
Developing a deep learning model to predict epilepsy recurrence in patients with focal cortical dysplasia type III
Introduction
In 1971, Taylor first described focal cortical dysplasia (FCD), which is characterized by disordered neurons in white matter, dyslamination, and aberrant balloon cells, and can induce drug-resistant or intractable epilepsy. Approximately half (46.5%) of drug-resistant epileptic patients have pathologically confirmed abnormalities (1,2). The International League Against Epilepsy (ILAE) suggested the first international categorization of FCD in 2011 (3,4). Type III FCD is characterized by cortical dyslamination anomalies linked with a primary lesion, and these anomalies are typically localized in the same cortical region/lobe. FCD type III is categorized as FCD type I adjacent to another major lesion. FCD type III comprises 4subgroups based on the pathophysiology of the accompanying lesions, specifically hippocampal sclerosis (FCD type IIIa), low-grade developmental epilepsy-associated tumors (LDEATs) (FCD type IIIb), vascular lesions (FCD type IIIc), and scar/hypoxic damage (FCD type IIId). (1,5,6).
Smaller lesions and even magnetic resonance (MR)-negative FCDs are also recognized, given advances in magnetic resonance imaging (MRI) technology and postprocessing approaches (7,8). Especially on T2-weighted fluid attenuated inversion recovery (T2W FLAIR), cortical and subcortical hypersignals are more readily detectable (9,10). Technological advancements in neuroimaging have increased the incidence of identified lesions, but MR-negative FCD neuroimaging cases still occur. Neuroimaging abnormalities in individuals with FCD type I and/or type II have been reported, including expanded/atrophic cortex, gray-white matter undifferentiated junctions, and hyperintensity alterations (11,12). The imaging findings of the principal lesions are obvious. Regarding low-grade epilepsy-associated tumors (LEATs), the differential diagnosis of ganglioglioma (GG) and dysembryoplastic neuroepithelial tumor (DNT) prior to surgery can be difficult. Both exhibit well-defined masses with cystic or solid constituents, some of which may be entirely cystic or consist solely of local leukoaraiosis (13). FCD type IIIc largely consists of cerebral cavernous malformation (CCM) and arteriovenous malformations. Typical imaging findings of CCM are ‘popcorn-like’ changes. The molecular pathology and biological mechanism of FCD type III remain unclear (8,14-16).
The definition of medication-resistant seizure is failure of adequate trials of 2 tolerable and correctly designed and administered anticonvulsant drug regimens (17). Surgery is considered a better treatment for medication-resistant disease. Studies on the efficacy of epilepsy surgery based on retrospective series of cases are available, and many of these studies investigated pathogenic subgroups in depth and risk factors for prognosis of FCD type III (12,18). MRI-negative FCDs and incomplete resection are risk factors for poor prognosis. Despite accurate focused positioning on the scalp and invasive electroencephalogram (EEG) surveillance, individuals with a detectable lesion have a more favorable prognosis than those with a negative MRI result on presurgical monitoring (19,20). Epileptogenic networks are better able to describe the complexity of epilepsy dynamics and realistically convey the distribution of epileptogenic abnormalities in the brain (21,22). Following full resection of the epileptogenic zone delineated by EEG and MRI, most patients exhibit a positive prognosis (23). In recent years, structural MRI scans have been used to automatically detect focal epileptic abnormalities, which immediately detect structural anomalies using a combined approach of postprocessing and artificial intelligence (24). Conventional MRI postprocessing includes surface-based classification (SBC) approaches (25), voxel-based morphometry (VBM) (26), and voxel-based MRI morphometry analysis program (MAP) (27). Combining MR image morphology and tissue signal intensity characteristics to develop neural network classifications can aid in the diagnosis of the FCD. In terms of medical image interpretation, this approach performs better than radiologists using conventional methods. We assumed that the combination of cortical dysplasia and associated lesions may have deep-learning imaging features that are difficult to detect visually and affect prognosis. Based on the assumption, we proposed a preoperative prediction model of seizure recurrence to provide referable information for individual risk of FCD type III-related refractory epilepsy and to determine adjuvant therapy, thereby improving postoperative management of patients. The purpose of this work was to explore the potential value of deep learning algorithms for predicting FCD type III-related refractory epilepsy recurrence based on MRI imaging data. The following article is presented in accordance with the TRIPOD reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-276/rc).
Methods
Participants
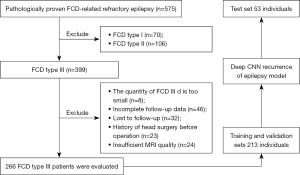
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of the Third Affiliated Hospital of Southern Medical University, and individual consent for this retrospective analysis was waived. From December 2015 to October 2019, 575 individuals with pathology-confirmed FCD-related refractory epilepsy form Guangdong 999 Brain Hospital were included in this trial. In all, 266 patients with FCD type III were initially included, with 213 individuals comprising the training and validation sets [median age: 13 years; interquartile range (IQR), 6–20 years] and 53 individuals comprising the internal test set (median age: 12 years; IQR, 7–20 years). The inclusion criteria were as follows: (I) a diagnosis of refractory epilepsy, intractable epilepsy, or drug-resistant epilepsy; and (II) patients were diagnosed with FCD type III using postoperative pathological examination. Figure 1 depicts the study flow. Profiles and medical records from the 266 included patients were extracted and saved in online databases. Data of baseline clinical characteristics, neuroimaging results, surgical technique, pathologic categories, and prognosis were collected. After 1 year, the prognosis for seizure was noted. Patients were classified as free of disabling seizures (Class I) or as experiencing recurrent seizures (Class II–VI) based on Engel’s classification result at the latest review (24). Complete resection was defined as: combined with the results of preoperative evaluation and intraoperative electrocorticography, the abnormal firing cortices found using EEG were removed as much as possible.
MRI data sets and preprocessing
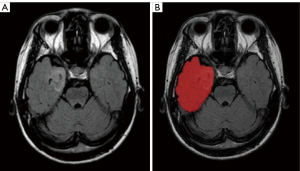
We used an 8-channel head coil for scanning on a 1.5T scanner (Philips Gyro Scan Inter; Philips, Amsterdam, The Netherlands). Each patient underwent an MRI examination using the procedures outlined in Table S1, which are routine brain imaging techniques. MR volumes of axial T2W FLAIR were collected from all cases. An example depicting the region of interest (ROI) is shown in Figure 2. All original images were reviewed and manually delineated by 2 readers for each transverse direction generated by the T2W FLAIR series by making use of the software ITK-SNAP (version 3.8; http://www.itksnap.org). An ROI was defined as the entire lobe/s where the combined lesion is located. The intraclass correlation coefficient (ICC) was computed for evaluating the degree of consistency and reliability that existed between the 2 observers.
The dataset was split into training, validation, and testing sets at a ratio of 7:1:2. Since the total number of the datasets was 266, we divided our cohort into 186, 27, and 53 cases for the training, validation, and testing sets, respectively. To avoid overfitting, random rotates were added to the training database to improve accuracy (ACC) (±10° in x, y coordinates), translations (±10 voxels in x, y coordinates), and flipping (x coordinates). The augmentation method permitted us to further extend the volume of our training set. Prior to feeding the training data into the neural network, it was supplemented for each epoch. The only set that was augmented was the training set, not the validation or testing sets. Real-time data augmentation was conducted to reduce memory consumption.
All MR images were resized to 1×1×3 mm using bicubic interpolation. Affine spatial registration (using the FSL package, https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) was applied to reduce the effect of spatial inconsistencies on the neural network. The median intensity was deducted and then divided by the interquartile intensity for the normalization of image intensities. Finally, the images were cropped to a size of 128×128×16 pixels (px).
Predictive model of FCD seizure recurrence
As shown in Figure 3, we proposed a classification model with a multiscale receptive field module (blue box), squeeze-and-excitation module (red box), and skip connection. A multiscale receptive field module embeds multiscale information and aggregates features on a range of various receptive fields to achieve improvements in performance. Furthermore, a squeeze-and-excitation module clearly describes the interrelations between the channels of its convolutional characteristics to enhance the quality of the outputs generated by every block. ResNet (28) and DenseNet (29) have already demonstrated impressive outcomes in classification tasks using skip connections. Here, a skip connection between each block was established, which enables feature reuse and achieves better performance with fewer parameters and computational costs.

In particular, our network has 2 inputs: 1 is the preprocessed original image, and the other is the ROI outlined by clinicians. In this way, more weight can be given to the brain regions that may be related to seizure recurrence (that is, the ROI outlined by clinicians), and we can help the network to judge whether a patient might achieve seizure-free status after surgery.
Model architecture
As shown in Figure 3, our classification architecture consisted of 3 blocks, a global average pooling layer, and a fully convolutional layer. Each block contained 2 multiscale receptive field modules with a stride of 1, 1 multiscale receptive field module with a stride of 2 (i.e., down-sampling), and 1 squeeze-and-excitation module.
Multi-scale receptive field module
As shown in Figure 4A, a multi-scale receptive field framework was adopted to enlarge the receptive field of convolutional filters and to combine the multiscale spatial details and context information. Convolution filters with the sizes of 1×1×1, 3×3×3, 5×5×5, and 7×7×7 were used. Such aggregation assembles feature representation between corresponding stages through the ‘coarse’ and ‘fine’ parts.

Squeeze-and-excitation module
After the feature extraction by multiscale receptive field module, we further used a squeeze-and-excitation module to reconsider the previously obtained features’ calibration. In other words, a squeeze-and-excitation module was used to automatically determine the value of each attribute channel via learning and also to boost the beneficial attributes based on the priority while suppressing the unusable attributes for the assigned mission. It consists of 3 operations (Figure 4B): squeeze [global average pooling layer (Global AvgPool)], excitation (2 fully convolutional layers and LeakyReLU), and reweighting (sigmoid and scale).
Squeeze
In general, every convolutional filter works with a local receptive field, and the result of the convolutional filters is incapable of utilizing context data from outside this field. Therefore, the global average pooling was used to ‘squeeze’ global spatial data into a channel descriptor. As illustrated in Figure 4B, attributes were compressed along spatial dimensions [height × width × depth (H×W×D)], and every 3-dimensional (3D) attribute channel was converted to a real number (1×1×1). This real number possesses a global receptive field, which shows how the response on the characteristic channel is distributed globally. Officially, a statistic is formed via decreasing the preceding outcome attributes U through their spatial dimensions H×W×D, whereby the c-th element of z is determined by the following formula:
Excitation
The ‘squeeze’ operation is followed by an ‘excitation’ operation that sought to truly grasp channel-wise dependencies to benefit from the data gathered.
Here, we created a bottleneck structure with 2 fully connected layers to describe the correlation between channels and produce equal amounts of weights as the input attributes. Initially the attribute dimension was lowered to C/r, and then, upon the stimulation of LeakyReLU, it was promoted via a fully connected layer to the original dimension C. Compared with sigmoid and ReLU, LeakyReLU can effectively avoid bilateral or unilateral saturated problems and reduce the occurrence of the vanishing gradients. Here, r is the reduction ratio. This has the following benefits versus the straightforward use of a fully connected layer: (I) it has greater nonlinearity and can accommodate complex channel correlations better; and (II) it substantially minimizes the quantity of parameters and calculations with the parameter of r. Then the output feature s can be represented by:
where δ stands for the LeakyReLU activation, and and represent the weights in 2 fully connected layers, separately.
Reweighting
Following attribute selection, the output weight of ‘excitation’ was considered the significance of every attribute channel, and the preceding attributes were then multiplied channel by channel to accomplish the recalibration of the unique features in the channel dimension. Then, a sigmoid gate was used to acquire a normalized weight between 0 and 1, and finally, a scale operation was employed to weight the normalized weight according to the attributes of each channel. The final output x is:
Training loss
Our model utilized a cross-entropy loss function between the actual labeling and the outcome. The weighted cross-entropy function was utilized to adjust for the imbalanced volume class between seizure relapse and seizure-free sections.
Implementation
The classification network presented earlier was programmed in Python 3.6 (Python Software Foundation, Fredericksburg, VA, USA) using the open-source Pytorch tools. On a graphics processing unit (GPU)-optimized workstation with a single NVIDIA GeForce GTX Titan X (12GB, Maxwell architecture; NVIDIA, Santa Clara, CA, USA), training was conducted. The following settings were established: an Adam optimizer with a learning rate of 1e−4, a weight decay of 1e−4, and a mini-batch size of 4. Initialization of convolution kernels was performed utilizing a Kaiming uniform initializer. A technique of lowering the learning rate gradually was employed, lowering it by 0.25*total epochs, 0.25*total epochs, and 0.75*total epochs at a pace of 0.1.
Model performances evaluation and statistical analysis
Patients who previously experienced seizures or those who were seizure-free were used to evaluate the performance of the algorithm using the ACC classification. Supplementary performance data for sensitivity (SEN), specificity (SPE), positive predictive value (PPV), and negative predictive value (NPV) were also reported. Throughout the training stage, a 5-fold cross-validation paradigm was utilized. According to this paradigm, 80% of the inputs were randomized into the training and validation cohort, and 20% of the inputs were used for testing. After that, the procedure was performed 5 times until every single component in the whole training dataset was utilized for validation at least once. The final results below are reported for the cumulative validation set statistics across the entire training dataset.
Results
Clinical data
We obtained 266 patients’ MRI images from the image archiving and communication systems at Guangdong 999 Brain Hospital. Table 1 shows the clinical profiles of all patients.
Table 1
| Characteristic | Value |
|---|---|
| Age at epilepsy onset, median [IQR] | 12.5 [6–19] |
| Children (≤12 years), n (%) | 122 (45.9) |
| Gender, n (%) | |
| Female | 95 (35.7) |
| Male | 171 (64.3) |
| Febrile seizures, n (%) | 42 (15.8) |
| Histology, n (%) | |
| FCD IIIa | 183 (68.8) |
| FCD IIIb | 61 (22.9) |
| FCD IIIc | 22 (8.3) |
| FCD MRI negative, n (%) | 36 (13.5) |
| Lesion location, n (%) | |
| Temporal | 229 (86.1) |
| Extemporal | 12 (4.5) |
| Multiple lesion | 25 (37.9) |
| Incomplete resection | 40 (15.0) |
| Outcome, n (%) | |
| Class I | 179 (67.3) |
| Class II–IV | 87 (37.2) |
FCD, focal cortical dysplasia; MRI, magnetic resonance imaging; IQR, interquartile range.
Performance of deep learning model
The hypothesized prediction model developed required a training time of 24 hours. The average test time for the epilepsy prediction system to judge whether a patient could achieve a seizure-free condition after surgery was only 0.5 seconds using the already trained networks.
Table 2 compares the SEN, SPE, PPV, and NPV for detecting seizure recurrence or seizure-free status through employing various inputs of the network as suggested here, such as using the original image, utilizing the ROI, and merging the original image with the ROI. All experiments estimated the SEN and SPE ranging from 84.47% to 90.86% and from 96.24% to 97.21%, respectively, and PPV and NPV ranging from 90.78% to 93.64% and from 92.52% to 95.72%, respectively. With combined inputs, the highest overall diagnostic performance for predicting the recurrence of epilepsy could be achieved.
Table 2
| Variables | Accuracy | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|---|
| Original image only | 92.83% | 84.47% | 97.21% | 93.34% | 92.52% | 96.22% |
| ROI only | 92.45% | 84.92% | 96.24% | 90.78% | 92.95% | 94.76% |
| Combined | 94.71% | 90.86% | 96.63% | 93.64% | 95.72% | 97.17% |
PPV, positive predictive value; NPV, negative predictive value; AUC, area under the curve; ROI, region of interest.
Table 3 demonstrates the performance of many networks using inputs from integrated data. The results produced by our suggested network were superior to those achieved by DenseNet and ResNet. Figure 5 depicts the ROC performance of various inputs and that of various networks with integrated data.
Table 3
| Variables | Accuracy | Sensitivity | Specificity | PPV | NPV | AUC |
|---|---|---|---|---|---|---|
| DenseNet | 93.21% | 92.30% | 93.13% | 87.60% | 96.01% | 95.17% |
| ResNet | 92.45% | 82.57% | 97.35% | 92.93% | 92.08% | 94.54% |
PPV, positive predictive value; NPV, negative predictive value; AUC, area under the curve.

Discussion
In this study, we trained a predictive system by utilizing a convolutional neural network (CNN) to retrieve high-order attributes from an MR image of the head. This study's deep learning approach provided the best performance for predicting the recurrence status of epilepsy, which indicated that this predictive system can possibly be applied in clinical settings. To the best of our knowledge, this is a novel usage of deep learning for predicting the likelihood of epilepsy relapse in patients who have FCD type III-associated refractory epilepsy.
We used T2W FLAIR to train and test the network because the sequence was more sensitive in detecting subtle lesions. In many instances, an epileptogenic zone involves the ‘normal-appearing’ perilesional cortex (22,30,31). Epileptic networks are arranged within the irritative zone, which can encompass the real seizures zone, even beyond bounds of the apparent lesion or extend it to further remote zones. Therefore, as many of the ROI were mapped out as possible. The information around the epileptogenic focus must be gathered from the entire volumes, whereas the data on the epileptogenic focus can be acquired from the ROI. The 2 types of data described above were included in the integrated input. To avoid the deep CNN model focusing on irrelevant parts of the brain image, we included the outlined ROI input in the CNN model and gave greater weight to the brain regions that may be related to seizure recurrence. This innovative strategy increases confidence in the deep neural networks’ capacity to forecast. We can infer that the improvement of our proposed method is based on 2 factors: (I) data augmentation, specifically, increasing the training set, and (II) giving more weight to the brain regions that may be related to seizure recurrence.
In our study, the deep learning model successfully predicted the postoperative relapse of epilepsy. In contrast to existing visual assessment with partial neuroimaging features, our deep learning algorithm was able to reach a definite conclusion by analyzing the holistic attributes of the original images, which may incorporate the effects of a number of different anatomic zones and histopathologies.
Regarding its predictive power, this deep learning algorithm treats the raw brain scans as a volume made up of pixels, and this quantification of MRI data is preferable and more reliable and precise than a qualitative approach to diagnosis based on images.
The average test time for the epilepsy predictive system to judge whether a patient can achieve freedom from seizures after surgery was only 0.5 seconds using the trained networks, which means that it has the potential to be beneficial as a quick tool for preoperative assessment. Furthermore, the outstanding performance of the proposed model could help physicians to recognize the risk of seizure recurrence and optimize postoperative clinical surveillance.
This study has several limitations. First, it was a retrospective study, and the outcomes reflect the specifics of the included sample. More extensive and prospective research is needed prior to clinical application of the approach. Moreover, some patients’ follow-up periods were not long enough. There is a risk of overfitting, and multicenter external validation is required. Third, subgroup analysis should be undertaken, no patients were recruited to perform an external test set, and it is likely that 1.5T MRI scanners underestimate the assessment of FCD type III.
Acknowledgments
Thanks to the pathologist’s (Li Hainan) assistance in confirming the postoperative pathology and the neurologists’ (Kai Peng and Haixuan Yang) interpretation and assessment of the EEG results. We also wish to thank the patients for their participation in this research.
Funding: This work was supported in part by the National Natural Science Foundation of China (No. 62276122), and Guangzhou Science and Technology Project (No. 202102020297).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-276/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-276/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Board of the Third Affiliated Hospital of Southern Medical University, and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Tahta A, Turgut M. Focal cortical dysplasia: etiology, epileptogenesis, classification, clinical presentation, imaging, and management. Childs Nerv Syst 2020;36:2939-47. [Crossref] [PubMed]
- Taylor DC, Falconer MA, Bruton CJ, Corsellis JA. Focal dysplasia of the cerebral cortex in epilepsy. J Neurol Neurosurg Psychiatry 1971;34:369-87. [Crossref] [PubMed]
- Najm IM, Sarnat HB, Blümcke I. Review: The international consensus classification of Focal Cortical Dysplasia - a critical update 2018. Neuropathol Appl Neurobiol 2018;44:18-31. [Crossref] [PubMed]
- Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, Palmini A, et al. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia 2011;52:158-74. [Crossref] [PubMed]
- Giulioni M, Martinoni M, Marucci G. About focal cortical dysplasia (FCD) type IIIa. Epilepsy Res 2014;108:1955-7. [Crossref] [PubMed]
- Kun Y, Zejun D, Jian Z, Feng Z, Changqing L, Xueling Q. Surgical histopathologic findings of 232 Chinese children cases with drug-resistant seizures. Brain Behav 2020;10:e01565. [Crossref] [PubMed]
- Wang S, Tang Y, Aung T, Chen C, Katagiri M, Jones SE, Prayson RA, Krishnan B, Gonzalez-Martinez JA, Burgess RC, Najm IM, Alexopoulos AV, Wang S, Ding M, Wang ZI. Multimodal noninvasive evaluation in MRI-negative operculoinsular epilepsy. J Neurosurg 2019;132:1334-44. [Crossref] [PubMed]
- Jayalakshmi S, Nanda SK, Vooturi S, Vadapalli R, Sudhakar P, Madigubba S, Panigrahi M. Focal Cortical Dysplasia and Refractory Epilepsy: Role of Multimodality Imaging and Outcome of Surgery. AJNR Am J Neuroradiol 2019;40:892-8. [Crossref] [PubMed]
- Feng C, Zhao H, Tian M, Lu M, Wen J. Detecting focal cortical dysplasia lesions from FLAIR-negative images based on cortical thickness. Biomed Eng Online 2020;19:13. [Crossref] [PubMed]
- Jafari-Khouzani K, Elisevich K, Wasade VS, Soltanian-Zadeh H. Contribution of Quantitative Amygdalar MR FLAIR Signal Analysis for Lateralization of Mesial Temporal Lobe Epilepsy. J Neuroimaging 2018;28:666-75. [Crossref] [PubMed]
- Widdess-Walsh P, Kellinghaus C, Jeha L, Kotagal P, Prayson R, Bingaman W, Najm IM. Electro-clinical and imaging characteristics of focal cortical dysplasia: correlation with pathological subtypes. Epilepsy Res 2005;67:25-33. [Crossref] [PubMed]
- Blumcke I, Spreafico R, Haaker G, Coras R, Kobow K, Bien CG, et al. Histopathological Findings in Brain Tissue Obtained during Epilepsy Surgery. N Engl J Med 2017;377:1648-56. [Crossref] [PubMed]
- Wang X, Deng D, Zhou C, Li H, Guan X, Fang L, Cai Q, Wang W, Zhou Q. Focal Cortical Dysplasia Type III Related Medically Refractory Epilepsy: MRI Findings and Potential Predictors of Surgery Outcome. Diagnostics (Basel) 2021;11:2225. [Crossref] [PubMed]
- Luyken C, Blümcke I, Fimmers R, Urbach H, Elger CE, Wiestler OD, Schramm J. The spectrum of long-term epilepsy-associated tumors: long-term seizure and tumor outcome and neurosurgical aspects. Epilepsia 2003;44:822-30. [Crossref] [PubMed]
- Wang DD, Piao YS, Blumcke I, Coras R, Zhou WJ, Gui QP, Liu CC, Hu JX, Cao LZ, Zhang GJ, Lu DH. A distinct clinicopathological variant of focal cortical dysplasia IIId characterized by loss of layer 4 in the occipital lobe in 12 children with remote hypoxic-ischemic injury. Epilepsia 2017;58:1697-705. [Crossref] [PubMed]
- Palkopoulou M, Bakola E, Foliadi M, Stefanidis P, Acquaviva PT. Cerebral cavernous malformation in a patient with pontine hemorrhage: A case study. Clin Pract 2020;10:1211. [Crossref] [PubMed]
- Kwan P, Arzimanoglou A, Berg AT, Brodie MJ, Allen Hauser W, Mathern G, Moshé SL, Perucca E, Wiebe S, French J. Definition of drug resistant epilepsy: consensus proposal by the ad hoc Task Force of the ILAE Commission on Therapeutic Strategies. Epilepsia 2010;51:1069-77. [Crossref] [PubMed]
- Fauser S, Essang C, Altenmüller DM, Staack AM, Steinhoff BJ, Strobl K, Bast T, Schubert-Bast S, Stephani U, Wiegand G, Prinz M, Brandt A, Zentner J, Schulze-Bonhage A. Long-term seizure outcome in 211 patients with focal cortical dysplasia. Epilepsia 2015;56:66-76. [Crossref] [PubMed]
- Korsakova MB, Kozlova AB, Arkhipova NA, Shishkina LV, Vlasov PA, Troshina EM. Features of ictal and interictal electrical activity in assessment of the epileptogenic zone in children with focal cortical dysplasias. Zh Vopr Neirokhir Im N N Burdenko 2019;83:90-7. [Crossref] [PubMed]
- Chassoux F, Devaux B, Landré E, Turak B, Nataf F, Varlet P, Chodkiewicz JP, Daumas-Duport C. Stereoelectroencephalography in focal cortical dysplasia: a 3D approach to delineating the dysplastic cortex. Brain 2000;123:1733-51. [Crossref] [PubMed]
- Aubert S, Wendling F, Regis J, McGonigal A, Figarella-Branger D, Peragut JC, Girard N, Chauvel P, Bartolomei F. Local and remote epileptogenicity in focal cortical dysplasias and neurodevelopmental tumours. Brain 2009;132:3072-86. [Crossref] [PubMed]
- Shen D, Wu G, Suk HI. Deep Learning in Medical Image Analysis. Annu Rev Biomed Eng 2017;19:221-48. [Crossref] [PubMed]
- Krsek P, Kudr M, Jahodova A, Komarek V, Maton B, Malone S, Miller I, Jayakar P, Resnick T, Duchowny M. Localizing value of ictal SPECT is comparable to MRI and EEG in children with focal cortical dysplasia. Epilepsia 2013;54:351-8. [Crossref] [PubMed]
- Wieser HG, Blume WT, Fish D, Goldensohn E, Hufnagel A, King D, Sperling MR, Lüders H, Pedley TA. ILAE Commission Report. Proposal for a new classification of outcome with respect to epileptic seizures following epilepsy surgery. Epilepsia 2001;42:282-6. [Crossref] [PubMed]
- Besson P, Bernasconi N, Colliot O, Evans A, Bernasconi A. Surface-based texture and morphological analysis detects subtle cortical dysplasia. Med Image Comput Comput Assist Interv 2008;11:645-52.
- Pail M, Mareček R, Hermanová M, Slaná B, Tyrlíková I, Kuba R, Brázdil M. The role of voxel-based morphometry in the detection of cortical dysplasia within the temporal pole in patients with intractable mesial temporal lobe epilepsy. Epilepsia 2012;53:1004-12. [Crossref] [PubMed]
- Aung TT, Murayi R, Aung T, Murakami H, Jones SE, Alexopoulos A, Najm I, Wang ZI. Prospective use of MRI post-processing in the surgical treatment of MRI-negative orbitofrontal epilepsy. J Neurol Sci 2020;414:116828. [Crossref] [PubMed]
- Huang G, Liu Z, Pleiss G, Maaten LV, Weinberger KQ. Convolutional Networks with Dense Connectivity. IEEE Trans Pattern Anal Mach Intell 2022;44:8704-16. [Crossref] [PubMed]
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 27-30 June 2016; Las Vegas, NV, USA. IEEE, 2016.
- Rossini L, Garbelli R, Gnatkovsky V, Didato G, Villani F, Spreafico R, Deleo F, Lo Russo G, Tringali G, Gozzo F, Tassi L, de Curtis M. Seizure activity per se does not induce tissue damage markers in human neocortical focal epilepsy. Ann Neurol 2017;82:331-41. [Crossref] [PubMed]
- Seifer G, Blenkmann A, Princich JP, Consalvo D, Papayannis C, Muravchik C, Kochen S. Noninvasive approach to focal cortical dysplasias: clinical, EEG, and neuroimaging features. Epilepsy Res Treat 2012;2012:736784. [Crossref] [PubMed]


