
Precise localization of microvascular invasion in hepatocellular carcinoma based on three-dimensional histology-MR image fusion: an ex vivo experimental study
Introduction
Hepatocellular carcinoma (HCC) is the second most lethal tumor and the fourth leading cause of cancer-related death worldwide (1,2). Although significant progress has been made in the comprehensive and individualized treatment of liver cancer, the postoperative 5-year recurrence rate is still as high as 70% (3,4). Studies have identified a relationship between a high recurrence rate and microvascular invasion (MVI), which has been repeatedly demonstrated to be an independent risk factor for the postoperative recurrence of HCC (5-7). A previous study showed that MVI can increase the recurrence rate of HCC by 4.4 times (8).
However, MVI can only be confirmed by postoperative histopathology, and its preoperative imaging features cannot be observed. Thus far, a large number of clinical studies have been conducted on the construction of a prediction model for MVI of HCC through extracting the radiomics features of the tumor and peritumoral region (9-11). However, some shortcomings to this approach remain: (I) the status of MVI can only be speculated upon based on tumor and peritumoral imaging features, and (II) these models can only predict the presence or absence of MVI and cannot clarify its specific location or degree and thus cannot provide more effective decision-making information for preoperatively guiding the precise radical treatment of HCC (such as local surgical resection and thermal ablation). Consequently, an effective clinical method for accurately diagnosing MVI before surgery remains lacking.
Medical image registration and fusion, in which spatial coordinates are matched by overlaying or correlating data from 2 sectional imaging techniques, enables us to combine the advantages of diverse imaging procedures and provide more diagnostic details to physicians (12). For example, ultrasound-computed tomography (CT)/magnetic resonance (MR) image fusion consists of a navigation system and a positioning system to detect, characterize, and monitor interventions (13). Therefore, we envisaged registering and fusing 3-dimensional (3D) histological image data and volumetric radiological data using image registration and fusion technology to achieve a real correspondence between pathological structures and radiological features. This involves identifying the location of MVI on radiological images through the marking of the MVI region on histological images. We intend to extract radiomics features of the MVI region and develop a diagnostic model for MVI in HCC in the near future. In this study, we established an animal model for the presence of MVI in rabbit VX2 liver tumors, which obtained MR volume images and serially sectioned histological images of the liver in vitro. By using image fusion technology, we evaluated the feasibility and accuracy of 3D histology-MR fusion imaging. Following this, the technique was used to accurately locate the MVI region on MR images, thus laying a foundation for the next step of extracting the radiomics features of MVI.
Methods
Animals and VX2 liver tumor models
The animal experiments were conducted at Zhuhai BesTest Bio-Tech Co., Ltd. (Zhuhai, China). Experiments were performed under a project license (No. L202104001) granted by the institutional ethics board of Zhuhai BesTest Bio-Tech Co., Ltd., in compliance with institutional guidelines for the care and use of animals. Eleven New Zealand white rabbits (one donor rabbit and ten recipient rabbits), aged from 3 to 4 months and weighing from 2 to 3 kg, were included in the model construction. Rabbits were purchased from Huadong Xinhua Experimental Animal Farm (Guangzhou, China). The VX2 tumor tissue was provided by Zhuhai BesTest Bio-Tech Co., Ltd., and stored in liquid nitrogen (−196 ℃).
The rabbit model of the hepatic VX2 tumor was established according to the literature (14,15). All rabbits were anesthetized with 0.1 mL/kg of intramuscular injections of xylazine hydrochloride (Dunhua Shengda Animal Medicine Co., Ltd., Qingdao, China), along with 0.1 mL/kg of tiletamine hydrochlorid and zolazepam hydrochloride (Virbac Inc., Carros, France). The VX2 tumor tissues was quickly thawed at 37 ℃ for approximately 5 min. Then, they were placed in a sterile plate containing normal saline, cut into approximately 0.5-mm3 pieces for tumor tissue suspension, and injected into the vastus muscles of both hind legs (0.5 mL for each side) of the donor rabbit. An ultrasound examination was performed to monitor the tumor growth consecutively in the legs, and the donor rabbit was euthanized with an air embolism when the tumors reached approximately 2 cm in diameter. The tumors in the thigh of the donor rabbit were then harvested. The peripheral tumor tissue, which had grown rapidly, was cut into cubes approximately 1-mm3 in size under sterile conditions.
The VX2 tumor fragments were implanted into the livers of the 10 recipient rabbits. We performed a percutaneous puncture in the subxiphoid area, penetrated a 16-G needle into the left lobe parenchyma of the liver under ultrasonographic guidance, pulled out the needle core, inserted a single 1-mm3 VX2 tumor fragment into the needle sheath, inserted the needle core to push the tumor fragments into the liver parenchyma, immediately sealed the puncture with absorbent gelatin sponge, and then pulled out the needle and gently compressed the area manually for 3 min. Penicillin was injected intramuscularly for 3 days after the operation to prevent infection. Continuous monitoring of tumor growth to 2–3 cm by ultrasound showed that the animal model of HCC had been successfully constructed. The tumor growth time and size, along with the rabbit weight, were recorded periodically.
Specimen processing
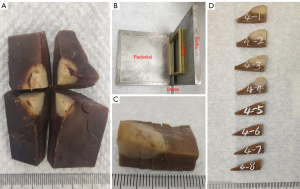
After the rabbit VX2 liver tumor model was successfully established, all rabbits were euthanized with air embolism after anesthesia. The whole liver lobe containing the tumor was fixed with 4% paraformaldehyde for more than 48 hours. Then, the liver lobe was divided into 4 specimens with the tumor at the center, and a small amount of marginal tissue was trimmed for each specimen (Figure 1A). Next, each specimen was used as a case for 3D histology-MR image fusion. The subsequent experiments, involving MR imaging (MRI), histology imaging, and image fusion processing, were carried out at The Fifth Affiliated Hospital of Sun Yat-sen University.
Ex vivo MRI protocol
MRI data were acquired with a 9.4-T MRI scanner (BioSpec94/30 USR, Bruker, Germany). According to the size of the specimen, we selected the appropriate rat body volume coil of 72 mm to maximize the image quality. Before scanning, we removed the specimen from 4% paraformaldehyde and hydrated it with 0.9% saline for 48 hours. For the MRI T2-weighted scan, the specimen was placed in a small plastic box containing fomblin (Perfluoro Polyether, Solvay, Italy) to isolate the air and preserve moisture. Axial sectional images of specimens parallel to the cut surface of the tumor were selected for the reference scan. T2-weighted axial MR images were generated using multislice turbo spin echo acquisition with a scan duration of 8 min and 48 s. The scan parameters were as follows: repetition time/time to echo (TR/TE) =3,300 ms/29 ms, resolution =94×94 µm2, field of view (FOV) =36×30 mm2, averages =2, slice thickness =0.8 mm with no gap, matrix size =384×320, excitation angle =90°, refocusing angle =180°, rapid imaging with refocused echo (RARE) factor =4, and bandwidth =33,333.3 Hz. The acquired MR images were stored in DICOM format.
Serial section histology
In an attempt to maintain a parallel orientation to the MRI axial sectional plane, each specimen was cut into parallel and equal-thickness tissue blocks using a homemade tool (including a baffle, a pedestal, two 3-mm thick frames, a 0.5-mm gap between the 2 frames, and a blade with a thickness of 0.5 mm) (Figure 1B,1C). All tissue blocks were marked and numbered consecutively (Figure 1D). An automatic tissue dehydrator was used to dehydrate the tissue blocks.
Next, the tissue blocks were embedded in the same marked orientation and shaped using a paraffin-embedding station and paraffin-embedding mold. Subsequently, each tissue wax block was intermittently serially sectioned to a 5-µm slice thickness with a 300-µm interval on a microtome, yielding regularly spaced and parallel tissue sections. The sections were stained with hematoxylin and eosin and then sections were scanned and digitized using a Pannoramic 250 Flash slide scanner (3DHistech, Budapest, Hungary) at ×20 magnification and visualized with CaseViewer 2.4 (3DHistech). To identify and locate the MVI, all sections were examined independently by 2 pathologists with more than 8 years of experience in HCC pathology. Any discrepancies were resolved by consensus with a third senior pathologist. In this study, only MVI lesions with maximum diameter of less than 1 mm and more than 1 mm from the main tumor were included for localization on MR images.
3D histology-MR image fusion
All pathological sections with whole-mount slide imaging (WSI) of each specimen were captured in a screenshot with CaseViewer software (3DHistech). The magnification of WSI was 1.0 times, and all screenshots were the same size, with a pixel of 300 dpi. According to the histological contours, 2D images were consecutively aligned with translation (section position), rotation (section orientation), and scaling (volume loss) functions using Photoshop 22.1.1 (Adobe Systems, San Jose, CA, USA). All matched histological images were stored in JPG format.
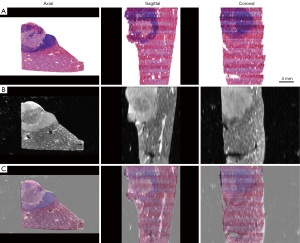
The histological images and their corresponding MR images for each specimen were imported into 3DSlicer 4.11.0 (http://www.slicer.org). The system automatically generated the 3D volume data and displayed the reconstructed 3D histology and MR images of the specimen in the axial, sagittal, and coronal planes on the screen. We chose histology images as the fixed volume and MR images as the moving volume for landmark registration in 3DSlicer software. Affine and nonrigid registrations of the moving image to the fixed image were performed to correct the positions, orientations, and volume loss and compensate for distortions due to resection and tissue slicing. A minimum of 10 pairs of landmarks on the histology images and MR images were handpicked by experienced radiologists in image fusion to serve as the reference standard for image registration. As shown in Figure 2, these distinctive landmarks include small vessel bifurcations, particular contours and anatomy, were identified within the liver.
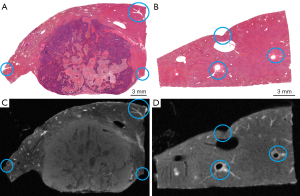
After coregistration, the effects of 3D histology-MR image fusion were evaluated by 2 authors (L.L. and J.C., radiologists with 8 and 5 years of experience with fusion imaging, respectively). Image fusion was defined as successful if the contours of the tissues, tumors, and vessels in the histological and MR images were subjectively judged to be well registered by the 2 radiologists independently. Image fusion was defined as unsuccessful if any disagreements occurred or both agreed that the registration was poor.
In addition, we picked 3 corresponding pairs of landmarks except to the 10 pairs of registered landmarks and recorded their spatial coordinates in the fixed (x1, y1, z1) and moving volumes (x2, y2, z2). The spatial distance (d) of each pair of landmarks was calculated by the following formula:
The average of the spatial distances of the 3 pairs of landmarks was used as the registration error of image fusion for a specimen.
Assessment of influencing factors
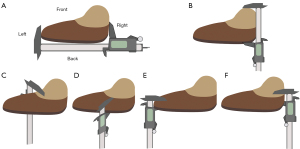
To assess the uniformity of the thickness of each tissue block after sampling and the uniformity of tissue shrinkage after dehydration, 50 tissue blocks were randomly selected. As depicted in Figure 3, the length and width of the tissue blocks were measured with a Vernier caliper, and the thickness was measured at intermediate positions in 4 directions (front, back, left, right) before and after dehydration. The thicknesses in the 4 directions and the thickness ratios between any 2 directions of 50 tissue blocks before and after dehydration were compared to assess the thickness uniformity. The shrinkage rates of the length, width, and thickness (average thickness in the 4 directions) after dehydration were calculated according to the following formula:
where R denotes the tissue shrinkage rate, and DB and DA denote the dimension of the tissue blocks before dehydration and after dehydration, respectively.
Statistical analysis
Statistical analysis was conducted using SPSS (version 25.0, IBM, NY, USA). The Shapiro-Wilk test was conducted to verify the normality of quantitative data. The mean ± standard deviation is used to express continuous variables that obeyed a normal distribution, the median (range) to express continuous variables that obeyed a skewed distribution, and percentage to express categorical variables. A paired t-test was used for comparisons before and after the intervention. Regarding the assessment of influencing factors, we conducted one-way analysis of variance (ANOVA) to compare the thickness, thickness ratio, and shrinkage rate of tissue blocks before and after dehydration. If the P value was less than 0.05, we further performed post hoc pairwise comparisons using Bonferroni correction, and the level of statistical significance was adjusted correspondingly.
Results
VX2 liver tumor formation in rabbits
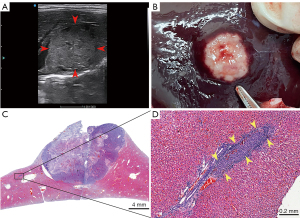
In this study, all rabbits successfully tolerated VX2 tumor inoculation. No mortality, implant-associated infection, or alterations in animal behavior were observed at any point in the experiment. The weights of the rabbits before intervention and after successful modeling were 2.6±0.4 and 2.8±0.4 kg, respectively, representing a statistically significant difference (P=0.03) and indicating that the weights of the rabbits continued to increase normally after tumor inoculation. The implanted tumor growth was monitored for 21 days, and all rabbits developed a single tumor (Figure 4A). The gross specimen and WSI confirmed that the VX2 liver tumor model was successfully established in the 10 rabbits (Figure 4B,4C), and MVI was present in the peritumoral histology (Figure 4D). The tumor was 2.6±0.2 cm in its maximum dimension and was located in the left hepatic lobe. Three rabbits had intrahepatic satellite nodules adjacent to the tumor, with a total of 5 nodules and a median maximum diameter of 0.50 (range, 0.15–0.55) cm.
3D histology-MR image fusion
A total of 40 specimens from 10 hepatic lobes containing tumors were collected from rabbits and included in this study. Each specimen was cut into 7 (range, 6–9) tissue blocks for a total of 285 tissue blocks. Each tissue block was cut into 8 (range, 7–9) sections for a total of 2,295 sections. All sections of a specimen were reconstructed into a 3D histology, which was registered and fused with the corresponding MR images.
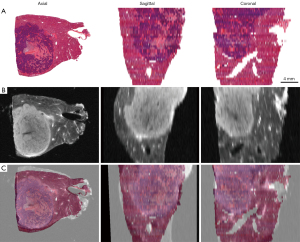
3D histology-MR image fusion was successfully performed in 39 specimens, and the success rate was 97.5% (39/40). All registration landmarks were fused with less than 1 mm of error in the 39 specimens, and the average registration error was 0.44±0.15 mm. Registration failed for 1 specimen because the tissue was significantly deformed; the registration error was 1.17 mm. Figure 5 shows the fusion of 3D histology and MR images in the axial, sagittal, and coronal planes, and the internal structures and histological contours were well registered. Figure 6 shows the specimen for which registration failed, as subjectively judged by 2 radiologists independently.
MVI localization on MRI
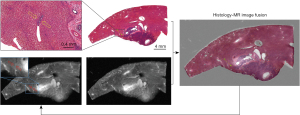
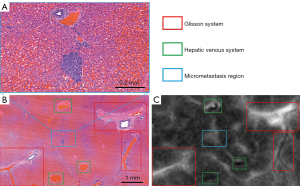
The histological results showed that MVI was found and identified in 8 of the 10 rabbits (i.e., the incidence of MVI was 80%) by the hepatic pathologists. MVI was detected in 20 of the 39 successfully registered specimens, resulting in a total of 166 MVI lesions, for which the median maximum diameter measured on pathological images was 0.29 (range, 0.08–0.96) mm. After the registration and fusion of 3D histology and MR images, the specific location of all MVI lesions on the MR images could be confirmed through the MVI displayed on the pathological images (Figure 7). As shown in Figure 8, the intrahepatic vascular structures were accurately registered on both histology and MR images, allowing micrometastasis to then be precisely localized on MR images. All MVI lesions showed as slightly hyperintense on the high-resolution MR T2-weighted images. The slight hyperintensity was similar to that of the main tumor but could not be distinguished from that of the surrounding vascular structures by naked-eye observations. Therefore, the above results will be used as the foundation for further research involving the extraction of the radiomics features of MVI.
Assessment of influencing factors
Table 1 shows the comparison of the thickness in 4 directions of 50 tissue blocks before and after dehydration. The average thicknesses of the 50 tissue blocks before dehydration were 3.37±0.13, 3.34±0.11, 3.38±0.13, and 3.37±0.11 mm for the front, back, left, and right directions, respectively, and there was no significant difference among the directions (P=0.38). Similarly, the average thicknesses after dehydration were 2.51±0.13, 2.49±0.16, 2.54±0.13, and 2.53±0.15 mm for the front, back, left, and right directions, respectively, and there was no significant difference among the directions (P=0.35). However, statistical analysis showed differences before and after dehydration (all P values <0.001). Table 2 shows the thickness ratios between any 2 directions of the 50 tissue blocks before and after dehydration, and none of the differences among the ratios were significant (both P values =0.09). The above results demonstrate that the thickness of the tissue block was uniform after sampling and after dehydration.
Table 1
| Direction | Before dehydration (mm) | After dehydration (mm) | P value |
|---|---|---|---|
| Front | 3.37±0.13 | 2.51±0.13 | <0.001 |
| Back | 3.34±0.11 | 2.49±0.16 | <0.001 |
| Left | 3.38±0.13 | 2.54±0.13 | <0.001 |
| Right | 3.37±0.11 | 2.53±0.15 | <0.001 |
| P value | 0.38 | 0.35 | NA |
Data are presented as the mean ± standard deviation. NA, not applicable.
Table 2
| Thickness ratio | Before dehydration | After dehydration |
|---|---|---|
| Front/back | 1.01±0.04 | 1.01±0.07 |
| Front/left | 1.00±0.05 | 0.99±0.05 |
| Front/right | 1.00±0.04 | 0.99±0.06 |
| Back/left | 0.99±0.03 | 0.98±0.05 |
| Back/right | 0.99±0.03 | 0.99±0.06 |
| Left/right | 1.01±0.04 | 1.01±0.06 |
| P value | 0.09 | 0.09 |
Data are presented as the mean ± standard deviation.
Table 3 shows the comparison of the shrinkage rates of 50 tissue blocks before and after dehydration. The mean length, width, and thickness of the 50 tissue blocks before dehydration were 20.97±2.70, 12.64±2.77, and 3.37±0.09 mm, respectively; after dehydration, the mean length, width, and thickness were 16.40±1.89, 9.75±1.99, and 2.52±0.11 mm, respectively. The average shrinkage rates of the 50 tissue blocks were 0.22±0.04, 0.22±0.05, and 0.25±0.03 for the length, width, and thickness, respectively, and the differences among them were statistically significant (P<0.001). Post hoc pairwise comparisons with Bonferroni correction showed that the shrinkage rate of the thickness of the tissue block was higher than that of its length and width (P<0.001 and P=0.003), but there was no significant difference in the shrinkage rate between the length and width (P=0.73).
Table 3
| Dimension | Before dehydration (mm) | After dehydration (mm) | Shrinkage rate |
|---|---|---|---|
| Length | 20.97±2.70 | 16.40±1.89 | 0.22±0.04*** |
| Width | 12.64±2.77 | 9.75±1.99 | 0.22±0.05** |
| Thickness | 3.37±0.09 | 2.52±0.11 | 0.25±0.03 |
Data are presented as the mean ± standard deviation. ***, length vs. thickness (P<0.001); **, width vs. thickness (P<0.05).
Discussion
In the clinical setting, pathology has retained its status as the definitive diagnostic modality for diseases. The imaging attributes demonstrate a strong association with the histopathological features of the disease. The main objective of medical practitioners is to continuously explore and interpret the imaging manifestations of human diseases through pathology. This may contribute to achieving a correct preoperative diagnosis and improving the radiological diagnosis. In most previous studies, the imaging features of a class of lesions were summarized and grouped according to pathological findings (16-18). The limitation of this method is that the pathological section does not always correspond to the imaging planes.
Recently, WSI has become widespread in oncology studies based on the development of histological slide digitization and computational image processing. WSI can be used to acquire a full high-resolution digital slide image of a histological section and has been applied in various scientific fields (19-21). Yu et al. (21) created a novel pathological examination method called image-matching digital macroscopic histological slides (IDS). The results of their study showed that the macroscopic histological slides could be matched with the corresponding imaging data from preoperative MRI (T2-weighted imaging) to obtain the MVI positions in IDS. However, similar to other studies (22,23), these studies also had some limitations. First, a small number of WSIs were limited to 2D registration, and correspondence with 3D imaging could not be established. Second, the imaging plane and pathological sections were only coarsely matched and contrasted, and an accurate image registration method was not implemented.
Due to the tedious fabrication procedures for the pathological sections, the registration and fusion of 3D pathological and radiological images have been less commonly reported. Commandeur et al. (24) registered the prostate 3D WSI to the preoperative MRI from 3 patients with prostate cancer. The results showed that accurate matching between 3D WSI and MRI was achieved with a mean final error in landmark positioning of 4.90±1.34 mm. Although MR images of the organ in vivo were used, the accuracy of the registration was not sufficient for clinical application. In our study, we registered and fused 3D WSI hepatic histology to ex vivo MRI, which produced successful matches in 39 specimens, yielding success rate of 97.5%. Most importantly, all registration landmarks were fused with less than 1 mm of error for the 39 specimens, and the mean final error was only 0.44±0.15 mm, which is significantly better than the results from studies reported in a literature review (25). A single case, with a registration error of 1.17 mm, was judged to be a failure. Therefore, we recommend defining successful 3D histology-MR image fusion as a registration error of less than 1 mm, which is narrower than that of other studies that have defined less than 2 mm as the standard (26,27).
This study employed high-resolution MRI with a resolution of 94 µm and a slice thickness of 0.8 mm to localize MVI. Despite a slightly higher signal intensity, the radiological characteristics of MVI lesions could not be distinguished from those of surrounding tissue structures due to their small size. This finding is consistent with the current clinical reality that MVI cannot be diagnosed through imaging alone. Our study aimed to establish a methodology for accurately locating MVI on imaging to enable future extraction of radiomics features from MVI. However, the median maximum diameter of MVI lesions was only 0.29 mm, and the average registration error of image fusion was 0.44 mm, leaving the possibility open that MVI lesions could be missed. Moreover, if only the MVI lesion is delineated as the region of interest (ROI), it may be too small for valuable radiomics features to extracted. To address these challenges, we recommend delimiting the ROI of the surrounding 1-mm area with the center of the located MVI lesion, thereby creating a 3D ROI with a diameter of 2 mm that encompasses the MVI lesion within the range of allowable registration error. Then, all the radiomics features of the ROI containing MVI can extracted, and the radiomics signature of MVI can screened by an artificial intelligence algorithm to finally construct the MVI diagnostic model.
The New Zealand white rabbits used in this study are large relative to other extant animals and are currently the most commonly used for developing experimental animal models of liver cancer (28). The rabbit liver VX2 tumor is convenient for imaging observation and suitable for imaging research of human HCC (29). The present results showed that the rabbit liver VX2 tumor grew rapidly and had a high tumor-forming rate. Moreover, it showed a similar blood supply and occurrence of MVI to human HCC. In this study, the incidence of MVI was higher than that reported by Wang et al. (30) (80% vs. 64%), and this may be attributable to the large size of the tumor and the large number of pathological sections. However, the hepatic architecture of rabbits is characterized by its comparatively thin structure and susceptibility to significant alterations in morphology and positioning due to visceral compression, particularly by the stomach. Furthermore, the impact of cardiac and respiratory movements adds further complexity to this issue. Therefore, to control for influencing factors, we chose to perform ex vivo imaging registration.
Sampling has the highest variability among the series of processes required for making histological slides. If the tissue blocks obtained are irregular, then complete and accurate 3D histology cannot be derived from the serial sections. Therefore, we used a homemade tool to make parallel and equal-thickness tissue blocks of the liver. The results showed that there was no significant difference among the thicknesses of the tissue blocks in the 4 directions, indicating that the tissue blocks had uniform thickness. Thus, the above results could meet the requirements for this experiment. Furthermore, the histological serial sections of the entire liver lobe made in this study were evenly spaced, similar to the slice thickness or spacing on consecutive CT/MRI tomograms. The spacing of pathological sections in this study was smaller than that of MRI, allowing precise registration to the MR images. Tissue shrinkage is another factor affecting the registration accuracy after dehydration. The results included a high shrinkage rate for tissue thickness, indicating that thinner tissue is more dehydrated. Although the shrinkage rate was not consistent, the registration accuracy was not greatly affected. The reason for this may be that the transformations and corrections were performed during the registration process.
To the best of our knowledge, this is the first study on 3D histology-MR image fusion of the liver. The results of our study indicate that preliminary methodological exploration for accurately matching 3D hepatic histology with MRI is feasible and accurate. The accurate coregistration of histology and MRI data could potentially improve the accuracy of diagnosis, treatment planning, and monitoring of treatment response. Additionally, our work may have implications for the development of new imaging biomarkers for HCC, which could ultimately improve patient outcomes, and may be helpful in imaging-based differential diagnosis in future clinical work. Moreover, by incorporating histology into the 3D reconstruction, we could also analyze the spatial distribution of the tumor and vessels and the correlation between vascularization, proliferation, hypoxia, and aggressiveness. In addition, this technology can also be applied to other relatively fixed organs (such as the brain and prostate) and other imaging modalities (such as ultrasound, CT, and MRI with other sequences).
This study had the following limitations. First, a large number of pathological sections were generated, and thus subsequent image processing required substantial manpower. However, automation of tissue processing and artificial intelligence-based image processing may improve the efficiency of our technique and save considerable manpower in the future. Second, we intermittently serially sectioned the tissue samples with a 300-µm interval, producing notable pathological tissue loss. Slide-free, 3D histology via hepatic tissue clearing and target structure labeling will hopefully become available in the future, providing more comprehensive histological information and reducing the tedious processes of routine pathological sectioning. Finally, and most importantly, image fusion was performed with ex vivo rather than in vivo livers. Unlike in vivo livers, ex vivo livers are not perfused with blood. In future investigations, we nevertheless expect that image fusion of preoperative imaging and postoperative pathological imaging will be carried out for the peritumoral local ROIs in human livers.
Conclusions
In this study, we successfully and precisely registered 3D histology with MR images for an isolated liver tumor model and demonstrated that this technique is feasible and accurate. Then, we accurately identified the specific location of MVI on MR images, laying a foundation for the next step of extracting the radiomics features of MVI.
Acknowledgments
Funding: This work was supported by
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-220/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Experiments were performed under a project license (No. L202104001) granted by the institutional ethics board of Zhuhai BesTest Bio-Tech Co., Ltd., in compliance with institutional guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Villanueva A. Hepatocellular Carcinoma. N Engl J Med 2019;380:1450-62. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Department of Medical Administration, National Health and Health Commission of the People’s Republic of China. Guidelines for diagnosis and treatment of primary liver cancer in China (2019 edition). Chin J Hepatol 2020;28:112-28.
- Vitale A, Peck-Radosavljevic M, Giannini EG, Vibert E, Sieghart W, Van Poucke S, Pawlik TM. Personalized treatment of patients with very early hepatocellular carcinoma. J Hepatol 2017;66:412-23. [Crossref] [PubMed]
- Wang H, Wu MC, Cong WM. Microvascular invasion predicts a poor prognosis of solitary hepatocellular carcinoma up to 2 cm based on propensity score matching analysis. Hepatol Res 2019;49:344-54. [Crossref] [PubMed]
- Large-Scale A. Multicenter Study. Oncologist 2019;24:e1476-88. Zhang XP, Wang K, Wei XB, Li LQ, Sun HC, Wen TF, Chai ZT, Chen ZH, Shi J, Guo WX, Xie D, Cong WM, Wu MC, Lau WY, Cheng SQ An Eastern Hepatobiliary Surgery Hospital Microvascular Invasion Scoring System in Predicting Prognosis of Patients with Hepatocellular Carcinoma and Microvascular Invasion After R0 Liver Resection:. [Crossref] [PubMed]
- Zhang XP, Zhou TF, Wang ZH, Zhang F, Zhong CQ, Hu YR, Wang K, Chai ZT, Chen ZH, Wu MC, Lau WY, Cheng SQ. Association of Preoperative Hypercoagulability with Poor Prognosis in Hepatocellular Carcinoma Patients with Microvascular Invasion After Liver Resection: A Multicenter Study. Ann Surg Oncol 2019;26:4117-25. [Crossref] [PubMed]
- Miyata R, Tanimoto A, Wakabayashi G, Shimazu M, Nakatsuka S, Mukai M, Kitajima M. Accuracy of preoperative prediction of microinvasion of portal vein in hepatocellular carcinoma using superparamagnetic iron oxide-enhanced magnetic resonance imaging and computed tomography during hepatic angiography. J Gastroenterol 2006;41:987-95. [Crossref] [PubMed]
- Li L, Wu C, Huang Y, Chen J, Ye D, Su Z. Radiomics for the Preoperative Evaluation of Microvascular Invasion in Hepatocellular Carcinoma: A Meta-Analysis. Front Oncol 2022;12:831996. [Crossref] [PubMed]
- Jiang C, Zhao L, Xin B, Ma G, Wang X, Song S. (18)F-FDG PET/CT radiomic analysis for classifying and predicting microvascular invasion in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Quant Imaging Med Surg 2022;12:4135-50. [Crossref] [PubMed]
- Wang X, Sun Y, Zhou X, Shen Z, Zhang H, Xing J, Zhou Y. Histogram peritumoral enhanced features on MRI arterial phase with extracellular contrast agent can improve prediction of microvascular invasion of hepatocellular carcinoma. Quant Imaging Med Surg 2022;12:1372-84. [Crossref] [PubMed]
- Abi-Jaoudeh N, Kruecker J, Kadoury S, Kobeiter H, Venkatesan AM, Levy E, Wood BJ. Multimodality image fusion-guided procedures: technique, accuracy, and applications. Cardiovasc Intervent Radiol 2012;35:986-98. [Crossref] [PubMed]
- Zhong-Zhen S, Kai L, Rong-Qin Z, Er-Jiao X, Ting Z, Ao-Hua Z, Shu-Fang Y, Xu-Qi H. A feasibility study for determining ablative margin with 3D-CEUS-CT/MR image fusion after radiofrequency ablation of hepatocellular carcinoma. Ultraschall Med 2012;33:E250-5. [Crossref] [PubMed]
- Yi HM, Cai BH, Ai X, Li KY, Zhang W. Establishment of Rabbit Liver VX2 Tumor Model Using Percutaneous Puncture Inoculation of Tumor Fragment Guided and Evaluated by Ultrasonography. Curr Med Sci 2019;39:820-4. [Crossref] [PubMed]
- Cheng Z, Zhang S, Wang L, Huang Z, Wang P, Zhu H, Wei Z, Zhou S. Ultrasound-guided percutaneous implantation of rabbit VX2 carcinoma, using a coaxial technique and gelfoam pellet injection combination to establish a rabbit liver tumor model. Diagn Interv Radiol 2022;28:103-7. [Crossref] [PubMed]
- Cui Y, Yang X, Du X, Zhuo Z, Xin L, Cheng X. Whole-tumour diffusion kurtosis MR imaging histogram analysis of rectal adenocarcinoma: Correlation with clinical pathologic prognostic factors. Eur Radiol 2018;28:1485-94. [Crossref] [PubMed]
- Takita H, Takeshita T, Shimono T, Tanaka H, Iguchi H, Hashimoto S, Kuwae Y, Ohsawa M, Miki Y. Cystic lesions of the parotid gland: radiologic-pathologic correlation according to the latest World Health Organization 2017 Classification of Head and Neck Tumours. Jpn J Radiol 2017;35:629-47. [Crossref] [PubMed]
- Li Q, Fan X, Huang XT, Luo TY, Chu ZG, Chen L, Zhang ZW, Li YQ, Wu JQ, Ouyang Y, Lv FJ, Ma CH. Tree-in-bud pattern in central lung cancer: CT findings and pathologic correlation. Lung Cancer 2015;88:260-6. [Crossref] [PubMed]
- Niazi MKK, Parwani AV, Gurcan MN. Digital pathology and artificial intelligence. Lancet Oncol 2019;20:e253-61. [Crossref] [PubMed]
- Hanna MG, Parwani A, Sirintrapun SJ. Whole Slide Imaging: Technology and Applications. Adv Anat Pathol 2020;27:251-9. [Crossref] [PubMed]
- Yu HM, Wang K, Feng JK, Lu L, Qin YC, Cheng YQ, Guo WX, Shi J. Hepatol Int 2022;16:381-95. [Crossref] [PubMed]
- Metzger GJ, Kalavagunta C, Spilseth B, Bolan PJ, Li X, Hutter D, Nam JW, Johnson AD, Henriksen JC, Moench L, Konety B, Warlick CA, Schmechel SC, Koopmeiners JS. Detection of Prostate Cancer: Quantitative Multiparametric MR Imaging Models Developed Using Registered Correlative Histopathology. Radiology 2016;279:805-16. [Crossref] [PubMed]
- Kalavagunta C, Zhou X, Schmechel SC, Metzger GJ. Registration of in vivo prostate MRI and pseudo-whole mount histology using Local Affine Transformations guided by Internal Structures (LATIS). J Magn Reson Imaging 2015;41:1104-14. [Crossref] [PubMed]
- Commandeur F, Acosta O, Simon A, Mathieu R, Fautrel A, Gnep K, Haigron P, de Crevoisier R. Prostate whole-mount histology reconstruction and registration to MRI for correlating in-vivo observations with biological findings. Annu Int Conf IEEE Eng Med Biol Soc 2015;2015:2399-402. [Crossref] [PubMed]
- Alyami W, Kyme A, Bourne R. Histological Validation of MRI: A Review of Challenges in Registration of Imaging and Whole-Mount Histopathology. J Magn Reson Imaging 2022;55:11-22. [Crossref] [PubMed]
- Li K, Su Z, Xu E, Huang Q, Zeng Q, Zheng R. Evaluation of the ablation margin of hepatocellular carcinoma using CEUS-CT/MR image fusion in a phantom model and in patients. BMC Cancer 2017;17:61. [Crossref] [PubMed]
- Liu Y, Yao D, Zhai Z, Wang H, Chen J, Wu C, Qiao H, Li H, Shi Y. Fusion of multimodality image and point cloud for spatial surface registration for knee arthroplasty. Int J Med Robot 2022;18:e2426. [Crossref] [PubMed]
- Doemel LA, Santana JG, Savic LJ, Gaupp FML, Borde T, Petukhova-Greenstein A, Kucukkaya AS, Schobert IT, Hamm CA, Gebauer B, Walsh JJ, Rexha I, Hyder F, Lin M, Madoff DC, Schlachter T, Chapiro J, Coman D. Comparison of metabolic and immunologic responses to transarterial chemoembolization with different chemoembolic regimens in a rabbit VX2 liver tumor model. Eur Radiol 2022;32:2437-47. [Crossref] [PubMed]
- Zhang W, Lowerison MR, Dong Z, Miller RJ, Keller KA, Song P. Super-Resolution Ultrasound Localization Microscopy on a Rabbit Liver VX2 Tumor Model: An Initial Feasibility Study. Ultrasound Med Biol 2021;47:2416-29. [Crossref] [PubMed]
- Wang Z, Wang W, Liu GJ, Yang Z, Chen LD, Huang Y, Li W, Xie XY, Lu MD, Kuang M. The role of quantitation of real-time 3-dimensional contrast-enhanced ultrasound in detecting microvascular invasion: an in vivo study. Abdom Radiol (NY) 2016;41:1973-9. [Crossref] [PubMed]


