
Nomogram based on quantitative dynamic contrast-enhanced magnetic resonance imaging, apparent diffusion coefficient, and clinicopathological features for early prediction of pathologic complete response in breast cancer patients receiving neoadjuvant chemotherapy
Introduction
Breast cancer is currently the most common malignant tumor in females, accounting for 11.7% of all malignant tumors. Moreover, breast cancer is one of the leading causes of death in women (1,2). Neoadjuvant chemotherapy (NACT) is the standard of care for locally advanced breast cancer. It is mainly used for reducing the primary tumor size and downstaging of disease, which facilitates breast conservation (3,4). Patients achieving pathologic complete response (pCR) after NACT show lower rates of distant recurrence and longer disease-free survival (5-7); the improved outcomes are particularly observed in patients with human epidermal growth factor receptor 2 (HER2)+ breast cancer and those with the more aggressive triple negative subtypes (5,8,9). The reported pCR rates range from 6% to 45% depending on the tumor subtype and the NACT regimen (10-13). Therefore, early detection of NACT non-responders is a key imperative in order to spare them from ineffective therapies and their potential side effects, and to facilitate personalized alterations in treatment regimen.
The National Comprehensive Cancer Network Clinical Practice Guidelines in Oncology-Breast Cancer (version 2.2022) recommend magnetic resonance imaging (MRI) for assessment of breast cancer response to NACT. Compared with mammography and ultrasonography, MRI offers higher soft-tissue resolution. Moreover, MRI not only delineates the morphological characteristics of lesions, but also reflects the pathophysiological state of the tissue through dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) and diffusion weighted imaging (DWI). In a previous study, DCE-MRI parameters were found to predict the efficacy of NACT, but most of these are semi-quantitative parameters. A few studies have focused on the use of quantitative DCE-MRI parameters combined with perfusion and diffusion quantitative parameters to identify early predictors of pCR in breast cancer patients after NACT (14-21).
The pCR rate differs in different tumor subtypes. Moreover, several factors such as estrogen receptor (ER) negativity, high Ki67, low T stage, and high histologic grade have been shown to be associated with pCR. To better predict pCR, an increasing number of clinical studies have developed clinical-pathological variables-based nomogram models, in which complex logistic regression models were transformed into simple and visual graphical models that include multiple risk factors. Therefore, several studies have developed nomograms based on clinical-pathologic variables to predict pCR (22-25). However, there is a paucity of well-designed nomograms for predicting the probability of pCR. Therefore, the purpose of this study was to develop nomogram incorporating clinical-pathologic variables, quantitative DCE-MRI parameters, and ADC values obtained before and after 2 cycles of NACT to estimate the probability of pCR in breast cancer patients after NACT. We present this article in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-869/rc).
Methods
Patients
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of Fujian Provincial Hospital (No. K2021-05-007, May 2019). Informed consent was obtained from all subjects involved in the study. In total, 215 patients with suspected breast cancer between July 2019 and January 2022 were prospectively collected in our hospital. The inclusion criteria were: (I) no needle biopsy, radiotherapy, or chemotherapy before MRI examination; (II) availability of complete MRI review data before NAT and within 3 weeks after NAT2 cycle treatment, with good image quality; (III) availability of complete pathological data; (IV) provision of written informed consent by patients before examination. The exclusion criteria are presented in Figure 1.

Neoadjuvant chemotherapy regimen
NACT was formulated in accordance with the institution’s standard regimen (11). The chemotherapy regimen was intravenously administered every 3 weeks for 6–8 cycles. The chemotherapy regimens of 74 female patients were as follows: 21 patients received TCbHP regimen (docetaxel, carboplatin, trastuzumab, pertuzumab); 12 patients received AC-THP regimen (epirubicin, cyclophosphamide, paclitaxel, trastuzumab, pertuzumab); 12 patients received AC-T regimen (epirubicin, cyclophosphamide, paclitaxel); 11 patients received TAC regimen (paclitaxel or docetaxel, epirubicin, cyclophosphamide); 9 patients received AC regimen (epirubicin, cyclophosphamide); 3 patients received AC-TP regimen (epirubicin, cyclophosphamide, paclitaxel, carboplatin); 2 patients received AC-H regimen (Epirubicin, cyclophosphamide, trastuzumab); 2 patients received TP regimen (paclitaxel, carboplatin); and 1 patient received THP regimen (paclitaxel, trastuzumab, pertuzumab) and 1 patient received AT regimen (epirubicin, paclitaxel).
Image acquisition and analysis
All MR examinations were performed in a 3.0T MR (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany) with 18-channel dual breast-dedicated phase-controlled surface coil. All patients were scanned in the prone position, with breasts naturally suspended in the coil. The sequences included T1WI (TR/TE =6.03/2.82 ms, thickness =0.9 mm, number of slices =160, bandwidth =300 Hz/Px, FOV read =340 mm, FOV phase =100%, matrix size =403×448), fat saturation T2WI (TR/TE =3,730/69 ms, thickness =4 mm, number of layers =35, bandwidth =246 Hz/Px, FOV read =340 mm, FOV phase =100%, matrix size =384×384, averages =2, concatenations =2), and dynamic contrast enhanced MRI (DCE-MRI) (TR/TE =4.03/1.33 ms, thickness =1.5 mm, number of slices =112, bandwidth =1,120 Hz/Px, FOV read =350 mm, FOV phase =100%, matrix size =259×320, measurements 36, volume: 350 mm × 350 mm × 1.5 mm × 112, scan time =343 s). The parameters of multiple b-value DWI sequences were TR/TE =5,700/62 ms, layer thickness =4 mm, number of layers =35, bandwidth =2,024 Hz/Px, FOV read =340 mm, FOV phase =60%, matrix size =114×190; b-values =0, 30, 50, 80, 120, 160, 200, 500, 1,000, 1,500, 2,000 s/mm2, averages =1, 1, 1, 1, 1, 1, 1, 1, 2, 2, 3; scan time =308 s. The contrast agent used was gadopentetate meglumine injection (Magnevist, 0.2 mmol/kg; GE Healthcare), which was injected through the dorsal vein of the hand through a high-pressure syringe at an injection rate of 1.5–2.0 mL/s; 15–20 mL of normal saline was injected to flush the connecting tube of the residual contrast agent.
Image analysis
Data post-processing was performed by two radiologists with 10 and 3 years of experience in breast tumor MRI diagnosis, respectively. On the Siemens workstation (Siemens Syngo.Via MR workspace), the solid part with the largest early enhancement in the lesion area was selected and the region of interest (ROI) was manually delineated, with a size of 27–193 mm2, avoiding areas with obvious necrosis, cystic degeneration, and/or liquefaction. We used the two compartments Tofts model in Tissue 4D module of Siemens Syngo.Via workstation. This model estimated three parameters, including Ktrans (transfer constant), Kep (rate constant), and Ve (relative extravascular extracellular space). In this work, the Tofts model was used as the pharmacokinetic model, and the population-based arterial input function preset was based on gamma-variate approximations (26-29). Rate of change in Ktrans, Kep, and Ve after 2 cycles of NACT was calculated as follows: ΔKtrans%, ΔKep%, ΔVe% = [(parameter value after 2 cycles of NACT minus parameter value of baseline map)/parameter value of baseline map] ×100%. The DWI images were post-processed using the Siemens workstation (body diffusion toolbox), and b=0 and 1,000 s/mm2 were calculated to obtain apparent diffusion coefficient (ADC) (30). On the ADC map, an ROI with a size of 29–203 mm2 was delineated on the largest layer of the solid component of the lesion displaying the lowest ADC-values, avoiding areas with obvious necrosis, cystic degeneration, and liquefaction. If the lesion showed complete response after 2 cycles of NACT, the ROI was delineated at the same level and location where the lesion was in pre-NACT images. All the above parameters were independently measured twice by two physicians respectively, and the average value was taken. ADC change rate after NACT2 cycle: ΔADC% = [(ADC value after 2 cycles of NACT minus baseline ADC value)/baseline ADC value] ×100%. Tumor size is the largest tumor diameter measured on early post-contrast images (120 s after contrast injection). Rate of change in tumor size ΔDmax% = [(maximum tumor diameter after 2 cycles of NACT minus maximum tumor diameter at baseline)/maximum tumor diameter at baseline] ×100%.
Histopathologic analysis
The status of ER, progesterone receptor (PR), HER2, and Ki67 index were determined by immunohistochemical examination (IHC) of breast cancer tissue collected from pre-NACT needle core biopsy. ER and PR are collectively referred to as hormone receptors (HR). According to the 2021 version of the Chinese Society of Clinical Oncology (CSCO) guidelines, HR positive status (i.e., ER or PR positive) is defined as nuclear immunostaining of ≥1% tumor cells. HR-negative status (i.e., ER and PR negative) is defined as nuclear immunostaining of <1% tumor cells. Positive HER2 expression was defined as HER2 membrane immunostaining score of 3+; if the membrane immunostaining score is 2+, in situ hybridization test is required to prove HER2 gene amplification. Definition of Ki67 positivity: ≥20% of tumor cell nuclei stained for Ki67. After 6–8 cycles of NACT, the surgically removed breast tissue and regional lymph nodes were fixed, then embedded in paraffin, and the specimens were cut into thin slices, stained with hematoxylin and eosin, and examined by a pathologist with several years of experience in breast tumor diagnosis. The residual situation of tumor cells was observed. According to the 2021 version of the CSCO guidelines (21), pCR is defined as no invasive carcinoma in the primary breast (ductal carcinoma in situ may be present) and regional lymph node negative, that is, the primary tumor is Miller-Payne (MP) grade 5 and lymph node negative, or residual tumor burden [residual cancer burden (RCB)] evaluation system grade 0.
Statistics
SPSS v25.0 and R software (4.0.0) were used for statistical analysis. The normality of distribution and homogeneity of variance of continuous variables were assessed by Shapiro-Wilk test and Levene variance test, respectively. Normally-distributed continuous variables were expressed as mean ± standard deviation (SD), and between-group differences were assessed using two independent samples t-test. Non-normally distributed continuous variables were expressed as median (upper and lower quartiles) and compared using the Mann-Whitney U test. Categorical variables were described as frequency (percentage) and the between-group differences were assessed using Pearson’s Chi-squared and continuous-corrected Chi-squared test. Variables associated with P values <0.10 in univariate analysis were included in the multivariate binary logistic regression analysis by stepwise method to establish the best combined prediction model. Taking the pathological pCR results as the gold standard, receiver operating characteristic (ROC) curve analysis was performed to evaluate the predictive value of univariate and combined model to predict pCR after NACT, and the area under the ROC curve (AUC) was calculated. The cutoff value was chosen according to the maximum Youden index. DeLong’s test or Bootstrap test was used to compare the differences between univariate and combined models. A nomogram model for predicting pCR was established based on a multivariate logistic regression model. Nomogram performance was assessed using AUC and calibration plots. A calibration plot is a graphical assessment of the predictive power by comparing the observed probabilities with nomogram-predicted probabilities. The ROC curve of the predictive model was plotted and the AUC was calculated to evaluate the predictive ability.
Results
Clinicopathological characteristics
A total of 74 female patients with stages II–III breast cancer were included. The mean age of patients was 49.7±10.2 years (range, 27–73 years). Data pertaining to all clinicopathological variables were collected before NACT. The pathological diagnosis was invasive ductal carcinoma in 70 cases (Figure 2), invasive lobular carcinoma in 3 cases, and metaplastic carcinoma in 1 case. Luminal A (5/74, 6.8%), Luminal B (38/74, 51.4%), HER2+ (21/74, 28.4%), Basal-like (10/74, 13.5%). HR-negative patients (16/25, 64.0%), HER2-positive patients (18/25, 72.0%) and Ki67-positive patients (24/25, 96.0%) in the pCR group were all significantly more than those in the non-pathologic complete response (npCR) group (P=0.006, P=0.002, P=0.043, respectively). There was no significant difference between pCR and npCR groups with respect to age or lesion morphology (Table 1).

Table 1
| Characteristics | Total numbers (n=74) | npCR group (n=49) | pCR group (n=25) | t/χ2 | P value |
|---|---|---|---|---|---|
| Age* (years) | 49.7±10.2 | 50.4±9.9 | 48.6±10.9 | −0.687 | 0.496 |
| Lesion morphology** | 0.003 | 0.959 | |||
| Mass-like | 53 (71.62) | 35 (71.43) | 18 (72.00) | ||
| Non-mass-like | 21 (28.38) | 14 (28.57) | 7 (28.00) | ||
| HR** | 7.581 | 0.006 | |||
| Negative | 31 (41.89) | 15 (30.61) | 16 (64.00) | ||
| Positive | 43 (58.11) | 34 (69.39) | 9 (36.00) | ||
| HER2** | 9.243 | 0.002 | |||
| Negative | 39 (52.70) | 32 (65.31) | 7 (28.00) | ||
| Positive | 35 (47.30) | 17 (34.69) | 18 (72.00) | ||
| Ki67*** | 4.108 | 0.043 | |||
| Negative | 14 (18.92) | 13 (26.53) | 1 (4.00) | ||
| Positive | 60 (81.08) | 36 (73.47) | 24 (96.00) |
Data are presented as mean ± standard deviation or n (%). *, independent two-sample t-test; **, Pearson χ2 test; ***, continuous corrected χ2 test. P values: comparisons between pCR and npCR. pCR, pathologic complete response; HR, hormone receptor; HER2, human epidermal growth factor 2.
Comparison of DCE-MRI quantitative parameters and ADC values between pCR and npCR groups by univariate analysis
Before NACT, The Ktrans value in the pCR group [0.29 (0.24, 0.32) min−1] was significantly higher than that in the npCR group [0.24 (0.20, 0.29) min−1 (P=0.033)]. The Kep value in the pCR group [0.96 (0.85, 1.20) min−1] was slightly higher than that in npCR group [0.83 (0.64, 1.04) min−1]; however, the difference was not statistically significant (P=0.052). There were no significant differences between the pCR and npCR groups with respect to Ve or ADC values (P=0.172 and P=0.622, respectively).
After 2 cycles of NACT, significant difference of ΔKtrans%, ΔKep%, and ΔADC% were found in pCR group, compared to npCR group (P≤0.001). These findings suggested the potential predictive value of ΔKtrans%, ΔKep%, and ΔADC% for pCR. No significant difference of ΔDmax% and ΔVe% was observed between the pCR and npCR groups (P>0.05) (Table 2).
Table 2
| Parameters | npCR (n=49) | pCR (n=25) | t/U | P value |
|---|---|---|---|---|
| Ktrans** (min−1) | 0.24 (0.20, 0.29) | 0.29 (0.24, 0.32) | 426 | 0.033 |
| Kep** (min−1) | 0.83 (0.64, 1.04) | 0.96 (0.85, 1.20) | 442 | 0.052 |
| Ve** | 0.28 (0.22, 0.37) | 0.25 (0.20, 0.34) | 732 | 0.172 |
| ADC* (×10−3 mm2/s) | 0.78±0.14 | 0.80±0.11 | −0.49 | 0.622 |
| ΔDmax%** (%) | −24.31 (−8.9, −33.1) | −26.79 (−11.1, −37.4) | 467 | 0.101 |
| ΔKtrans%* (%) | −13±0.46 | −62±0.16 | 6.68 | <0.001 |
| ΔKep%** (%) | −8.12 (−35.21, 20.54) | −42.34 (−68.76, −28.83) | 907 | 0.001 |
| ΔVe%** (%) | 7.23 (−20.01, 30.23) | −16.45 (−46.91, 34.67) | 723 | 0.210 |
| ΔADC%** (%) | 9.23 (−4.41, 25.52) | 44.56 (31.72, 61.73) | 234 | <0.001 |
Data are presented as mean ± standard deviation or median (upper and lower quartiles). *, independent two-sample t-test; **, Mann-Whitney U test. DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging; ADC, apparent diffusion coefficient; Ktrans, transfer constant; Kep, rate constant; Ve, relative extravascular extracellular space; pCR, pathologic complete response; NACT, neoadjuvant chemotherapy.
Intra- and interclass correlation analysis of the DCE-MRI quantitative parameters and ADC
The intraclass and interclass correlation coefficients (CCs) were used to analyze the consistency of the results of two measurements by the same physician and the first measurement by the two physicians. The values of ICC range from 0 to 1: ≥0.75 indicates good consistency, 0.50≤ ICC <0.75 indicates general consistency, and <0.50 indicates poor consistency. The results showed that Ktrans and Ve exhibited better consistency, compared with Kep and ADC (Table 3).
Table 3
| Variables | Ktrans | Kep | Ve | ADC |
|---|---|---|---|---|
| Interclass CC | 0.837 | 0.589 | 0.885 | 0.665 |
| Intraclass CC | 0.91 | 0.867 | 0.755 | 0.736 |
DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging; ADC, apparent diffusion coefficient; CC, correlation coefficient; Ktrans, transfer constant; Kep, rate constant; Ve, relative extravascular extracellular space.
Development of nomogram models
Clinicopathological variables and DCE-MRI quantitative parameters associated with P values <0.10 in the univariate analysis were included. After stepwise multivariate binary logistic regression analysis, the results showed that pre-NACT HER2, Ki67, and Ktrans were independent predictors of pCR. The HER2-Ki67-Ktrans based nomogram model 1 was established (Figure 3). Each predictive factor was awarded a score based on the upper scale line. The probability of pCR was shown when the total score was summed up from all the predictors. Its equation is ln(p/1−p) = −6.403 + 2.03 × HER2 + 2.536 × Ki67 + 8.965 × Ktrans (HER2 positive =1, Ki67 positive =1, HER2 negative =0, Ki67 negative =0).

Similarly, Ki67, ΔADC%, and ΔKtrans% were identified as independent predictors of pCR after 2 cycles of NACT. The Ki67-ΔADC%-ΔKtrans% based nomogram model 2 was established (Figure 4) and its equation is ln(p/1−p) = 2.433 × Ki67 + 2.562 × ΔADC% + −4.737 × ΔKtrans% (Ki67 positive =1, Ki67 negative =0).

Performance of pCR prediction by single-parameter and nomogram model
Among all single-parameters obtained before NACT and after 2 cycles of NACT, ΔKtrans% had the highest AUC value (area under the receiver operating characteristic curve): 0.818 (cutoff −36.894), suggesting the highest predictive performance of ΔKtrans%. The diagnostic sensitivity, specificity, positive predictive value, negative predictive value, and accuracy of ΔKtrans% were 100.0%, 69.4%, 62.5%, 100.0%, and 79.7%, respectively (Tables 4,5 and Figures 5,6).
Table 4
| Parameters | AUC | Sensitivity | Specificity | Positive predictive value | Negative predictive value | Positive likelihood ratio | Negative likelihood ratio | Odds ratio | Youden Index | Accuracy | Cutoff | P value# |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ktrans | 0.652 (0.520, 0.784) | 0.680 (0.465, 0.851) | 0.653 (0.504, 0.783) | 0.500 (0.324, 0.676) | 0.800 (0.644, 0.909) | 1.960 (1.226, 3.133) | 0.490 (0.267, 0.899) | 4.000 (1.434, 11.157) | 0.333 (0.107, 0.559) | 0.662 (0.543, 0.768) | 0.269 | <0.001* |
| Kep | 0.639 (0.505, 0.773) | 0.680 (0.465, 0.851) | 0.633 (0.483, 0.766) | 0.486 (0.314, 0.660) | 0.795 (0.635, 0.907) | 1.851 (1.174, 2.919) | 0.506 (0.275, 0.931) | 3.660 (1.318, 10.164) | 0.313 (0.085, 0.540) | 0.649 (0.529, 0.756) | 0.911 | <0.001* |
| Nomogram1 | 0.833 (0.736, 0.930) | 0.800 (0.593, 0.932) | 0.755 (0.611, 0.867) | 0.625 (0.437, 0.789) | 0.881 (0.744, 0.960) | 3.267 (1.924, 5.546) | 0.265 (0.119, 0.589) | 12.333 (3.803, 40.000) | 0.555 (0.357, 0.753) | 0.770 (0.658, 0.860) | 98.527 | – |
Data are presented as value (95% CI). #, compared with Nomogram1; *, Delong test. ROC, receiver operating characteristic; pCR, pathologic complete response; NACT, neoadjuvant chemotherapy; AUC, area under the receiver operating characteristic curve; CI, confidence interval; Ktrans, transfer constant; Kep, rate constant.
Table 5
| Parameters | AUC | Sensitivity | Specificity | Positive predictive value | Negative predictive value | Positive likelihood ratio | Negative likelihood ratio | Odds ratio | Youden Index | Accuracy | Cutoff | P value# |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔADC% | 0.809 (0.711, 0.907) | 0.800 (0.593, 0.932) | 0.776 (0.634, 0.882) | 0.645 (0.454, 0.808) | 0.884 (0.749, 0.961) | 3.564 (2.044, 6.214) | 0.258 (0.116, 0.573) | 13.818 (4.213, 45.318) | 0.576 (0.380, 0.771) | 0.784 (0.673, 0.871) | 26.509 | 0.048* |
| ΔKtrans% | 0.818 (0.723, 0.913) | 1.000 (0.863, 1.000) | 0.694 (0.546, 0.817) | 0.625 (0.458, 0.773) | 1.000 (0.897, 1.000) | 3.267 (2.143, 4.979) | 0.028 (0.002, 0.437) | 113.516 (6.486, 1986.724) | 0.694 (0.565, 0.823) | 0.797 (0.688, 0.882) | –36.894 | 0.041** |
| ΔKep% | 0.740 (0.625, 0.856) | 0.800 (0.593, 0.932) | 0.653 (0.504, 0.783) | 0.541 (0.369, 0.705) | 0.865 (0.712, 0.955) | 2.306 (1.498, 3.549) | 0.306 (0.136, 0.688) | 7.529 (2.401, 23.613) | 0.453 (0.247, 0.659) | 0.703 (0.585, 0.803) | –26.668 | 0.009** |
| ΔVe% | 0.590 (0.436, 0.744) | 0.560 (0.349, 0.756) | 0.694 (0.546, 0.817) | 0.483 (0.294, 0.675) | 0.756 (0.605, 0.871) | 1.829 (1.059, 3.159) | 0.634 (0.392, 1.025) | 2.885 (1.065, 7.813) | 0.254 (0.020, 0.487) | 0.649 (0.529, 0.756) | –14.512 | <0.001** |
| Nomogram2 | 0.900 (0.829, 0.970) | 0.920 (0.740, 0.990) | 0.837 (0.703, 0.927) | 0.742 (0.554, 0.881) | 0.953 (0.842, 0.994) | 5.635 (2.958, 10.733) | 0.096 (0.025, 0.363) | 58.938 (11.531, 301.246) | 0.757 (0.608, 0.905) | 0.865 (0.765, 0.933) | 104.803 | – |
Data are presented as value (95% CI). #, compared with Nomogram2; *, Delong test; **, Bootstrap test. ROC, receiver operating characteristic; pCR, pathologic complete response; NACT, neoadjuvant chemotherapy; AUC, area under the receiver operating characteristic curve; ADC, apparent diffusion coefficient; CI, confidence interval; Ktrans, transfer constant; Kep, rate constant; Ve, relative extravascular extracellular space.


The predictive performance of nomogram model 2 was significantly higher compared to the other single-parameter [AUC =0.900 (0.829, 0.970), P<0.001] (Table 5). The AUC value of nomogram model 2 was slightly higher than that of nomogram model 1 (0.900 and 0.833, respectively), but the difference was not statistically significant (P=0.174). nomogram model 1 was found to be the best model for predicting pCR before NACT, compared to Ktrans and Kep [AUC =0.833, 0.652, and 0.639, respectively) (Table 4). nomogram model 2 was the best model [AUC =0.900, P<0.05) for predicting pCR after 2 cycles of NACT, compared to other single-parameters (AUC values of ΔADC%, ΔKtrans%, ΔKep%, and ΔVe% were 0.809, 0.818, 0.740, and 0.590, respectively) (Table 5).
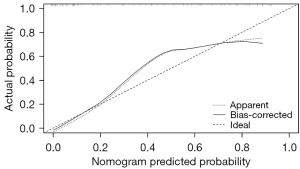
The calibration plots from both nomogram models showed a good fit of the calibration prediction curve with the ideal curve, suggesting a high predictive value of both models (Figures 7,8). In both nomogram models, the predicted pCR showed a good agreement with the observed pCR. Importantly, nomogram model 2 had a smaller mean absolute error of the agreement, compared with nomogram model 1 (0.017 vs. 0.051). This suggested that nomogram model 2 is a better tool to predict pCR with a better agreement with the observed pCR.

Discussion
The ability to accurately predict post-NACT pCR in patients with breast cancer can provide a distinct leverage during treatment decision-making and help improve patient prognosis. In this study, HR negative, HER2 positive, and Ki67 positive status were associated with pCR. Before NACT, higher Ktrans value was associated with pCR; after 2 cycles of NACT, higher absolute values of ΔKtrans%, ΔKep%, and ΔADC% were associated with pCR. Notably, we developed two nomograms based on the clinicopathological features, quantitative DCE-MRI parameters, and ADC values, before NACT and after 2 cycles of NACT, respectively. Both HER2-Ki67-Ktrans based nomogram model 1 (pre-NACT) and Ki67-ΔADC%-ΔKtrans% based nomogram model 2 (after 2 cycles of NACT) showed good predictive ability for pCR, which was better than that achieved with using quantitative DCE-MRI parameters alone.
pCR rate differs based on the clinicopathological status. In previous studies, high-grade tumor, HER2-positive, high Ki67, and ER negative status were associated with a higher likelihood of achieving pCR after NACT (8,31-33). Consistent with this, in our study, HR-negative, HER2-positive, and high Ki67 status before NACT showed a significant association with pCR after NACT.
However, in clinical practice, patients with the same tumor histological type and stage often show different treatment outcomes even if they receive the same NACT. Therefore, there is a need to include functional imaging combined with magnetic resonance imaging parameters to further improve the predictive performance. Quantitative DCE-MRI can help assess the exchange of contrast agent between blood vessels and intercellular spaces, and evaluate tissue perfusion and the integrity of the vascular endothelium within the tissue. Currently, the most commonly used pharmacokinetic model is the two-compartment model proposed by Tofts et al. (26) in 1991. The two compartments are the vascular space and the interstitial space [or extravascular extracellular space (EES)]. The model includes the following three parameters (27): (I) Ktrans (transfer constant): refers to volume transfer constant of gadolinium from blood plasma to the EES, which is related to microangiogenesis and vascular permeability (28); (II) Kep (rate constant): rate constant gadolinium reflux from the EES back into the vascular system; (III) Ve (extravascular extracellular volume): volume of the EES per unit volume of tissue (i.e., the amount of “space” available within the interstitium for accumulating gadolinium; it is a ratio: Ktrans/Kep), indicating the degree of cell necrosis and cellularization. The larger the extracellular space volume of the extracellular tissue, the lower the degree of cellularization or the greater the degree of cell necrosis. Among the pre-NACT quantitative DCE-MRI parameters in this study, higher Ktrans value showed a significant association with pCR. Wu et al. (28) also reported a strong correlation between Ktrans and tumor microvascular density (Pearson R2=0.451, P<0.05). Therefore, a higher Ktrans value indicates greater tumor microvascular density and vascular permeability, which leads to an accumulation of the therapeutic drug in the lesion. This facilitates the disintegration and necrosis of tumor cells, thereby achieving pCR. However, the predictive performance of Ktrans in our study was low [AUC =0.652) before NACT. There was no significant difference in Kep and Ve values between pCR and npCR groups, which is consistent with the results of most studies (14,34).
After 2 cycles of NACT treatment, there was no significant difference between pCR and npCR groups with respect to ΔDmax% (P>0.05), which indicated no changes in tumor size after 2 cycles of NACT. Notably, higher absolute values of ΔKtrans% and ΔKep% after 2 cycles of NACT were significantly associated with pCR (P<0.001). This suggested that quantitative ΔKtrans% and ΔKep% after 2 cycles of NACT can more readily reflect the pathophysiological changes occurring at an earlier stage than morphological changes, and can be used as early predictors of pCR. ΔVe% was not found to predict pCR, which may be related to the disintegration and necrosis of tumor cells and the formation of local fibrous tissue in the extracellular space after NACT (35).
DWI reflects the cell density and tissue microstructure based on the diffusion of water in tissue. The ADC value increases with decrease in the tumor cell density during NACT. In our study, absolute ΔADC% value after 2 cycles of NACT in the pCR group was significantly higher than that in the npCR group. Similar results have been reported previously (36). The pre-treatment ADC value was not found to predict pCR, which is also consistent with previous studies (19,20).
According to the univariate analysis and stepwise multivariate binary logistic regression analysis, the independent predictors before NACT were HER2, Ki67, and Ktrans, and those after 2 cycles of NACT were Ki67, ΔADC%, and ΔKtrans%. Two nomograms were developed based on different predictors obtained before and after 2 cycles of NACT. Both nomograms showed good predictive discrimination [AUC =0.833 and 0.900, before and after 2 cycles of NACT nomograms, respectively) and calibration ability (mean absolute error of the agreement =0.051 and 0.017, before and after 2 cycles of NACT nomograms, respectively). This indicated a better predictive ability of nomogram after 2 cycles of NACT. This study combined the clinicopathological characteristics and MR quantitative parameters to improve the prediction efficacy compared with the univariate analysis. It also further verified that the clinical-imaging-pathological multidisciplinary participation is needed to evaluate the efficacy of NACT in breast cancer patients.
Nevertheless, some limitations of our study should be acknowledged. First, the impact of different molecular subtypes of breast cancer, different quadrant, density, or depth of breast on the predictive efficacy for post-NACT pCR could not be analyzed due to the limited sample size. Second, this study was based on a single-center cohort, and the nomogram was not validated in an external cohort. Third, whether the earlier time point (e.g., after 1 cycle of NACT) could predict pCR worth our further investigation. Finally, this study selected the solid component of the largest level of the tumor to manually delineate the ROI, which may have introduced an element of selection bias. Since the overall lesion was not measured, it may not be representative of the overall tumor. Further multi-center studies with larger patient cohorts are required to verify the pCR prediction performance of nomogram model based on the full-volume VOI of the lesion.
Conclusions
Nomograms based on HER2, Ki67, and Ktrans and Ki67 pre-NACT, ΔKtrans%, and ΔADC% after 2 cycles of NACT can help predict pCR in breast cancer and inform individualized treatment decision-making.
Acknowledgments
We thank Medjaden Inc. for scientific editing of this manuscript.
Funding: This work was supported by the China International Medical Foundation (No. Z-2014-07-1912-23, to M He), and the Natural Science Foundation of Fujian Province, China (No. 2020J011057, to M He).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-869/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-869/coif). M He reports that this work was supported by the China International Medical Foundation (No. Z-2014-07-1912-23), and the Natural Science Foundation of Fujian Province, China (No. 2020J011057). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Fujian Provincial Hospital (No. K2021-05-007, May 2019). Informed consent was obtained from all subjects involved in the study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Henley SJ, Ward EM, Scott S, Ma J, Anderson RN, Firth AU, Thomas CC, Islami F, Weir HK, Lewis DR, Sherman RL, Wu M, Benard VB, Richardson LC, Jemal A, Cronin K, Kohler BA. Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer 2020;126:2225-49. [Crossref] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Burstein HJ, Curigliano G, Thürlimann B, Weber WP, Poortmans P, Regan MM, Senn HJ, Winer EP, Gnant MPanelists of the St Gallen Consensus Conference. Customizing local and systemic therapies for women with early breast cancer: the St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021. Ann Oncol 2021;32:1216-35. [Crossref] [PubMed]
- Halberg AK, Gravesen CD, Cold S, Jensen JD. Neoadjuvant chemotherapy for primary operable breast cancer. Dan Med J 2020; [PubMed]
- Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 2014;384:164-72. [Crossref] [PubMed]
- Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: meta-analysis of individual patient data from ten randomised trials. Lancet Oncol 2018;19:27-39. [Crossref] [PubMed]
- Eremin J, Cowley G, Walker LG, Murray E, Stovickova M, Eremin O. Women with large (≥3 cm) and locally advanced breast cancers (T3, 4, N1, 2, M0) receiving neoadjuvant chemotherapy (NAC: cyclophosphamide, doxorubicin, docetaxel): addition of capecitabine improves 4-year disease-free survival. Springerplus 2015;4:9. [Crossref] [PubMed]
- Blanco Sánchez A, Yébenes L, Berjón A, Hardisson D. Evaluation of pathological response to neoadjuvant chemotherapy in breast cancer: correlation with molecular phenotype. Rev Esp Patol 2021;54:8-16. [PubMed]
- Myers SP, Ahrendt GM, Lee JS, Steiman JG, Soran A, Johnson RR, McAuliffe PF, Diego EJ. Association of Tumor Molecular Subtype and Stage with Breast and Axillary Pathologic Complete Response After Neoadjuvant Chemotherapy for Breast Cancer. Ann Surg Oncol 2021;28:8636-42. [Crossref] [PubMed]
- Bonnefoi H, Litière S, Piccart M, MacGrogan G, Fumoleau P, Brain E, et al. Pathological complete response after neoadjuvant chemotherapy is an independent predictive factor irrespective of simplified breast cancer intrinsic subtypes: a landmark and two-step approach analyses from the EORTC 10994/BIG 1-00 phase III trial. Ann Oncol 2014;25:1128-36. [Crossref] [PubMed]
- von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012;30:1796-804. [Crossref] [PubMed]
- Zhang X, Wang D, Liu Z, Wang Z, Li Q, Xu H, Zhang B, Liu T, Jin F. The diagnostic accuracy of magnetic resonance imaging in predicting pathologic complete response after neoadjuvant chemotherapy in patients with different molecular subtypes of breast cancer. Quant Imaging Med Surg 2020;10:197-210. [Crossref] [PubMed]
- Zambetti M, Mansutti M, Gomez P, Lluch A, Dittrich C, Zamagni C, Ciruelos E, Pavesi L, Semiglazov V, De Benedictis E, Gaion F, Bari M, Morandi P, Valagussa P, Luca G. Pathological complete response rates following different neoadjuvant chemotherapy regimens for operable breast cancer according to ER status, in two parallel, randomized phase II trials with an adaptive study design (ECTO II). Breast Cancer Res Treat 2012;132:843-51. [Crossref] [PubMed]
- Kim Y, Kim SH, Song BJ, Kang BJ, Yim KI, Lee A, Nam Y. Early Prediction of Response to Neoadjuvant Chemotherapy Using Dynamic Contrast-Enhanced MRI and Ultrasound in Breast Cancer. Korean J Radiol 2018;19:682-91. [Crossref] [PubMed]
- Drisis S, Metens T, Ignatiadis M, Stathopoulos K, Chao SL, Lemort M. Quantitative DCE-MRI for prediction of pathological complete response following neoadjuvant treatment for locally advanced breast cancer: the impact of breast cancer subtypes on the diagnostic accuracy. Eur Radiol 2016;26:1474-84. [Crossref] [PubMed]
- Marinovich ML, Sardanelli F, Ciatto S, Mamounas E, Brennan M, Macaskill P, Irwig L, von Minckwitz G, Houssami N. Early prediction of pathologic response to neoadjuvant therapy in breast cancer: systematic review of the accuracy of MRI. Breast 2012;21:669-77. [Crossref] [PubMed]
- Stevens W, Farrow IM, Georgiou L, Hanby AM, Perren TJ, Windel LM, Wilson DJ, Sharma N, Dodwell D, Hughes TA, Dall BJ, Buckley DL. Breast tumour volume and blood flow measured by MRI after one cycle of epirubicin and cyclophosphamide-based neoadjuvant chemotherapy as predictors of pathological response. Br J Radiol 2021;94:20201396. [Crossref] [PubMed]
- Cho N, Im SA, Park IA, Lee KH, Li M, Han W, Noh DY, Moon WK. Breast cancer: early prediction of response to neoadjuvant chemotherapy using parametric response maps for MR imaging. Radiology 2014;272:385-96. [Crossref] [PubMed]
- Xu HD, Zhang YQ. Evaluation of the efficacy of neoadjuvant chemotherapy for breast cancer using diffusion-weighted imaging and dynamic contrast-enhanced magnetic resonance imaging. Neoplasma 2017;64:430-6. [Crossref] [PubMed]
- Richard R, Thomassin I, Chapellier M, Scemama A, de Cremoux P, Varna M, Giacchetti S, Espié M, de Kerviler E, de Bazelaire C. Diffusion-weighted MRI in pretreatment prediction of response to neoadjuvant chemotherapy in patients with breast cancer. Eur Radiol 2013;23:2420-31. [Crossref] [PubMed]
- Choi BB. Effectiveness of ADC Difference Value on Pre-neoadjuvant Chemotherapy MRI for Response Evaluation of Breast Cancer. Technol Cancer Res Treat 2021;20:15330338211039129. [Crossref] [PubMed]
- Rouzier R, Pusztai L, Delaloge S, Gonzalez-Angulo AM, Andre F, Hess KR, Buzdar AU, Garbay JR, Spielmann M, Mathieu MC, Symmans WF, Wagner P, Atallah D, Valero V, Berry DA, Hortobagyi GN. Nomograms to predict pathologic complete response and metastasis-free survival after preoperative chemotherapy for breast cancer. J Clin Oncol 2005;23:8331-9. [Crossref] [PubMed]
- Keam B, Im SA, Park S, Nam BH, Han SW, Oh DY, Kim JH, Lee SH, Han W, Kim DW, Kim TY, Park IA, Noh DY, Heo DS, Bang YJ. Nomogram predicting clinical outcomes in breast cancer patients treated with neoadjuvant chemotherapy. J Cancer Res Clin Oncol 2011;137:1301-8. [Crossref] [PubMed]
- Colleoni M, Bagnardi V, Rotmensz N, Viale G, Mastropasqua M, Veronesi P, Cardillo A, Torrisi R, Luini A, Goldhirsch A. A nomogram based on the expression of Ki-67, steroid hormone receptors status and number of chemotherapy courses to predict pathological complete remission after preoperative chemotherapy for breast cancer. Eur J Cancer 2010;46:2216-24. [Crossref] [PubMed]
- Xu W, Chen X, Deng F, Zhang J, Zhang W, Tang J. Predictors of Neoadjuvant Chemotherapy Response in Breast Cancer: A Review. Onco Targets Ther 2020;13:5887-99. [Crossref] [PubMed]
- Tofts PS, Kermode AG. Measurement of the blood-brain barrier permeability and leakage space using dynamic MR imaging. 1. Fundamental concepts. Magn Reson Med 1991;17:357-67. [Crossref] [PubMed]
- Tang L, Wang XJ, Baba H, Giganti F. Gastric cancer and image-derived quantitative parameters: Part 2-a critical review of DCE-MRI and (18)F-FDG PET/CT findings. Eur Radiol 2020;30:247-60. [Crossref] [PubMed]
- Wu Y, Yan Y, Gao X, Yang L, Li Y, Guo X, Xie J, Wang K, Sun X. Gd-encapsulated carbonaceous dots for accurate characterization of tumor vessel permeability in magnetic resonance imaging. Nanomedicine 2019;21:102074. [Crossref] [PubMed]
- Parker GJ, Roberts C, Macdonald A, Buonaccorsi GA, Cheung S, Buckley DL, Jackson A, Watson Y, Davies K, Jayson GC. Experimentally-derived functional form for a population-averaged high-temporal-resolution arterial input function for dynamic contrast-enhanced MRI. Magn Reson Med 2006;56:993-1000. [Crossref] [PubMed]
- Woodhams R, Matsunaga K, Iwabuchi K, Kan S, Hata H, Kuranami M, Watanabe M, Hayakawa K. Diffusion-weighted imaging of malignant breast tumors: the usefulness of apparent diffusion coefficient (ADC) value and ADC map for the detection of malignant breast tumors and evaluation of cancer extension. J Comput Assist Tomogr 2005;29:644-9. [Crossref] [PubMed]
- Petrelli F, Viale G, Cabiddu M, Barni S. Prognostic value of different cut-off levels of Ki-67 in breast cancer: a systematic review and meta-analysis of 64,196 patients. Breast Cancer Res Treat 2015;153:477-91. [Crossref] [PubMed]
- Kontzoglou K, Palla V, Karaolanis G, Karaiskos I, Alexiou I, Pateras I, Konstantoudakis K, Stamatakos M. Correlation between Ki67 and breast cancer prognosis. Oncology 2013;84:219-25. [Crossref] [PubMed]
- Esserman LJ, Berry DA, DeMichele A, Carey L, Davis SE, Buxton M, et al. Pathologic complete response predicts recurrence-free survival more effectively by cancer subset: results from the I-SPY 1 TRIAL--CALGB 150007/150012, ACRIN 6657. J Clin Oncol 2012;30:3242-9. [Crossref] [PubMed]
- Padhani AR, Hayes C, Assersohn L, Powles T, Makris A, Suckling J, Leach MO, Husband JE. Prediction of clinicopathologic response of breast cancer to primary chemotherapy at contrast-enhanced MR imaging: initial clinical results. Radiology 2006;239:361-74. [Crossref] [PubMed]
- Minarikova L, Bogner W, Pinker K, Valkovič L, Zaric O, Bago-Horvath Z, Bartsch R, Helbich TH, Trattnig S, Gruber S. Investigating the prediction value of multiparametric magnetic resonance imaging at 3 T in response to neoadjuvant chemotherapy in breast cancer. Eur Radiol 2017;27:1901-11. [Crossref] [PubMed]
- Ramírez-Galván YA, Cardona-Huerta S, Elizondo-Riojas G, Álvarez-Villalobos NA. Apparent Diffusion Coefficient Value to Evaluate Tumor Response After Neoadjuvant Chemotherapy in Patients with Breast Cancer. Acad Radiol 2018;25:179-87. [Crossref] [PubMed]


