
Automated segmentation of the human supraclavicular fat depot via deep neural network in water-fat separated magnetic resonance images
Introduction
In 1902, Japanese biologist Hatai first observed interscapular and supraclavicular brown adipose tissue (BAT) in human fetuses (1). It was long believed that most of the mitochondria, which drive thermogenesis in brown adipocytes, disappear after reaching adulthood and that the BAT would become functionally and morphologically similar to white adipose tissue (WAT). However, many recent studies discovered the presence of active BAT in a significant portion of healthy adults using 18F-fluorodeoxyglucose positron emission tomography (18F-FDG PET) (2,3). In addition to its thermo-regulatory role, postprandial BAT activation triggers glucose uptake and triglyceride clearance (4,5), and thus can improve insulin resistance. Furthermore, postprandial BAT activation was found to induce satiation (6). BAT activation is thus considered as a potential therapeutic target for the treatment of obesity (7). Some works reckon that BAT activation may even have more benefits, such as having a role in bone health and density (8). A steadily growing community of researchers aim at examining BAT presence and activity with various imaging modalities including PET, ultrasound (US), and magnetic resonance imaging (MRI) (9). Facing the large amount of current and future study data, we not only need an easy, robust, and reproducible way to automatically extract the BAT-containing region within the supraclavicular fossa, but also a method to quantify BAT accurately in order to enable the evaluation of possible associations between BAT and clinical parameters.
While BAT in rodents is linked with a specific anatomical region, namely the interscapular region, BAT studies in human adults have revealed a more heterogeneous tissue composition comprising both BAT, and WAT in different perivascular regions (10), and under constant influence by seasonal and hormonal changes (11). Many studies had already been conducted exploiting magnetic resonance (MR) contrast mechanisms to characterize BAT presence and to detect BAT activation, both in rodents and humans (9). Chemical-shift based water-fat imaging techniques acquire gradient echoes at different echo times (TEs) to resolve the water-fat separation, rendering a proton density fat fraction (PDFF) map. Due to the clinical availability of this technique, it has become a popular tool in the research of BAT with MRI.
There is controversy about the correlation between the MRI-based fat fraction in a given region of interest (ROI) scanned during rest and the BAT presence measured with PET during BAT activation (12-15). Yet, some MR-only studies revealed significant relative change of PDFF during BAT activation paradigms in comparison to baseline PDFF measurements (16-19). The BAT ROI selection was guided by simultaneously or previously acquired PET images revealing voxels with glycolysis (20,21).
In case of MR-only studies, however, diverse segmentation rules have been followed: In one study (18), a small ROI around the dorsal scapular artery was drawn, another study (19) defined a fat depot restricted by the trapezius, the sternocleidomastoid muscle, and the clavicle. In two studies (16,17), a larger ROI was chosen and divided into PDFF intervals for further analysis. Thus, the comparability between different pure MR studies is hampered by different segmentation rules. Automatic segmentation following consistent rules would circumvent a potential bias from different segmentation rules.
So far, fully automated segmentation algorithms of BAT have been only reported in rodents (22,23). In human anatomy, only a semi-automatic multi-atlas segmentation (MAS) has been proposed (24). However, the lateral borders of the segmentation mask did not follow any anatomical structure but were defined by a geometric cylinder. MAS conducts segmentation through registering a set of atlases to the target image. The corresponding labels of those atlases are then directly transferred from the atlas space to the target space using the same obtained parameters, and further integrated as the final segmentation, which is time-consuming and requires high computational costs (25). In addition to a lack of a consistent anatomical border definition for BAT studies, there is also a lack of consensus on the optimal PDFF threshold used for defining potential BAT regions. While it has been shown that PDFF in infants of the supraclavicular fossa can be as low as 17% (26), MR studies in adults have been using the threshold of PDFF as 30% (16-19) or 50% (14,27,28) to exclude adjacent voxels originating from muscles or vessels after manual annotation. Being directly adjacent to bones like the scapula and the clavicle, the bone signal can be easily misinterpreted as BAT, as bone marrow exhibits similar values on the PDFF maps.
Deep learning methods, as one of the most advanced machine learning approaches, have the advantage of being able to learn salient feature representations automatically and efficiently from the data. They avoid the effort of feature-engineering in conventional machine learning methods to design useful hand-crafted features and therefore make the learning process data-driven (29). For image segmentation, convolutional neural networks (CNNs) have achieved significant success in segmenting previously unseen images (30,31). Fully convolutional networks (FCNs) (32) were developed to perform semantic segmentation using a convolution and deconvolution architecture. The U-Net (33), equipped with skip-connections was one such architecture proposed for medical image segmentation. Already widely utilized in medical image analysis, U-Net-like models recently still show state-of-the-art performance in medical image analysis (34-36).
In this work, we leverage the deep CNN architecture for the automated segmentation of the supraclavicular fat depot for the purpose of human BAT analysis based on chemical-shift based water-fat MR images. To efficiently encode the multi-modal information and 3D context information from MRI scans and mimic the workflow of physicians for characterizing BAT, we jointly leveraged two-dimensional (2D) and three-dimensional (3D) CNN architecture to build the network (termed as BAT-Net). Our developed deep-learning-based BAT-Net can provide consistent quantitative evaluation among subjects and therefore support further investigations. Experiment results demonstrated that the BAT-Net outperformed widely-utilized 2D U-Net-like networks (33) and 3D U-Net-like networks (37). Our code is available for download at: https://github.com/BMRRgroup/BATNet.
Methods
Data
MRI data
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Study protocols and procedures were approved by the Ethics Committee of the Faculty of Medicine of the Technical University of Munich, Germany (Number 165/16 S). Informed consent was given by all individual participants. A total of 50 healthy subjects (demographics information in Table 1) underwent MR scans of the neck region on a 3T Ingenia scanner (Philips Healthcare, Best, Netherlands) [a subcohort from the following study (38)]. No intervention for BAT activation was performed. A combination of a head and neck coil, anterior and posterior coil arrays inside the scanner table was used. A 3D multi-echo gradient echo sequence with bipolar readouts was used in free breathing with following sequence parameters: 6 echoes, repetition time (TR) =12 ms, TE =1.24 ms, ΔTE =1 ms, flip angle =5°, bandwidth =1,413 Hz/pixel, 268×200×93 acquisition matrix size, field of view (FOV) =400×300×140 mm3 sensitivity encoding (SENSE) with R=2.5 in antero-posterior (AP) direction and scan time of 4 min 16 s. The reconstruction matrix size was (initially 288×216×93, before zero-filling 72 lines in AP direction), with voxel size 1.389×1.389×1.5 mm3. The acquired multi-echo gradient echo imaging data were processed online on the scanner using the fat quantification routine of the MR vendor. The technique first performs a phase error correction and then a complex-based water-fat decomposition using a pre-calibrated seven-peak fat spectrum and a single T2* to model the T2* decay of the signal with TE. The PDFF map was computed as the ratio of the fat signal over the sum of fat and water signals. The magnitude discrimination approach was used to reduce noise bias in areas with low signal-to-noise ratio (SNR), which results in PDFF values above 100% and below 0% (39). In addition, water and fat images were reconstructed. These four image modalities made up four separate channel inputs in the deep learning model (40).
Table 1
| Characteristics | Value |
|---|---|
| N | 50 |
| Gender, n [%] | |
| Female | 34 [68] |
| Male | 16 [32] |
| Normally distributed, mean ± SD [range] | |
| Weight (kg) | 71.8±17.6 [51–118] |
| Height (cm) | 171.7±9.3 [156.7–195.0] |
| Not normally distributed, median [range] | |
| Age (years) | 36 [22–77] |
| BMI (kg/m2) | 24.3 [17.4–39.4] |
BMI, body mass index; SD, standard deviation.
Manual segmentation of the human supraclavicular fat depot was done based on fat contrast images following coherent anatomical rules. The initial raw manual segmentation included adjacent muscle tissue, vessels, and slightly signal from cortical bone. In a successive pre-processing step, pixels with PDFF values lower than a threshold are removed to exclude muscle and vessel signal. Pixels with T2* lower than a certain threshold are removed to exclude bone signal (details in section “Pre-processing”).
Manual annotation rules
For the training of our network, ground truth labels were generated based on coherent segmentation rules for depicting the cervical, supraclavicular, and axillary fat depot. As these three are connected, and especially the border between axillary and supraclavicular fat depot is molten, we refer to it as supraclavicular fat depot. In our work, radiological scientists with 4 to 8 years of experience in working with MR body imaging, together with a board-certified radiologist, defined anatomical landmarks for the segmentation in order to obtain reproducible anatomical borders to define the supraclavicular fossa with potential BAT presence. The segmentation rules were derived both from previously published segmentation rules in MR studies (16-19), as well as published PET images (20,21), that reveal anatomical areas with the highest likelihood of BAT glycolysis. Perivascular fat signal in all acquired axial slices was included, starting from the chin until 30 cm below. Subcutaneous fat tissue and all bones were excluded. The tissue of interest was further narrowed down by excluding interscapular fat and paravertebral fat, leaving us with cervical (= within the neck), supraclavicular (= above the clavicle) and axillary fat pockets, as illustrated in Figure 1 of (41). These fat depots follow the carotid artery, the subclavian artery as well as the axillary artery, respectively.
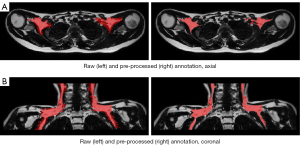
Following lateral exclusion criteria were defined based on the coronal view (Figure S1):
- Fat, laterally located of the touch point of the supraspinatus muscle with the trapezius, was excluded;
- Fat, laterally located from the coracoid process, or the collum scapulae, was excluded.
Following medial exclusion criteria were defined based on the axial view (Figure S1):
- Fat, medially located from the scalene muscles (superior parts of the scanned region) or the serratus anterior muscle (inferior parts of the scanned region), was excluded.
Following dorsal exclusion criteria were defined:
- Fat, dorsally located from the levator scapulae muscle, was excluded. This was for excluding the fat pocket surrounding the dorsal scapular artery.
An example of annotation applying these rules is shown in Figure S1.
Deep learning model
Overview
To take advantage of 3D information and to prevent over-fitting, we proposed a hybrid 2D and 3D approach for segmentation of supraclavicular fat depot for quantitative evaluation of BAT (termed as BAT-Net). The overview of the framework is illustrated in Figure 2, which presents the structure of the segmentation network and briefly describes the function of each component. The main pipeline consists of a preliminary segmentation stage and a fine-tuning segmentation stage. In the preliminary segmentation stage, we developed three combining 2D U-Net-like CNNs to mimic the workflow of physicians, who are used to review images slice-by-slice in axial, coronal, and sagittal planes to delineate areas with potential BAT. The three combining 2D CNN networks were designed for encoding the context information from the axial, coronal, and sagittal dimensions, respectively. Considering there exists four different MRI modalities, i.e., water, fat, PDFF and T2* (detailed information can be found in section “Data”), we inputted MRI slices of each modality as four channels to leverage the multi-modality information. In the fine-tuning segmentation stage, a shallow 3D CNN network was developed to synthesize predictions of three-combining 2D networks and the original four-modality MRI scans to predict the final segmentation result.

The details of the proposed methods are described in the following sections respectively. The U-Net-like 2D segmentation is presented in section “Three-combining 2D segmentation network”. The 3D fusion network is presented in section “3D fusion network”. Finally, the loss function of the BAT-Net is described in section “Loss function”.
Three-combining 2D segmentation network
Widely regarded as an elegant FCN architecture for image segmentation, U-Net-like architectures have been used in a great amount of biomedical segmentation applications (33,42-44). In this study, we used three combining 2D U-Nets to extract informative latent features in water/fat-contrast MRI, as well as quantitative PDFF and T2* maps using axial, coronal, and sagittal planes simultaneously.
The detailed architecture of the U-Net is illustrated in Figure S2. It consists of convolutional layers, max pooling operation, batch normalization (45), transposed convolution, concatenation operation and activation functions organized as a down-sampling path including three repeated encoder stacks and an up-sampling path including three repeated decoder stacks. In each decoder-encoder pair of the U-Net architecture, feature maps in the encoder part are cropped and concatenated to the corresponding feature maps in the decoder part via the skip-connection (concatenation) operation, which allows U-Net to better utilize the features with higher spatial resolution from shallow layers of the encoder part directly to the layers of the decoder part for segmentation. With this setting, U-Net is able to effectively and efficiently capture local information and high-level global information to achieve high accuracy in segmentation tasks.
In this work, the developed U-Net has four down-sampling and up-sampling processes. Each encoder stack consists of two 3×3 convolutions together with a rectified linear unit (ReLU) activation function and a batch normalization after each convolution and a 2×2 max pooling operation with stride of 2 for down-sampling at the end of the encoder. Each decoder stack consists of a transposed convolution with kernel size 2×2 and a stride of 2 for up-sampling, a concatenation operation to fuse feature maps from the encoder stack into the corresponding decoder stack, and two 3×3 convolutions followed by a ReLU and a batch normalization. At the end of the U-Net, a 1×1 convolution with sigmoid activation is applied to map learnt features to the segmentation probability map.
3D fusion network
After the three-combining 2D U-Net, we obtained three segmentation results with utilizing comprehensive 2D context information in the axial, coronal, and sagittal planes, respectively. The 3D fusion network is proposed to merge these predictions and original multi-modal MRI scans for conducting the fine-tuning segmentation. As shown in the Figure 3, the 3D fusion network is a 3D FCN (32). Considering that the preliminary segmentation results can be used to provide references during the fine-tuning segmentation stage with 3D fusion network, and to reduce computing load, the 3D fusion network is designed to be a shallow FCN, which is easier to converge. The 3D fusion network, leveraging 3D context information for obtaining a fine-tuning segmentation, consists of four 3D convolution modules and a final 3D 1×1×1 convolution layer with a softmax activation function to map 3D feature maps to the final segmentation prediction. Each 3D convolution module has a 3×3×3 convolution layer with batch normalization and leaky ReLU activation function (46).

Loss function
In medical image segmentation, one of the challenges is class imbalance between the foreground and the background in the dataset. In this work, the supraclavicular fossa occupies a relatively small part of the MRI scans. Therefore, in this work, we leverage the Dice loss as the loss function, which was initially proposed in (47) and then has been proven to be well adaptable to the data imbalance tasks (48-50). To address possible class imbalance problems, we also considered loss functions such as the generalized dice loss (50), the focal loss (51), the Tversky loss (52) and their combinations. However, as we did not find any obvious improvement of segmentation performance on our data (Table S1), we kept it simple with the standard dice loss. To be specific, let P(xi) denote the prediction probability of a voxel xi and G(xi) be the corresponding ground truth at the same voxel. The dice loss can be defined as:
where X represents the training images, ε denotes a small term to prevent any division by 0 issues.
Pre-processing
MRI scans including PDFF, T2* maps and the corresponding manual annotations were pre-processed before being fed into neural networks. The pre-processing pipelines are illustrated in Figure 4. Figures 1,5 illustrate the comparison of images and manual labels before and after pre-processing.
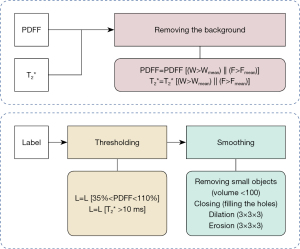
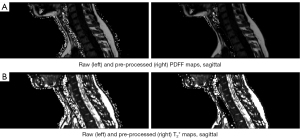
Image pre-processing (PDFF map, T2* map)
The background was removed in both PDFF maps and T2* maps by applying a foreground mask. This foreground mask was obtained slice-wise by calculating the mean value of the water and the fat image respectively and thresholding below the mean value. These two separate masks were combined by the logical or operator. Thus, voxels with water signal under the mean threshold and mean fat signal per slice were regarded as background.
Label pre-processing
During the manual segmentation procedure of the entire supraclavicular fossa, it would be dramatically time-consuming to annotate each voxel in the 3D volume belonging to the supraclavicular fossa. Therefore, the annotator was allowed to draw rough pre-annotations including adjacent muscle tissue and vessels. Subsequently, a set of well-designed pre-processing rules were applied to process the obtained rough raw manual pre-annotations into finely adjusted segmentation, which are then regarded as the ground truth.
Specifically, after the supraclavicular fossa regions have been delineated roughly using ITK-SNAP (version 3.8.0), the irrelevant surrounding non-fatty tissues were then removed. As shown in Figure 4, the detailed pre-processing includes following two steps: in the first step, voxel-wise masks are applied referring to PDFF maps (annotation regions satisfy PDFF >35% and PDFF <110%) and T2* maps (annotation regions satisfy T2* >10 ms). The PDFF threshold of 35% was chosen based on literature values (14,16-19,27,28) combined with empiric experience with our data. As bone exhibits much lower T2* values than fat, the additional T2* maps can help to distinguish fat from bone signal. The T2* threshold was thus chosen based on literature reports on T2* values of cortical bone, being consistently shorter than 10 ms (53-55). After that, in the second step, smoothing operations including removal of small disconnected regions (voxel volume <100), filling of holes, dilation (3×3×3 filter) and an erosion (3×3×3 filter) step were performed.
Evaluation
Dice similarity coefficient (DSC) (56) is a popular evaluation criteria applied in many segmentation tasks and therefore is adopted in this work, which is defined as:
where |X| and |Y| in our case denote the segmentation prediction of the proposed BAT-Net and the ground truth label (pre-processed manual segmentation annotation), respectively. Since the label data are Boolean, it can also be written as the formulation on the right-hand side, where TP, FP, and FN represent true positive, false positive, and false negative. All experiment results in this paper are evaluated by DSC.
Aside from DSC, precision score and recall score are also applied to obtain an extended evaluation with focus on FP and FN, respectively. The precision and recall are defined as:
In addition, to evaluate the quantitative agreement of segmentations with the ground truth, Bland-Altman plots (57) were applied on the volume (mL) and the mean PDFF (%) of the predictions of the proposed BAT-Net and the ground-truth labels. For a more accurate quantitative evaluation, thresholding steps in section “Label pre-processing” were applied here again to exclude the voxels that were added by smoothing operations.
Experimental setup
Experiments were conducted on a server with the Linux operating system (Ubuntu 16.04), which was equipped with Intel Core i7-6700 CPU, 32 GB RAM, an NVIDIA Quadro GPU (24 GB RAM) and an NVIDIA Titan-Xp GPU (12 GB RAM).
The dataset was randomly divided into a developing set (80%) and a test set (20%, 10 subjects). When dividing the data, the BMI was considered such that both the developing set and the test set has included subjects with all ranges of BMIs and the mean BMIs for developing and test sets are very close, 24.4 and 24.1 kg/m2, respectively. The developing set was further randomly split into training set and a validation set in a ratio of 4:1 to develop and train the proposed BAT-Net. The proposed model was implemented with Python and the Keras library (58). Adam optimizer (59) was utilized for training the network with an initial learning rate (lr) =5×10−4. Due to the large scale of the 3D fusion parameters, the batch size of 3D fusion net was 16, while that of 2D U-nets was 128. To avoid over-fitting (60) and save training times, we applied an early stopping strategy during the training phase, which means the training process will be stopped if the validation loss does not decrease after certain epochs. The patience of early-stopping was 20 for all the networks. In addition, rather than saving the model in the last epoch, we automatically saved the model which performed best on the validation set.
We conducted comparison experiments between our proposed BAT-Net and 2D U-Net/3D U-Net on the same training, validation, and test sets. The 3D U-Net used in this experiment has three down-sampling and up-sampling blocks. The encoder and decoder stacks have very similar structure as in a 2D U-Net, with 3D convolutional kernels of size 3×3×3. The comparison with 2D U-Net was implemented in three different slicing planes, denoted as 2D U-Net (axial), 2D U-Net (coronal), and 2D U-Net (sagittal).
Training based on raw annotation labels vs. pre-processed labels
At the beginning, we compared two different supervision strategies [using the baseline network architecture 2D U-Net (axial)]: (I) using the raw annotation before pre-processing as ground truth to let the network learn the original human-defined annotations and then using the pre-processing steps as described in section “Label pre-processing” as a post-processing step to refine the segmentation prediction and obtain the final segmentation. (II) Using the fine-tuned labels after pre-processing as in section “Label pre-processing” to enable the network to directly learn the processed labels.
Inter-rater evaluation
We evaluated the annotation difference between different raters (after pre-processing), 12 randomly selected subjects from the dataset were independently annotated by an additional rater and the inter-rater consistency was evaluated by DSC and Bland-Altman plots (57) on the volume (mL) and mean PDFF (%) of the annotation of two raters (after label processing).
Results
The experimental results are given in Table 2. The proposed BAT-Net achieved an average DSC of 0.878. The performance on DSC ranged from 0.853 to 0.920 with a standard deviation of 0.020. In general, the joint 2D-3D BAT-Net outperformed both the 2D and the 3D U-Net. We also conducted a 5-fold cross validation on all the proposed networks and performance of BAT-Net in the cross-validation was also superior to other compared methods (Table S2). In addition, we compared the proposed BAT-Net to other methods in terms of trainable parameters and operation times. The results of this comparison are presented in Table S3.
Table 2
| Method | Description | Evaluation metrics, mean ± SD | ||
|---|---|---|---|---|
| DSC | Precision | Recall | ||
| 2D U-Net (axial) | 2D U-Net with inputs sliced in the axial plane | 0.860±0.028 | 0.890±0.029 | 0.823±0.032 |
| 2D U-Net (coronal) | 2D U-Net with inputs sliced in coronal plane | 0.862±0.032 | 0.896±0.046 | 0.832±0.029 |
| 2D U-Net (sagittal) | 2D U-Net with inputs sliced in sagittal plane | 0.855±0.029 | 0.883±0.052 | 0.842±0.038 |
| 3D U-Net | 3D U-Net | 0.861±0.032 | 0.869±0.040 | 0.856±0.034 |
| BAT-Net | Three 2D U-Nets combining with 3D fusion net | 0.878±0.020 | 0.910±0.030 | 0.848±0.026 |
The test set is unified, consisting of 4-modality-MR-images. The evaluation metrics are the DSC, precision, and recall. SD, standard deviation; DSC, dice similarity coefficient; 2D, two-dimensional; 3D, three-dimensional; BAT, brown adipose tissue; MR, magnetic resonance.
Raw annotation vs. fine-tuned annotation
As mentioned in section “Training based on raw annotation labels vs. pre-processed labels”, we did an experiment on comparing training with raw annotation and with fine-tuned annotation on 2D U-Net. The result of this experiment is illustrated in Table 3. The performance of the first strategy is denoted as ‘raw annotation’ with unprocessed raw labels and the second strategy is denoted as ‘fine-tuned annotation’ with processed labels after the steps in section “Label pre-processing”. We found that the second training supervision strategy outperformed the first one with an average DSC of 0.860 compared to 0.805. Accordingly, we applied the ‘fine-tuned annotation’ strategy to all the segmentation models in the succeeding experiments.
Table 3
| Label | Mean | Max | Min | SD |
|---|---|---|---|---|
| Raw annotation | 0.805 | 0.855 | 0.766 | 0.033 |
| Finetuned annotation | 0.860 | 0.915 | 0.821 | 0.028 |
DSC is the performance metric. 2D, two-dimensional; SD, standard deviation; DSC, dice similarity coefficient.
BAT-Net vs. 2D U-Net
Among all three 2D U-Nets, the network with inputs in axial plane and that in coronal plane performed better than that in sagittal plane, with average dice score of 0.860, 0.862 and 0.855, respectively. As expected, the BAT-Net outperformed all individual 2D U-Nets with average dice score 0.878 (Table 2), which highlights the advantage of using a 3D fusion module to encode 3D context information in case of our BAT-Net design. Comparing the result predictions of BAT-Net with other networks, we found the BAT-Net makes fewer FPs than 2D U-Net (axial) and 2D U-Net (sagittal) and fewer FNs than 2D U-Net (coronal). An increase of both the precision score and the recall score was also consistent with these results.
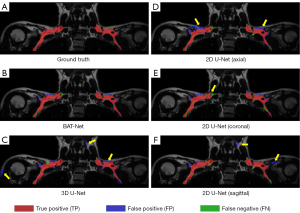
BAT-Net vs. 3D U-Net
Comparing the BAT-Net with 3D U-Net, we found a performance enhancement of 0.017 in average DSC (Table 2). In Figure 6, we compared the segmentation predictions of the BAT-Net and 3D U-Net with the ground truth annotation labels. It can be seen that the 3D U-Net makes more FP predictions (over segmentation problem), and thus, has a lower precision score. The FP predictions around the neck regions also indicate the limitation of the location recognition of the 3D U-Net. Similar results were found in the predictions of the subject with higher volume of supraclavicular fat depots, as shown in Figure S3.
PDFF and volume extraction
The Bland-Altman plots of the volume and mean PDFF of two raters are shown in Figure 7. The two raters annotated segmentations of similar volumes with a mean/minimum/maximum volume of 189/95.9/443 mL in case of rater 1 and 193/96.9/446 mL in case of rater 2. The mean absolute difference between rater 1 and rater 2 was −4.68 mL with minimum/maximum absolute differences of −3.24/8.88 mL except for one outlier. This outlier had a difference of −68.9 mL and was due to falsely annotated regions in an obese subject, such as subcutaneous fat by one of the raters.
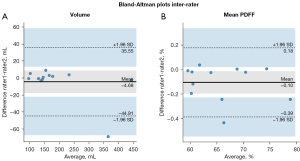
The resulting mean PDFF values in the segmented regions had a mean/minimum/maximum of 65.9/59.5/78.8% for rater 1 and 66.0/59.5/79.0% for rater 2. Mean PDFF values only varied by small amounts between raters 1 and 2 with the minimum/maximum absolute differences of −0.425/0.038% and a mean of −0.103%. Thus, except for the volume extraction in case of one outlier, both annotators’ segmentations were in good agreement with each other regarding segmented volume and mean PDFF inside the segmented regions. The inter-rater consistency evaluated with the average DSC was 0.966.
As shown in Figure 8, the volumes predicted by the network in the test set had a mean/minimum/maximum of 305/151/624 mL and the volumes of the manually segmented ground truth labels had a mean/minimum/maximum of 314/160/627 mL. In terms of absolute differences, there was a trend for smaller volumes predicted by the network with a mean difference between the outcome of the network and the ground truth labels of −9.20 mL and minimum/maximum volume differences of −19.8/6.96 mL.
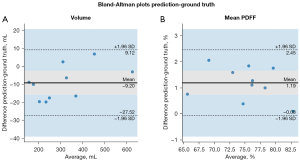
Mean PDFF values inside the areas predicted by the network had mean/minimum/maximum values of 75.7/66.0/82.7% while the ground truth mean/minimum/maximum PDFF values were 74.5/65.2/82.8%. There was also a trend for differences in PDFF. The mean PDFF of the network predicted regions was elevated with respect to the manually annotated ground truth segmentations in every subject in the test set. The mean absolute difference was 1.19% and minimum/maximum absolute differences were 0.098/2.06%. Therefore, there was a slight bias towards smaller volumes and higher mean PDFF values predicted by the network compared to the ground truth labels.
Discussion
In this paper, a joint 2D-3D CNN architecture, i.e., BAT-Net, was developed for the automatic segmentation of the supraclavicular fossa to quantify BAT volume and PDFF. The design of the proposed BAT-Net followed the principle of leveraging multi-modal information, encoding both global and local 3D context information with—in comparison to full 3D U-Net—relatively low computing resource and GPU memory consumption.
As shown in section “Raw annotation vs. fine-tuned annotation”, we observed a performance enhancement when using fine-tuned annotations, compared to raw annotation. This suggests that the proposed processing pipeline solves the problem of inconsistency of segmentation border among subjects in case of the raw annotation, which confuses the deep learning networks. Therefore, training based on pre-processed, ‘fine-tuned’ labels results in a better performance.
In the BAT segmentation task, the 3D location and context information are essential for recognizing the supraclavicular fat region. Conventional 2D convolutional segmentation network conducts slice-wise segmentation, which cannot learn the correlation and context information among slices. Besides, a performance gap was observed among 2D U-Nets with inputs in different slicing directions, which may suggest axial and coronal planes, compared with sagittal plane, provide a better view for a 2D U-Net to encode context information to segment supraclavicular fat depots.
Pure 3D convolutional segmentation networks with 3D kernels have the ability to directly encode 3D context information within the MRI scans, but these networks face the challenge of having significantly more parameters to be optimized and require more GPU memory during the training stage (61). In this study, the performance of 3D U-Net is even lower than one of the 2D U-Nets, which may be caused by following reasons: (I) the 3D U-Net has much more trainable parameters than the 2D U-Net, making it hard to converge to the optimal point with relatively limited medical training data; (II) 3D U-Nets need more GPU memory and a relatively small batch size was used during training (batch size =16), which may have caused the instability in the training (62); (III) the training samples were relatively scarce in the 3D settings, where each MRI volume is regarded as a sample instead of a slice in the 2D settings. All above-mentioned factors are prone to lead to over-fitting. In comparison, the proposed BAT-Net employs a shallow 3D fusion net to synthesize information and use three prediction maps of 2D U-Nets as references to obtain the fine-tuned segmentation, which faces a smaller risk for over-fitting.
A consistent result from the quantification method is essential in BAT analysis, especially when comparing inter-individual differences, or when observing intra-individual changes during activation studies. To achieve this requested quantification precision, reproducible labels are necessary. In this paper, the reproducibility of volume and mean PDFF extraction from a BAT segmentation task was evaluated for the first time. This was done by an inter-rater evaluation as well as comparing the network-predicted labels with a set of ground truth labels, with a high DSC of 0.966 in case of the inter-rater consistency. Nevertheless, one of the twelve subjects had clearly flawed annotations by one rater evident by a 68.9 mL difference at a total segmented volume of 331 mL, accounting for a difference of 20.8%. While the faulty annotation was not used to train the network, it reveals the possibility of flawed segmentations. However, the mean PDFF of the segmented region seems to be not influenced by annotation errors as the inter-rater difference in mean PDFF was negligible. It needs to be mentioned that manual segmentation in obese subjects, who exhibit an above-average amount of fat in the supraclavicular fossa, is challenging, as different fat depots are morphologically connected in the MRI images, and thus visually more difficult to be separated.
When comparing the ground truth labels to the network-predicted labels with respect to volume and mean PDFF, a small bias is observable in both cases. While the volume segmented by the network is smaller by an average of 9.20 mL in the test set, the mean PDFF is higher in each subject and is elevated by 1.19% on average. A possible explanation for the observed differences is that the network predicted masks included fewer voxels at the edges of the fat depots, where partial volume effects comprising muscle may occur. As these voxels affected by partial volume effect exhibit smaller PDFF values, mean overall mean PDFF decreases. However, no large deviations with regards to volume or mean PDFF are apparent in the test set. Therefore, the labels predicted by the network are in good agreement with the ground truth labels.
One limitation of this study is that we trained our model on 50 healthy volunteers including a limited number of obese subjects. The trained result may not be applicable to other cohorts, for instance with primarily obese subjects, or children. Furthermore, only data from one center were used. Training the model on datasets of multiple different centers may allow the network to learn domain-invariant features among centers and enhance its generalization ability. And, due to the data-driven nature of deep learning, collecting more training data would further improve the performance of the BAT-Net. Therefore, one future step is to collect more data and evaluate the BAT-Net in different medical centers.
Another limitation is that we did not benchmark all possible segmentation network architectures. To prove the concept and to match the expectation of clinical community, where simple and robust methods are more favorable for the potential of clinical translation, we only used the widely proven and robust U-Net-like convolutional network to build the supraclavicular fossa segmentation model.
It remains to be emphasized that the segmented anatomy of the supraclavicular fossa (including the cervical and axillary fat pockets) was chosen based on previous publications revealing active BAT. While the terms BAT, WAT, and beige adipose tissue refer to the cellular lineage and cellular function, the term ‘perivascular fat’ refers to an anatomical location surrounding vessels. In our study, we limited the segmented anatomy to the cervical, supraclavicular, and axillary fossa, where BAT, WAT, and beige adipocytes are present at the same time. It is not possible with MR yet, which has an image resolution of about 1 mm3, to morphologically differentiate between cells that have a diameter of tens of microns. However, active BAT can be also found next to the vertebrae, a location we did not include.
While we have trained the network based on locations with PDFF in a range of 35–110%, it will be easy for the end user of the code we share to choose the final range of PDFF values to be included for their analysis. As we did not define borders in the superior or inferior direction (we segmented all fat pockets surrounding the carotid, the subclavian and axillary artery), it will remain open to the end user to define the cranial/caudal border according to their preference. A possible landmark for the cranial border could be the vocal cord. The clavicle could be used as the inferior border, but a variability is to be expected depending on the patient’s shoulder positioning. Future designs of improved network architectures need to account for the potential problem of over-fitting to current annotation labels, since as of now, we only have a limited number of subjects with labels. Besides, one interesting future possibility for improving the segmentation performance can be leveraging unannotated data with semi-supervised or unsupervised methods to further enhance the performance of our method.
Conclusions
This paper proposed an artificial intelligence (AI) system on a deep neural network for segmenting the supraclavicular fossa for BAT quantification. Experimental results demonstrated the potential of deep learning in the automatic segmentation of supraclavicular fat depots. The proposed BAT-Net with the ability of extracting informative latent features can provide consistent segmentation among subjects since the architecture and the corresponding parameters are determined after training. The model integrated multi-modal MRI scans, 2D and 3D, local and global context information and showed high-level segmentation performance. Our AI system can provide consistent quantification results and has the potential to facilitate the research on BAT using MRI.
Acknowledgments
The authors would like to thank the volunteers who participated in the research and the authors would also like to acknowledge NVIDIA Corporation for the donation of a Titan XP GPU used for this research. The authors acknowledge support in manual annotation for inter-rater evaluation by Stella Näbauer.
Funding: The present work was supported by the German Research Foundation (FOR 5298: iMAGO-Personalized diagnostics for the treatment of obesity, Project number: 455422993), Philips Healthcare, the Else Kroener-Fresenius-Foundation, Bad Homburg, Germany, and the Helmholtz cross-program topic “Metabolic Dysfunction”.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-304/coif). DK receives research grant support from Philips Healthcare and consulting fees from the AMCA. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol and the standard operating procedures were approved by the ethical committee of the Technical University of Munich, Germany (Number 165/16 S). Written informed consent was obtained from subjects included in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hatai S. On the presence in human embryos of an interscapular gland corresponding to the so-called hibernating gland of lower mammals. Anat Anz 1902;21:369-73.
- van Marken Lichtenbelt WD, Vanhommerig JW, Smulders NM, Drossaerts JM, Kemerink GJ, Bouvy ND, Schrauwen P, Teule GJ. Cold-activated brown adipose tissue in healthy men. N Engl J Med 2009;360:1500-8. Erratum in: N Engl J Med 2009;360:1917. [Crossref] [PubMed]
- Cohade C, Osman M, Pannu HK, Wahl RL. Uptake in supraclavicular area fat ("USA-Fat"): description on 18F-FDG PET/CT. J Nucl Med 2003;44:170-6. [PubMed]
- Bartelt A, Bruns OT, Reimer R, Hohenberg H, Ittrich H, Peldschus K, Kaul MG, Tromsdorf UI, Weller H, Waurisch C, Eychmüller A, Gordts PL, Rinninger F, Bruegelmann K, Freund B, Nielsen P, Merkel M, Heeren J. Brown adipose tissue activity controls triglyceride clearance. Nat Med 2011;17:200-5. [Crossref] [PubMed]
- Fedorenko A, Lishko PV, Kirichok Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012;151:400-13. [Crossref] [PubMed]
- Li Y, Schnabl K, Gabler SM, Willershäuser M, Reber J, Karlas A, Laurila S, Lahesmaa M. U Din M, Bast-Habersbrunner A, Virtanen KA, Fromme T, Bolze F, O'Farrell LS, Alsina-Fernandez J, Coskun T, Ntziachristos V, Nuutila P, Klingenspor M. Secretin-Activated Brown Fat Mediates Prandial Thermogenesis to Induce Satiation. Cell 2018;175:1561-74.e12. [Crossref] [PubMed]
- Yoneshiro T, Saito M. Activation and recruitment of brown adipose tissue as anti-obesity regimens in humans. Ann Med 2015;47:133-41. [Crossref] [PubMed]
- Lee P, Brychta RJ, Collins MT, Linderman J, Smith S, Herscovitch P, Millo C, Chen KY, Celi FS. Cold-activated brown adipose tissue is an independent predictor of higher bone mineral density in women. Osteoporos Int 2013;24:1513-8. [Crossref] [PubMed]
- Wu M, Junker D, Branca RT, Karampinos DC. Magnetic Resonance Imaging Techniques for Brown Adipose Tissue Detection. Front Endocrinol (Lausanne) 2020;11:421. [Crossref] [PubMed]
- Hildebrand S, Stümer J, Pfeifer A. PVAT and Its Relation to Brown, Beige, and White Adipose Tissue in Development and Function. Front Physiol 2018;9:70. [Crossref] [PubMed]
- Au-Yong IT, Thorn N, Ganatra R, Perkins AC, Symonds ME. Brown adipose tissue and seasonal variation in humans. Diabetes 2009;58:2583-7. [Crossref] [PubMed]
- Holstila M, Pesola M, Saari T, Koskensalo K, Raiko J, Borra RJ, Nuutila P, Parkkola R, Virtanen KA. MR signal-fat-fraction analysis and T2* weighted imaging measure BAT reliably on humans without cold exposure. Metabolism 2017;70:23-30. [Crossref] [PubMed]
- Franz D, Karampinos DC, Rummeny EJ, Souvatzoglou M, Beer AJ, Nekolla SG, Schwaiger M, Eiber M. Discrimination Between Brown and White Adipose Tissue Using a 2-Point Dixon Water-Fat Separation Method in Simultaneous PET/MRI. J Nucl Med 2015;56:1742-7. [Crossref] [PubMed]
- Held C, Junker D, Wu M, Patzelt L, Mengel LA, Holzapfel C, Diefenbach MN, Makowski MR, Hauner H, Karampinos DC. Intraindividual difference between supraclavicular and subcutaneous proton density fat fraction is associated with cold-induced thermogenesis. Quant Imaging Med Surg 2022;12:2877-90. [Crossref] [PubMed]
- McCallister A, Zhang L, Burant A, Katz L, Branca RT. A pilot study on the correlation between fat fraction values and glucose uptake values in supraclavicular fat by simultaneous PET/MRI. Magn Reson Med 2017;78:1922-32. [Crossref] [PubMed]
- Coolbaugh CL, Damon BM, Bush EC, Welch EB, Towse TF. Cold exposure induces dynamic, heterogeneous alterations in human brown adipose tissue lipid content. Sci Rep 2019;9:13600. [Crossref] [PubMed]
- Abreu-Vieira G, Sardjoe Mishre ASD, Burakiewicz J, Janssen LGM, Nahon KJ, van der Eijk JA, Riem TT, Boon MR, Dzyubachyk O, Webb AG, Rensen PCN, Kan HE. Human Brown Adipose Tissue Estimated With Magnetic Resonance Imaging Undergoes Changes in Composition After Cold Exposure: An in vivo MRI Study in Healthy Volunteers. Front Endocrinol (Lausanne) 2020;10:898. [Crossref] [PubMed]
- Stahl V, Maier F, Freitag MT, Floca RO, Berger MC, Umathum R, Berriel Diaz M, Herzig S, Weber MA, Dimitrakopoulou-Strauss A, Rink K, Bachert P, Ladd ME, Nagel AM. In vivo assessment of cold stimulation effects on the fat fraction of brown adipose tissue using DIXON MRI. J Magn Reson Imaging 2017;45:369-80. [Crossref] [PubMed]
- Oreskovich SM, Ong FJ, Ahmed BA, Konyer NB, Blondin DP, Gunn E, Singh NP, Noseworthy MD, Haman F, Carpentier AC, Punthakee Z, Steinberg GR, Morrison KM. MRI Reveals Human Brown Adipose Tissue Is Rapidly Activated in Response to Cold. J Endocr Soc 2019;3:2374-84. [Crossref] [PubMed]
- Gifford A, Towse TF, Walker RC, Avison MJ, Welch EB. Human brown adipose tissue depots automatically segmented by positron emission tomography/computed tomography and registered magnetic resonance images. J Vis Exp 2015;52415. [PubMed]
- Ouwerkerk R, Hamimi A, Matta J, Abd-Elmoniem KZ, Eary JF, Abdul Sater Z, Chen KY, Cypess AM, Gharib AM, Proton MR. Spectroscopy Measurements of White and Brown Adipose Tissue in Healthy Humans: Relaxation Parameters and Unsaturated Fatty Acids. Radiology 2021;299:396-406. [Crossref] [PubMed]
- Bhanu Prakash KN, Verma SK, Yaligar J, Goggi J, Gopalan V, Lee SS, Tian X, Sugii S, Leow MK, Bhakoo K, Velan SS. Segmentation and characterization of interscapular brown adipose tissue in rats by multi-parametric magnetic resonance imaging. MAGMA 2016;29:277-86. [Crossref] [PubMed]
- Bhanu Prakash KN, Srour H, Velan SS, Chuang KH. A method for the automatic segmentation of brown adipose tissue. MAGMA 2016;29:287-99. [Crossref] [PubMed]
- Lundström E, Strand R, Forslund A, Bergsten P, Weghuber D, Ahlström H, Kullberg J. Automated segmentation of human cervical-supraclavicular adipose tissue in magnetic resonance images. Sci Rep 2017;7:3064. [Crossref] [PubMed]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans Pattern Anal Mach Intell 2013;35:611-23. [Crossref] [PubMed]
- Armstrong T, Ly KV, Ghahremani S, Calkins KL, Wu HH. Free-breathing 3-D quantification of infant body composition and hepatic fat using a stack-of-radial magnetic resonance imaging technique. Pediatr Radiol 2019;49:876-88. [Crossref] [PubMed]
- Jones TA, Wayte SC, Reddy NL, Adesanya O, Dimitriadis GK, Barber TM, Hutchinson CE. Identification of an optimal threshold for detecting human brown adipose tissue using receiver operating characteristic analysis of IDEAL MRI fat fraction maps. Magn Reson Imaging 2018;51:61-8. [Crossref] [PubMed]
- Franz D, Weidlich D, Freitag F, Holzapfel C, Drabsch T, Baum T, Eggers H, Witte A, Rummeny EJ, Hauner H, Karampinos DC. Association of proton density fat fraction in adipose tissue with imaging-based and anthropometric obesity markers in adults. Int J Obes (Lond) 2018;42:175-82. [Crossref] [PubMed]
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436-44. [Crossref] [PubMed]
- Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Communications of the ACM 2017;60:84-90. [Crossref]
- Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell 2017;39:640-51. [Crossref] [PubMed]
- Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2015:3431-40.
- Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2015: 18th International Conference. Munich: Springer International Publishing, 2015:234-41.
- Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306, 2021.
- Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: Redesigning Skip Connections to Exploit Multiscale Features in Image Segmentation. IEEE Trans Med Imaging 2020;39:1856-67. [Crossref] [PubMed]
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 2021;18:203-11. [Crossref] [PubMed]
- Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: learning dense volumetric segmentation from sparse annotation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2016: 19th International Conference. Athens: Springer International Publishing, 2016:424-32.
- Drabsch T, Holzapfel C, Stecher L, Petzold J, Skurk T, Hauner H. Associations Between C-Reactive Protein, Insulin Sensitivity, and Resting Metabolic Rate in Adults: A Mediator Analysis. Front Endocrinol (Lausanne) 2018;9:556. [Crossref] [PubMed]
- Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med 2007;58:354-64. [Crossref] [PubMed]
- Franz D, Diefenbach MN, Treibel F, Weidlich D, Syväri J, Ruschke S, Wu M, Holzapfel C, Drabsch T, Baum T, Eggers H, Rummeny EJ, Hauner H, Karampinos DC. Differentiating supraclavicular from gluteal adipose tissue based on simultaneous PDFF and T(2)* mapping using a 20-echo gradient-echo acquisition. J Magn Reson Imaging 2019;50:424-34. [Crossref] [PubMed]
- Karampinos DC, Weidlich D, Wu M, Hu HH, Franz D. Techniques and Applications of Magnetic Resonance Imaging for Studying Brown Adipose Tissue Morphometry and Function. Handb Exp Pharmacol 2019;251:299-324. [Crossref] [PubMed]
- Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans Med Imaging 2018;37:2663-74. [Crossref] [PubMed]
- Zeng Z, Xie W, Zhang Y, Lu Y. RIC-Unet: An improved neural network based on Unet for nuclei segmentation in histology images. IEEE Access 2019;7:21420-8.
- Guan S, Khan AA, Sikdar S, Chitnis PV. Fully Dense UNet for 2-D Sparse Photoacoustic Tomography Artifact Removal. IEEE J Biomed Health Inform 2020;24:568-76. [Crossref] [PubMed]
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: Proceedings of the 32nd International Conference on Machine Learning, PMLR. 2015:448-56.
- Agarap AF. Deep learning using rectified linear units (ReLU). arXiv preprint arXiv:1803.08375, 2018.
- Milletari F, Navab N, Ahmadi SA. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 2016 fourth international conference on 3D vision (3DV). IEEE, 2016:565-71.
- Li H, Jiang G, Zhang J, Wang R, Wang Z, Zheng WS, Menze B. Fully convolutional network ensembles for white matter hyperintensities segmentation in MR images. Neuroimage 2018;183:650-65. [Crossref] [PubMed]
- Drozdzal M, Chartrand G, Vorontsov E, Shakeri M, Di Jorio L, Tang A, Romero A, Bengio Y, Pal C, Kadoury S. Learning normalized inputs for iterative estimation in medical image segmentation. Med Image Anal 2018;44:1-13. [Crossref] [PubMed]
- Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. Deep Learn Med Image Anal Multimodal Learn Clin Decis Support (2017) 2017;2017:240-8.
- Yeung M, Sala E, Schönlieb CB, Rundo L. Unified Focal loss: Generalising Dice and cross entropy-based losses to handle class imbalanced medical image segmentation. Comput Med Imaging Graph 2022;95:102026. [Crossref] [PubMed]
- Nasalwai N, Punn NS, Sonbhadra SK, Agarwal S. Addressing the class imbalance problem in medical image segmentation via accelerated Tversky loss function. In: Advances in Knowledge Discovery and Data Mining: 25th Pacific-Asia Conference. Cham: Springer International Publishing, 2021:390-402.
- Du J, Bydder GM. Qualitative and quantitative ultrashort-TE MRI of cortical bone. NMR Biomed 2013;26:489-506. [Crossref] [PubMed]
- Weiger M, Wu M, Wurnig MC, Kenkel D, Boss A, Andreisek G, Pruessmann KP. ZTE imaging with long-T2 suppression. NMR Biomed 2015;28:247-54. [Crossref] [PubMed]
- Ma YJ, Jerban S, Jang H, Chang D, Chang EY, Du J. Quantitative Ultrashort Echo Time (UTE) Magnetic Resonance Imaging of Bone: An Update. Front Endocrinol (Lausanne) 2020;11:567417. [Crossref] [PubMed]
- Dice LR. Measures of the amount of ecologic association between species. Ecology 1945;26:297-302. [Crossref]
- Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986;1:307-10. [Crossref] [PubMed]
- Gulli A, Pal S. Deep learning with Keras. Birmingham: Packt Publishing Ltd., 2017.
- Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980, 2014.
- Li Z, Kamnitsas K, Glocker B. Overfitting of neural nets under class imbalance: Analysis and improvements for segmentation. In: Medical Image Computing and Computer Assisted Intervention (MICCAI) 2019: 22nd International Conference. Shenzhen: Springer International Publishing, 2019:402-10.
- Zheng Y, Liu D, Georgescu B, Nguyen H, Comaniciu D. 3D deep learning for efficient and robust landmark detection in volumetric data. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2015: 18th International Conference. Munich: Springer International Publishing, 2015:565-72.
- Smith SL, Kindermans PJ, Ying C, Le QV. Don't decay the learning rate, increase the batch size. arXiv preprint arXiv:1711.00489, 2017.


