapy. Ultimately, 177 samples were enrolled the following exclusion criteria were applied: (I) only MR images available (n=70); (II) only CT plain scans available (n=8); (III) history of surgery and/or chemotherapy prior to CT scans (n=6); (IV) multiple lesions (n=5); and (V) poor quality of CT scans (n=1). The patient characteristics are shown as in Table S3 in Appendix 2. A simplified 2-tiered Fuhrman grade system was also used in this experiment. In order to build and validate the developed radiomics models, 107 samples (low grade =67, high grade =40) were assigned into the training cohort, and 70 samples (low grade =38, high grade =32) were assigned into test cohort. Radiologists with 20 years of experience labeled the ROIs by delineating the outline of the entire tumor in all contiguous slices, and AIMS subsequently automatically created a corresponding peritumoral region by isotropically expanding 5 mm of the tumor in 3 dimensions (Figure 7).
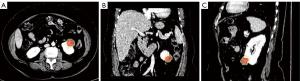
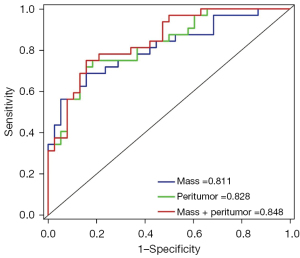
The statistical performance of 3 models on the test cohort is shown in Table 3 and Figure 8. The results intuitively showed that the models using peritumoral features achieved better performance than did the mass model, which demonstrated the effectiveness of the multiregion analysis provided by AIMS.
Table 3
| Model | AUC (95% CI) | Acc | Sen | Spe | Pre | MCC |
|---|---|---|---|---|---|---|
| Mass | 0.811 (0.706–0.914) | 0.743 | 0.687 | 0.789 | 0.733 | 0.417 |
| Peritumor | 0.828 (0.766–0.924) | 0.686 | 0.643 | 0.974 | 0.816 | 0.412 |
| Mass + peritumor | 0.848 (0.759–0.937) | 0.743 | 0.663 | 0.895 | 0.818 | 0.423 |
The data are presented as the mean value. AUC, area under the curve; CI, confidence interval; Acc, accuracy; Sen, sensitivity; Spe, specificity; Pre, precision; MCC, Matthews correlation coefficient.
Multimodality analysis for prostate cancer Gleason grading
Multimodality images are widely applied in radiological diagnosis and can significantly improve the diagnostic accuracy (46,47). In this experiment, we used AIMS to learn deep learning features from multimodality images and built deep learning-based models for distinguishing clinically significant prostate cancer (csPCa; Gleason score ≥7) from nonclinically significant prostate cancer (ncsPCa; Gleason score <7).
We retrospectively collected 301 patients who underwent prostate bpMRI scans with T2-weighting (T2W) and apparent diffusion coefficient (ADC) imaging at the Second Affiliated Hospital of Soochow University (Suzhou, China) between January 2017 and March 2020. Ultimately, 206 patients were enrolled after following exclusion criteria were applied: (I) lack of pathological results (n=52); (II) motion artifact corruption (n=31); and (III) mismatch between pathological results and bpMRI findings (n=12). The patient characteristics are shown in Table S4 in Appendix 2. The image dataset included 89 samples of csPCa and 117 samples of ncsPCa, and all the samples were confirmed by pathology reports. Subsequently, 164 samples (csPCa =75, ncsPCa =89) were assigned into the training cohort, and 42 samples (csPCa =14, ncsPCa =28) were assigned into the test cohort. Each lesion in the T2W images was pixel-wise labeled in all contiguous slices by a radiologist with 20 years of experience. Since some motion was present between T2W and ADC, we aligned T2W to ADC using an affine registration method integrated in AIMS for mapping lesion masks in T2W to their corresponding ADC images (show in Figure 9).

In order to reduce the time cost and computational burden, subimages were cropped from T2W and ADC images, which covered the whole lesion. Then, all the subimages were resampled to 80×80×16 pixels for model training. The pixel values in each subimage were normalized using Z-score. T2W and ADC images were further merged as a dual-channel image and input into DenseNet-169 network to build classification models. The network was trained with a 5-fold cross-validation on the training cohort, and the performance of the DenseNet-169 network was assessed on the test cohort. The details of classification results are shown in Table 4 and Figure 10, with the diagnostic performance of a radiologist with 13 years of experience included for comparison. It can be observed that the constructed DenseNet-169 model based on bpMRI performed better than did those based on single-parameter MRI and the radiologist.
Table 4
| Method | Modality | AUC (95% CI) | Acc | Sen | Spe | MCC |
|---|---|---|---|---|---|---|
| Radiologist | T2+ADC | 0.946 (0.877–1.00) | 0.952 | 0.929 | 0.964 | 0.883 |
| AIMS | T2 | 0.719 (0.555–0.864) | 0.762 | 0.571 | 0.857 | 0.684 |
| ADC | 0.857 (0.755–0.949) | 0.786 | 0.857 | 0.750 | 0.723 | |
| T2+ADC | 0.980 (0.947–1.00) | 0.953 | 1.000 | 0.929 | 0.903 |
The data are presented as the mean value. AUC, area under the curve; CI, confidence interval; Acc, accuracy; Sen, sensitivity; Spe, specificity; MCC, Matthews correlation coefficient; ADC, apparent diffusion coefficient; T2, T2-weighted; AIMS, artificial intelligence–assisted diagnosis modeling software.

Discussion
The development of a common software infrastructure for medical image analysis and computer-assisted diagnosis is a highly demanded, urgent need. Some classical numerical optimization toolkits that offer standardized implementations to build algorithm workflows for medical image analysis include NiftyNet (48) and ITK (30) for segmentation; Elastix (49), NiftyReg (50), Advanced Normalization Tools (ANTs) (51), and ITK (30) for registration; and VTK (30) for visualization. Some open-source software, including 3D Slicer (30), MITK (29), NifTK (52), ITK-SNAP (53), and MevisLab (https://www.mevislab.de/mevislab) provide standardized interfaces and loosely coupled frameworks with a GUI to build customized analysis workflow. However, these toolkits were developed for medical image processing (e.g., segmentation and registration) and not for machine learning and deep learning toolkits for medical image analysis.
Recently, the software infrastructure for general purpose machine learning has also progressed rapidly. In the field of radiomics analysis, PyRadiomics (25) is an open-source Python package used for radiomics feature extraction. scikit-learn (31), a well-known toolkit in Python used for machine learning, has a convenient and simple application programming interface for classification, regression, clustering, dimensionality reduction, model selection, and evaluation (54). The Cancer Imaging Phenomics Toolkit (CaPTk) (27,55) was developed as a cancer imaging phenomics toolkit for radiographic image analysis of cancer and currently focuses on brain, breast, and lung cancer. As it pertains to deep learning-based analysis, some deep learning platforms, including TensorFlow (56), TORCH (57), MONAI (https://monai.io/), Theano (58), Caffe (59), CNTK (60), and MatConvNet (61), have been developed in order to meet the growing demand for the training of customized deep learning models. Although these toolkits have been designed to lessen the requirement of expertise, optimizing the interrelated components of the complex workflows for researchers without programming capability or expertise in machine learning remains a considerable challenge. The generally nuanced deep learning framework (GaNDLF) (https://github.com/mlcommons/GaNDLF) provides an end-to-end solution involving segmentation, regression, and classification while producing robust deep learning models without requiring intimate knowledge of deep learning or coding experience. The federated tumor segmentation (FeTS) tool (28) was developed to enable federated training of a tumor subcompartment delineation model at several sites dispersed across the world without the need to share patient data. However, some limitations exist in these toolkits. First, they lack end-to-end application pipelines for radiomics and deep learning training and inference. Second, they do not include all the necessary applications (segmentation, registration) or end-to-end machine learning pipelines (radiomics-based and deep learning-based classification). Third, they remain difficult to operate for ordinary researchers without expertise in machine learning or programming.
In this work, AIMS was developed as an AIMS platform based on medical images and machine learning. The development of clinically relevant, medical imaging tools and the associated algorithms has been driven by a specific medical need involving an understanding of a researcher’s/clinician’s workflow. The main advantages of AIMS are as follows: (I) it is an all-in-one AIMS platform that includes both a radiomics and deep learning workflow. AIMS provides a comprehensive image segmentation package for tissue labeling, complete machine learning workflows for medical image analysis, and a variety of tools for medical image processing and visualization. (II) AIMS facilitates standardized machine learning processing procedures, such as image preprocessing, lesion labeling, classification training, evaluation, and performance comparison to cover the entire workflow of machine learning. The use of standardized approaches is a critical aspect of AIMS in creating reliable and reproducible models. In the radiomics workflow, AIMS provides standardized procedures of feature extraction, feature selection, model building, and statistical evaluation. In the deep learning-based workflow, AIMS provides some wide-used CNNs (DenseNet-169, DenseNet-121, and DenseNet-201) to train a deep learning classifier. (III) AIMS is simple and user-friendly and can be readily operated by users without machine learning expertise to rapidly and conveniently build customized machine learning models. Moreover, it offers well-designed GUI wizards to help researchers use each method. (IV) AIMS provides a flexible and powerful lesion-labeling toolkit for complex applications, such as multiphase, multiregion, and multimodality analysis. Specifically, AIMS facilitates semiautomatic segmentation, registration, morphological operations, and their combination to efficiently label images. The functionality and efficiency of AIMS was demonstrated in 3 independent experiments in radiation oncology, in which multiphase, multiregion, and multi-modality analyses were respectively performed. The AUC value of AIMS with multiphase analysis increased to 9.14% for ccRCC Fuhrman grading, demonstrating that the multiphase combined model could effectively improve the prediction performance (62). The AUC value of AIMS in multiregion analysis increased to 4.36% for ccRCC Fuhrman grading, demonstrating that the peritumoral delineation correctly captured the characteristics and heterogeneity of the malignancies (43). The AUC value of in with multimodality analysis increased to 14.35% for prostate cancer Gleason grading, demonstrating that bpMRI can be leveraged to detect and identify csPCa (63).
Lesion pixel-wise labeling is a time-consuming and subjective processing task, which is usually generated by manually delineating the outline of the entire lesion in all contiguous slices. Manual segmentation by experienced radiologists is often considered to the gold standard of lesion labeling. In order to meet the challenge of pixel-wise labeling in medical images, AIMS supplies a convenient and flexible segmentation toolkit, which incorporates classical semiautomatic methods, morphology methods, registration-based labeling methods, and deep learning-based automatic labeling methods. The semiautomatic methods include manual brush labeling, live wire-based delineation, threshold segmentation, hole filling, and connected domain extraction. Users can flexibly combine these algorithms into a semiautomatic segmentation pipeline to reduce manual intervention and guarantee the efficiency, consistency, and reproducibility of labeling work (64). A few previous radiation oncology studies have demonstrated that peritumoral regions can accurately and comprehensively reflect genetic alterations, heterogeneity, and tumor biology, significantly bolstering tumor malignancy grading (24,43). Therefore, peritumor delineation is another common labeling task. AIMS provides a morphology tool that can facilitate efficient peritumor labeling. Three basic methods-dilation, erosion, and subtraction-have been integrated into AIMS for the processing of binary mask images. Registration-based labeling methods are designed to be auxiliary tools for rapidly label lesions on multiseries images. AIMS supplies 2 options for registration: landmark-based rigid registration and volume-based rigid/affine registration. For landmark-based rigid registration, users should select at least 6 landmarks in both the moving image and fixed image, and the pair-wise landmarks in different images should be located on the same anatomical structures. For volume-based rigid/affine registration, AIMS automatically aligns the mask image to the unlabeled image via a rigid or affine spatial transform. Deep learning-based segmentation is a powerful tool for automatic lesion labeling. AIMS uses a residual UNet-based (65) segmentation training interface to build a customized segmentation model for a specific type of tissue or lesion. The default network consists of 5 encoding-decoding branches with 16, 32, 64, 128 and 256 channels, respectively. Each encoder block consists of 2 repeated residual units of 3×3×3 convolutions (with a stride of 2), instance normalization and parametric rectified linear unit (PReLU) activations (66). The decoder blocks have a similar architecture as the encoder blocks, using transpose convolutions with a stride of 2 for upsampling. The segmentation network is trained by minimizing the Dice loss function using the Adam optimizer (67) with a default learning rate of 10−3. The default number of training epochs is 100, and the batch size is set to 1. The size and dimension of input images, channel number, residual units, feature normalization, loss function, learning rate, batch size, and training epoch number can be set in a GUI wizard to adapt various segmentation tasks. The deep learning-based segmentation model can automatically generate initial lesion masks, and the initial masks can be refined conveniently by using the provided semiautomatic labeling tools. Consequently, the cost of manual labeling can be greatly reduced, which helps to accelerate the subsequent analysis.
The limitations of the current AIMS and our future work are discussed below. First, the class imbalance problem is highly common in medical diagnosis and has not yet been effectively addressed with AIMS. We will implement data resampling techniques to alleviate class imbalance (68). Currently, AIMS is able to tackle binary classification tasks conveniently, but it is not yet equipped with adequate methods for multiclass classification. We will incorporate more inherently multiclass methods for feature selection and classifier construction to further facilitate the use of AIMS in the future. Second, although the graphical training interface is provided for building a customized segmentation model, only the Residual UNet is supported, which may be insufficient for various medical image segmentation tasks. We will incorporate more advanced deep networks, such as nnU-Net (23); in addition, other well-verified segmentation models (69,70) will be deployed to speed up the labeling process. Furthermore, AIMS should include a text-data interface in the development of an image-text combined model, which would be an effective means to improving the classification performance. Third, while some deep learning computation is performed on Linux-based computers, the current AIMS version only supports installation on Windows systems, limiting AIMS’s cross-platform applicability. AIMS is dependent on PyRadiomics and MONAI for feature extraction and deep learning, both of which require the runtime environment for AIMS operation. Fourth, although the image datasets in the experiments came from multiple centers, the small sample size is a limitation in this work. More prospective and multicenter experimental research with a large sample size is necessary in future studies to empirically validate the performance of AIMS.
Conclusions
AIMS was developed as an artificial intelligence assisted diagnosis modeling software platform based on medical images and machine learning and is a new tool for radiologists and researchers to use in artificial intelligence-assisted diagnosis studies. All the experiments conducted in this study attested to the accessibility and efficiency of AIMS in radiation oncology.
Acknowledgments
Funding: This study was supported by
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-23-20/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-20/coif). YD is the founder of Suzhou Guoke Kangcheng Medical Technology Co., Ltd. Some technical assistance from the company was provided to this study. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013), and the experiment was approved by the institutional review board of The Second Affiliated Hospital of Soochow University and Suzhou Science & Technology Town Hospital. Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
1Qt: a cross-platform application development framework, https://www.qt.io/
2DCMTK: a collection of libraries and applications implementing large parts of the DICOM standard, https://www.dcmtk.org/en/
3MONAI: multiple open-source PyTorch-based frameworks for deep learning in medical image analysis. https://github.com/Project-MONAI
4Memorial Sloan-Kettering Cancer Center, New York; University of Pittsburgh/UPMC, Pittsburgh; Mayo Clinic, Rochester; University of North Carolina, Chapel Hill; National Cancer Institute, Bethesda, MD; M.D. Anderson Cancer Center, Houston TX; Roswell Park Cancer Institute, Buffalo, NY.
References
- Song Y, Zhang YD, Yan X, Liu H, Zhou M, Hu B, Yang G. Computer-aided diagnosis of prostate cancer using a deep convolutional neural network from multiparametric MRI. J Magn Reson Imaging 2018;48:1570-7. [Crossref] [PubMed]
- Manjunath BS, Ma WY. Texture features for browsing and retrieval of image data. IEEE Trans Pattern Anal Mach Intell 1996;18:837-42.
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, Zegers CM, Gillies R, Boellard R, Dekker A, Aerts HJ. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017;542:115-8. [Crossref] [PubMed]
- Zheng X, Yao Z, Huang Y, Yu Y, Wang Y, Liu Y, Mao R, Li F, Xiao Y, Wang Y, Hu Y, Yu J, Zhou J. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat Commun 2020;11:1236. [Crossref] [PubMed]
- Zeleznik R, Foldyna B, Eslami P, Weiss J, Alexander I, Taron J, et al. Deep convolutional neural networks to predict cardiovascular risk from computed tomography. Nat Commun 2021;12:715. [Crossref] [PubMed]
- Trajanovski S, Mavroeidis D, Swisher CL, Gebre BG, Veeling BS, Wiemker R, Klinder T, Tahmasebi A, Regis SM, Wald C, McKee BJ, Flacke S, MacMahon H, Pien H. Towards radiologist-level cancer risk assessment in CT lung screening using deep learning. Comput Med Imaging Graph 2021;90:101883. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Lee G, Lee HY, Park H, Schiebler ML, van Beek EJR, Ohno Y, Seo JB, Leung A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol 2017;86:297-307. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJ, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Bera K, Braman N, Gupta A, Velcheti V, Madabhushi A. Predicting cancer outcomes with radiomics and artificial intelligence in radiology. Nat Rev Clin Oncol 2022;19:132-46. [Crossref] [PubMed]
- Tian J, Dong D, Liu Z, Wei J. Chapter 1 - Introduction, in Radiomics and Its Clinical Application. In The Elsevier and MICCAI Society Book Series, Radiomics and Its Clinical Application, Academic Press; 2021:1-18.
- Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM 2017;60:84-90.
- Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak JAWM, van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017;42:60-88. [Crossref] [PubMed]
- Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. [Crossref] [PubMed]
- Shen W, Zhou M, Yang F, Yang C, Tian J. Multi-scale Convolutional Neural Networks for Lung Nodule Classification. Inf Process Med Imaging 2015;24:588-99. [Crossref] [PubMed]
- Nam JG, Park S, Hwang EJ, Lee JH, Jin KN, Lim KY, Vu TH, Sohn JH, Hwang S, Goo JM, Park CM. Development and Validation of Deep Learning-based Automatic Detection Algorithm for Malignant Pulmonary Nodules on Chest Radiographs. Radiology 2019;290:218-28. [Crossref] [PubMed]
- Yasaka K, Akai H, Kunimatsu A, Abe O, Kiryu S. Liver Fibrosis: Deep Convolutional Neural Network for Staging by Using Gadoxetic Acid-enhanced Hepatobiliary Phase MR Images. Radiology 2018;287:146-55. [Crossref] [PubMed]
- Naik N, Madani A, Esteva A, Keskar NS, Press MF, Ruderman D, Agus DB, Socher R. Deep learning-enabled breast cancer hormonal receptor status determination from base-level H&E stains. Nat Commun 2020;11:5727. [Crossref] [PubMed]
- Huang SG, Qian XS, Cheng Y, Guo WL, Zhou ZY, Dai YK. Machine learning-based quantitative analysis of barium enema and clinical features for early diagnosis of short-segment Hirschsprung disease in neonate. J Pediatr Surg 2021;56:1711-7. [Crossref] [PubMed]
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Zhou Z, Qian X, Hu J, Ma X, Zhou S, Dai Y, Zhu J. CT-based peritumoral radiomics signatures for malignancy grading of clear cell renal cell carcinoma. Abdom Radiol (NY) 2021;46:2690-8. [Crossref] [PubMed]
- van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. [Crossref] [PubMed]
- Song Y, Zhang J, Zhang YD, Hou Y, Yan X, Wang Y, Zhou M, Yao YF, Yang G. FeAture Explorer (FAE): A tool for developing and comparing radiomics models. PLoS One 2020;15:e0237587. [Crossref] [PubMed]
- Davatzikos C, Rathore S, Bakas S, Pati S, Bergman M, Kalarot R, et al. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J Med Imaging (Bellingham) 2018;5:011018. [Crossref] [PubMed]
- Pati S, Baid U, Edwards B, Sheller MJ, Foley P, Anthony Reina G, Thakur S, Sako C, Bilello M, Davatzikos C, Martin J, Shah P, Menze B, Bakas S. The federated tumor segmentation (FeTS) tool: an open-source solution to further solid tumor research. Phys Med Biol 2022;67: [Crossref] [PubMed]
- Nolden M, Zelzer S, Seitel A, Wald D, Müller M, Franz AM, Maleike D, Fangerau M, Baumhauer M, Maier-Hein L, Maier-Hein KH, Meinzer HP, Wolf I. The Medical Imaging Interaction Toolkit: challenges and advances: 10 years of open-source development. Int J Comput Assist Radiol Surg 2013;8:607-20. [Crossref] [PubMed]
- Pieper S, Lorensen B, Schroeder W, Kikinis R. The NA-MIC Kit: ITK, VTK, pipelines, grids and 3D slicer as an open platform for the Medical Image Computing community. 3rd IEEE International Symposium on Biomedical Imaging: Nano to Macro 2006:698-701.
- Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2012;12:2825-30.
- Zwanenburg A, Abdalah MA, Apte A, Ashrafinia S, Beukinga J, Bogowicz M, et al. Image biomarker standardisation initiative. Radiotherapy & Oncology 2018;127:543-4.
- Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. [Crossref] [PubMed]
- Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell 2005;27:1226-38. [Crossref] [PubMed]
- Pudil P, Novovičová J, Kittler J. Floating search methods in feature selection. Pattern Recogn Lett 1994;15:1119-25.
- Huang G, Liu Z, Van Der Maaten L, Weinberger K. Densely connected convolutional networks. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017:4700-8.
- Lai S, Sun L, Wu J, Wei R, Luo S, Ding W, Liu X, Yang R, Zhen X. Multiphase Contrast-Enhanced CT-Based Machine Learning Models to Predict the Fuhrman Nuclear Grade of Clear Cell Renal Cell Carcinoma. Cancer Manag Res 2021;13:999-1008. [Crossref] [PubMed]
- Nguyen K, Schieda N, James N, McInnes MDF, Wu M, Thornhill RE. Effect of phase of enhancement on texture analysis in renal masses evaluated with non-contrast-enhanced, corticomedullary, and nephrographic phase-enhanced CT images. Eur Radiol 2021;31:1676-86. [Crossref] [PubMed]
- Alimu P, Fang C, Han Y, Dai J, Xie C, Wang J, Mao Y, Chen Y, Yao L, Lv C, Xu D, Xie G, Sun F. Artificial intelligence with a deep learning network for the quantification and distinction of functional adrenal tumors based on contrast-enhanced CT images. Quant Imaging Med Surg 2023;13:2675-87. [Crossref] [PubMed]
- Becker A, Hickmann D, Hansen J, Meyer C, Rink M, Schmid M, Eichelberg C, Strini K, Chromecki T, Jesche J, Regier M, Randazzo M, Tilki D, Ahyai S, Dahlem R, Fisch M, Zigeuner R, Chun FK. Critical analysis of a simplified Fuhrman grading scheme for prediction of cancer specific mortality in patients with clear cell renal cell carcinoma--Impact on prognosis. Eur J Surg Oncol 2016;42:419-25. [Crossref] [PubMed]
- Novakovic J, Veljovic A. C-Support Vector Classification: Selection of kernel and parameters in medical diagnosis. 2011 IEEE 9th International Symposium on Intelligent Systems and Informatics; 2011:465-70.
- Kopp RP, Aganovic L, Palazzi KL, Cassidy FH, Sakamoto K, Derweesh IH. Differentiation of clear from non-clear cell renal cell carcinoma using CT washout formula. Can J Urol 2013;20:6790-7.
- Wang X, Zhao X, Li Q, Xia W, Peng Z, Zhang R, Li Q, Jian J, Wang W, Tang Y, Liu S, Gao X. Can peritumoral radiomics increase the efficiency of the prediction for lymph node metastasis in clinical stage T1 lung adenocarcinoma on CT? Eur Radiol 2019;29:6049-58. [Crossref] [PubMed]
- Zhu X, Dong D, Chen Z, Fang M, Zhang L, Song J, Yu D, Zang Y, Liu Z, Shi J, Tian J. Radiomic signature as a diagnostic factor for histologic subtype classification of non-small cell lung cancer. Eur Radiol 2018;28:2772-8. [Crossref] [PubMed]
- Hodgdon T, McInnes MD, Schieda N, Flood TA, Lamb L, Thornhill RE. Can Quantitative CT Texture Analysis be Used to Differentiate Fat-poor Renal Angiomyolipoma from Renal Cell Carcinoma on Unenhanced CT Images? Radiology 2015;276:787-96. [Crossref] [PubMed]
- Tian H, Ding Z, Wu H, Yang K, Song D, Xu J, Dong F. Assessment of elastographic Q-analysis score combined with Prostate Imaging-Reporting and Data System (PI-RADS) based on transrectal ultrasound (TRUS)/multi-parameter magnetic resonance imaging (MP-MRI) fusion-guided biopsy in differentiating benign and malignant prostate. Quant Imaging Med Surg 2022;12:3569-79. [Crossref] [PubMed]
- Hu L, Zhou DW, Guo XY, Xu WH, Wei LM, Zhao JG. Adversarial training for prostate cancer classification using magnetic resonance imaging. Quant Imaging Med Surg 2022;12:3276-87. [Crossref] [PubMed]
- Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G, Eaton-Rosen Z, Gray R, Doel T, Hu Y, Whyntie T, Nachev P, Modat M, Barratt DC, Ourselin S, Cardoso MJ, Vercauteren T. NiftyNet: a deep-learning platform for medical imaging. Comput Methods Programs Biomed 2018;158:113-22. [Crossref] [PubMed]
- Klein S, Staring M, Murphy K, Viergever MA, Pluim JP. elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196-205. [Crossref] [PubMed]
- Modat M, Ridgway GR, Taylor ZA, Lehmann M, Barnes J, Hawkes DJ, Fox NC, Ourselin S. Fast free-form deformation using graphics processing units. Comput Methods Programs Biomed 2010;98:278-84. [Crossref] [PubMed]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 2011;54:2033-44. [Crossref] [PubMed]
- Clarkson MJ, Zombori G, Thompson S, Totz J, Song Y, Espak M, Johnsen S, Hawkes D, Ourselin S. The NifTK software platform for image-guided interventions: platform overview and NiftyLink messaging. Int J Comput Assist Radiol Surg 2015;10:301-16. [Crossref] [PubMed]
- Yushkevich P, Piven J, Cody H, Ho S, Gee JC, Gerig G. User-guided level set segmentation of anatomical structures with ITK-SNAP. Insight J 2005;1:1-9.
- Buitinck L, Louppe G, Blondel M, Pedregosa F, Andreas C, Grisel O, Niculae V, Prettenhofer P, Gramfort A, Grobler J, Layton R, Vanderplas A, Joly A, Holt B, Varoquaux G. API design for machine learning software: experiences from the scikit-learn project. arXiv 2013:1309.0238.
- Pati S, Singh A, Rathore S, Gastounioti A, Bergman M, Ngo P, et al. The Cancer Imaging Phenomics Toolkit (CaPTk): Technical Overview. Brainlesion 2020;11993:380-94. [Crossref] [PubMed]
- Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv 2016:1603.04467.
- Collobert R, Kavukcuoglu K, Farabet C, Torch7: A MATLAB-like environment for machine learning. in Proceedings of the NIPS Workshop on Algorithms, Systems, and Tools for Learning at Scale (Big Learning); 2011.
- Bastien F, Lamblin P, Pascanu R, Bergstra J, Goodfellow I, Bergeron A, Bouchard N, Warde-Farley D, Bengio Y. Theano: new features and speed improvements. arXiv 2012;1211.5590.
- Jia Y, Shelhamer E, Donahue J, Karayev S, Long J, Girshick R, Guadarrama S, Darrell T. Caffe: Convolutional Architecture for Fast Feature Embedding. Proceedings of the 22nd ACM international conference on Multimedia 2014:675-8.
- Seide F, Agarwal A. CNTK: Microsoft's Open-Source Deep-Learning Toolkit. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 2016:2135.
- Vedaldi A, Lenc K. MatConvNet - Convolutional Neural Networks for MATLAB. Proceedings of the 23rd ACM international conference on Multimedia; 2014:689-92.
- Lin F, Cui EM, Lei Y, Luo LP. CT-based machine learning model to predict the Fuhrman nuclear grade of clear cell renal cell carcinoma. Abdom Radiol (NY) 2019;44:2528-34. [Crossref] [PubMed]
- Xu L, Zhang G, Shi B, Liu Y, Zou T, Yan W, Xiao Y, Xue H, Feng F, Lei J, Jin Z, Sun H. Comparison of biparametric and multiparametric MRI in the diagnosis of prostate cancer. Cancer Imaging 2019;19:90. [Crossref] [PubMed]
- Larue RT, Defraene G, De Ruysscher D, Lambin P, van Elmpt W. Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 2017;90:20160665. [Crossref] [PubMed]
- Jha D, Smedsrud P, Riegler M, Johansen D, Lange T, Halvorsen P, Johansen H. ResUNet++: An Advanced Architecture for Medical Image Segmentation. 2019 IEEE International Symposium on Multimedia; 2019:225-30.
- He K, Zhang X, Ren S, Sun J. Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification. Proceedings of the IEEE International Conference on Computer Vision (ICCV); 2015:1026-34.
- Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. International Conference on Learning Representations (ICLR); 2015.
- Lin C, Tsai CF, Lin WC. Towards hybrid over- and under-sampling combination methods for class imbalanced datasets: an experimental study. Arti Intel Rev 2023;56:845-63.
- Heller N, Isensee F, Maier-Hein KH, Hou X, Xie C, Li F, et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med Image Anal 2021;67:101821. [Crossref] [PubMed]
- Bilic P, Christ P, Li HB, Vorontsov E, Ben-Cohen A, Kaissis G, et al. The Liver Tumor Segmentation Benchmark (LiTS). Med Image Anal 2023;84:102680. [Crossref] [PubMed]


