
Free-breathing simultaneous water-fat separation and T1 mapping of the whole liver (SWALI) with isotropic resolution using 3D golden-angle radial trajectory
Introduction
In recent years, T1 mapping—especially the T1 change before and after magnetic resonance (MR) contrast administration of the liver—has been used for the diagnosis of liver disease (1,2), assessing liver fibrosis (3-6) and inflammation (6,7), and for evaluating liver function (8-10). To quantify the T1 of the liver, previous studies often used the variable flip angle technique (4,7-10) or the modified Look-Locker inversion recovery (MOLLI) sequence (1-3,5,6,8). However, these conventional liver T1 mapping techniques are usually performed under breath-holding conditions and have limited slice coverage or rely on multiple acquisitions (1-3,5,9,10). Therefore, a single-scan, free-breathing, whole-liver T1 quantitative technique is needed.
In addition, due to the different resonance frequencies between water and fat protons (~3.5 ppm), the accuracy of T1 quantification can be affected by the presence of fat in the liver, especially for patients with fatty liver. A possible way to solve this problem is to suppress the fat signal using fat saturation, inversion recovery (IR), or water excitation techniques (11-13). However, the fat signal also has diagnostic values under certain circumstances (14-16). Another idea is to separate water and fat by postprocessing the chemical shift-encoded (CSE) data (17-19). Recently, based on the Dixon method, a variety of improved methods for water-fat separation have been developed (20-24). Water-fat separation methods can eliminate the effects of the fat in T1 mapping and also allow the quantification of fat, which is useful for diagnosing fatty liver disease (14-16).
Several previous techniques were proposed that aimed to achieve water-fat separation and T1 quantification simultaneously. The MR fingerprinting technique (25,26) and the PROFIT1 (simultaneous proton density fat-fraction and R2* imaging with water-specific T1 mapping) technique (27) have been used to estimate multiparameters of the liver. However, these techniques were performed under breath-holding conditions with limited slice coverage (25-27). An imaging sequence for joint myocardial T1 mapping and fat–water separation was proposed (28). It can be performed under free-breathing conditions using a pencil beam navigator to track respiratory motion; however, using the navigator reduces the acquisition efficiency (28). The Multiecho Magnetization-Prepared Golden-Angle Radial Sparse Parallel (MP-Dixon-GRASP) technique using IR-prepared stack-of-stars acquisition was proposed for fat-water-separated T1 mapping of the liver (29). Moreover, multitasking multiecho (MT-ME) magnetic resonance imaging (MRI) has been proposed to assess the liver water-specific T1 and fat fraction (FF) (30). However, limited spatial resolution in the slice direction is a common problem faced by the stack-of-stars trajectory used in MP-Dixon-GRASP and MT-ME techniques (29,30).
Here, we propose a free-breathing technique that simultaneously achieves water-fat separation and T1 mapping for the whole liver with isotropic spatial resolution and present the results from testing its clinical feasibility in volunteers and patients.
Methods
Sequence design
The proposed simultaneous water-fat separation and T1 mapping of the whole liver (SWALI) sequence is implemented on a 3.0T MR scanner (Ingenia CX, Philips Healthcare, Best, the Netherlands). An IR preparation pulse is used to generate the T1 contrast (Figure 1A). After a short time Tgap, a series of excitation pulses are performed, and a series of multiecho radial spokes are acquired (Figure 1A). At the end of each shot, a time Tex is used to allow further recovery of the longitudinal magnetization (Figure 1A).
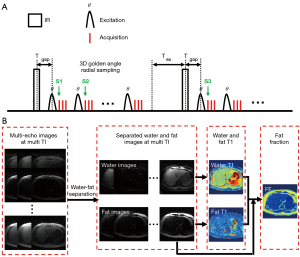
Radial spokes in the adjacent shots with the same inversion time (TI; S1 and S3 in Figure 1A) follow the consecutive three-dimensional (3D) golden-angle distribution (31). The azimuthal angle and polar angle increments (∆α and ∆β) between S1 and S3 (Figure 1A) are as follows:
and are two-dimensional (2D) golden-angle means, where and (31).
The azimuthal angle and polar angle increments between the adjacent radial spokes after the same IR (S1 and S2 in Figure 1A) are as follows:
M is the total number of shots, […] indicates that only a fractional part is kept.
Spokes after the same excitation pulse acquired at different echo times (TEs) share the same acquisition trajectory.
Assuming the longitudinal magnetization before IR is Mss, then the longitudinal magnetization before the first excitation pulse Mθ (1) is the following:
M0 is the equilibrium magnetization, and .
The longitudinal magnetization before the kth excitation pulse Mθ (k) is as follows:
where . After experiencing a series of the excitation pulses and the time Tex, the longitudinal magnetization before the next IR is as follows:
N is the number of excitation pulses in 1 shot (turbo field echo factor).
After several shots, the steady-state can be achieved, with . Thus, Mss can be calculated as follows:
The MR signal acquired after each of the kth excitation pulses Mxy (k) can be calculated as follows:
The scan parameters of the proposed SWALI sequence are as follows: field of view (FOV) =200×200×200 mm3 (2-fold oversampling), spatial resolution =2.5×2.5×2.5 mm3, matrix size =160×160×160, repetition time (TR) =10 ms, 3 unipolar echoes, first TE/ΔTE =1.02/1.40 ms, flip angle =8°, turbo field echo factor =175, Tgap =10 ms (minimum value allowed on the scanner), inversion recovery repetition time (IRTR) =2,000 ms, Tex =240 ms [IRTR − (Tgap + turbo field echo factor × TR)], the total number of shots =150, and scan duration =5 min.
Image reconstruction
The reconstruction was implemented in MATLAB (MathWorks, Inc., Natick, MA, USA). For each TE, a series of T1-weighted water-fat mixed images were reconstructed using a sliding window method with a temporal width of 25 spokes (temporal resolution =250 ms) (32-34). The reconstruction method with low-rank modeling and sparsity constraints was applied to further improve the image quality (35-37).
Water-fat separation and T1 mapping
The quantification process of T1 and FF is shown in Figure 1B. First, for the reconstructed multiecho images at each TI, water and fat are separated based on a multipeak fat model (24):
Where s (tn) is the MR signal at TE ; fB is the frequency shift caused by B0 field inhomogeneity; MW and MF are the amplitudes of the water and fat components, respectively; P is the number of fat spectrum peaks; fP is the frequency difference of the p th fat spectral peak relative to water (p=1, 2, …, P); αP is the relative amplitude for the p th fat spectral peak; and . In this study, the values of P, fP and αP were taken from the literature (20). s (tn) was obtained from the reconstructed multiecho images, and fB, MW and MF were fitted using a graph cut algorithm (20,38).
After separating the water and fat images at each TI, separated water and fat T1 mapping of the liver are estimated by fitting the signal intensities extracted from a series of T1-weighted water or fat images to the theoretically calculated signal {Eqs. [5-10]} using the nonlinear least squares algorithm (39).
Then, the equilibrium magnetization of the water (M0W) and fat (M0F) are calculated on the last TI image by treating water or fat T1 as the known values in Eqs. [5-10]. FF is defined as 100× M0F /(M0W + M0F). The magnitude discrimination method is used to reduce the noise bias at a low FF (40).
Phantom study
A total of 13 in-house water-fat phantoms with different T1 values (range, 100–1,000 ms) and FFs (range, 0–100%) were made with distilled and deionized water, agar (2.0% w/v), gadolinium diethylenetriamine pentaacetic acid (Gd-DTPA; concentration range, 1.85–0.07 mmol/L to produce different T1 values; Magnevist, Bayer Schering Pharma, Berlin, Germany), peanut oil, and sodium dodecyl sulfate (Sinopharm Chemical Reagent Co., Ltd., Shanghai, China) (41,42). Phantoms were scanned on a 3.0T MR scanner (Ingenia CX, Philips Healthcare). Apart from the proposed SWALI sequence, the commercial CSE-MRI method (mDIXON, Philips Healthcare) and 2D fat-suppressed (FS) IR-spin echo (SE) sequences were used as the gold standard of the FF and water T1 quantification. The scan parameters of CSE-MRI were FOV =380×264 mm2, spatial resolution =2.5×2.5 mm2, slice thickness =5 mm, TR =5.80 ms, 6 bipolar echoes, first TE/ΔTE =1.01/0.70 ms, flip angle =3°, and scan duration =10 s. FS IR-SE were scanned using the following parameters: FOV =200×140 mm2, spatial resolution =2.5×2.5 mm2, slice thickness =5 mm, TR/TE =10,000/9.0 ms, TI = (100, 200, 300, 400, 500, 600, 700, 800, 900, 1,000, 1,500, 2,000) ms, and scan duration =9 min 40 s per TI. The total scan time of the gold standard sequences was 1 h 56 min and 10 s. In addition, to investigate the effect of fat on T1 quantification, the MOLLI (43) sequence was performed to estimate the water-fat compound T1 of the phantom with the following scan parameters: FOV =300×300 mm2, spatial resolution =2×2 mm2, slice thickness =10 mm, TR/TE =1.98/0.89 ms, flip angle =20°, no fat suppression, and scan duration =11 s.
In vivo study
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Tsinghua University, and informed consent was obtained from all individual participants. The study recruited 9 healthy volunteers, 2 patients with fatty liver, and 3 patients with hepatocellular carcinoma (HCC). All scans were performed on a 3.0-T MR scanner (Ingenia CX, Philips Healthcare) using a 16-channel torso coil and a 16-channel posterior coil. Healthy volunteers and patients were instructed to breathe normally during the SWALI sequence acquisition.
Because the total scan time of FS IR-SE is too long and can be easily affected by respiratory motion, only CSE-MRI and MOLLI (43) sequences were used for FF and T1 quantification comparison in healthy volunteers and patients with fatty liver. The CSE-MRI sequence was performed under breath-holding conditions with the same parameters as those used in the phantom study. Three axial slices at different locations were imaged using 3 MOLLI sequences. Each MOLLI sequence was performed in 1 breath-hold under the following scan parameters: FOV =380×380–400×400 mm2, spatial resolution =2×2 mm2, slice thickness =5 mm, TR/TE =1.98/0.89 ms, flip angle =20°, no fat suppression, and scan duration =11 s. Three square regions of interest (ROIs; ~100 mm2 each) were manually drawn on each MOLLI image, resulting in 9 liver ROIs for each participant. Corresponding liver ROIs were also manually drawn on SWALI and CSE-MRI images on the slices at the same location as MOLLI. To evaluate the reproducibility of the proposed SWALI sequence, a repeated SWALI scan was acquired in 5 healthy volunteers and 2 patients with fatty liver using the same imaging protocol.
Patients with HCC underwent a gadolinium ethoxybenzyl diethylenetriamine pentaacetic acid (Gd-EOB-DTPA)-enhanced MR scans. During the MR scan, 0.025 mmol/kg of Gd-EOB-DTPA (Primovist; Bayer Schering Pharma) was injected at a rate of 2 ml/s. The SWALI sequence was performed precontrast and 15 min after the contrast administration. The difference between the pre- and postcontrast water T1 (ΔT1) was calculated as 100 × (T1pre − T1post)/T1pre pixel by pixel, which could reflect the liver function (9). Nine square ROIs (~100 mm2 each) were drawn on liver parenchyma, and the mean precontrast FF and T1, the mean postcontrast FF and T1, and the mean ΔT1 within each ROI were averaged.
Statistical analysis
In the phantom study, the mean and standard deviation of the FF and water T1 of each phantom estimated by SWALI were calculated and compared with those obtained using the CSE-MRI and FS IR-SE using Pearson or Spearman correlation and linear regression analysis.
For healthy volunteers and patients with fatty liver, a paired t-test or Wilcoxon signed-rank test was used to compare the mean FF or the mean T1 within each ROI estimated by the SWALI sequence with that estimated using CSE-MRI or MOLLI. Pearson or Spearman correlation and linear regression analyses were also performed to compare the SWALI and reference sequences. In the reproducibility study, the FF and water T1 from the second scan were compared with those from the first SWALI scan using the intraclass correlation coefficient (ICC). For patients with HCC, the differences between the pre- and postcontrast FF and the pre- and postcontrast T1 were compared using a paired t-test or Wilcoxon signed-rank test, as appropriate.
A P value less than 0.05 was considered statistically significant. All statistical analyses were performed in SPSS 23.0 (IBM Corp, Armonk, NY, USA).
Results
Phantom study
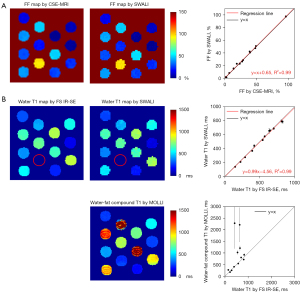
The FF and water T1 maps of the phantom estimated with the gold standard sequences and the proposed SWALI sequence are shown in Figure 2. The mean FF and water T1 values within each tube estimated by the SWALI sequence show good agreement with those estimated with CSE-MRI [correlation coefficient (r) =0.998; P<0.001; R2=0.99] and FS IR-SE (r=0.997; P<0.001; R2=0.99). In tubes with high FFs, the water-fat compound T1 estimated using MOLLI was longer than the water T1 estimated using FS IR-SE (Figure 2B).
In vivo study
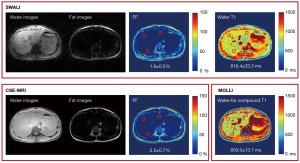
All 9 healthy volunteers (7 males and 2 females, age 34.7±10.8 years) and 2 patients with fatty liver (2 males, age 25.5±3.5 years) underwent MR acquisitions. The separated water and fat images, FF maps, and T1 maps of 1 healthy volunteer are shown in Figure 3. Similar FF and T1 distributions were found when using the SWALI and compared sequences (Figure 3). Figure 4 shows the separated water and fat images and quantitative maps of 1 patient with fatty liver (mean FF =18.9%). The water T1 map estimated using the SWALI sequence was different from the water-fat compound T1 map estimated using MOLLI (Figure 4).
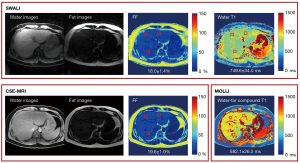
For all healthy volunteers and patients with fatty liver, the comparison of FF and water T1 between the proposed SWALI sequence and the reference sequences are shown in Table 1. Results from the SWALI sequence showed no significant difference from those estimated using CSE-MRI in FF quantification (P=0.53) with a r of 0.989 (P<0.001) in the correlation analysis and an R2 value of 0.98 in the linear regression analysis. The mean FF calculated using the SWALI and CSE-MRI techniques were 2.6%±1.3% vs. 2.5%±0.7% in healthy volunteers (n=9). The mean FF calculated using the SWALI and CSE-MRI techniques were 27.5%±9.5% vs. 27.0%±8.7% in patients with fatty liver (n=2). In T1 quantification, a slightly higher T1 was obtained by the proposed SWALI sequence than that by MOLLI without a significant difference (775.9±71.0 vs. 768.4±61.5 ms; P=0.10) in healthy volunteers (n=9), with a r of 0.821 (P<0.001) in the correlation analysis and an R2 value of 0.68 in the linear regression analysis. The water T1 estimated using the SWALI sequence was significantly lower than the water-fat compound T1 estimated by MOLLI (778.1±53.8 vs. 1,093.9±230.1 ms; P<0.001) in patients with fatty liver (n=2).
Table 1
| Participants | FF (%) | Water T1 (ms) | |||||
|---|---|---|---|---|---|---|---|
| SWALI | CSE-MRI | P | SWALI | MOLLI | P | ||
| Healthy volunteers | 2.6±1.3 | 2.5±0.7 | 0.53 | 775.9±71.0 | 768.4±61.5 | 0.10 | |
| Patients with fatty liver | 27.5±9.5 | 27.0±8.7 | 778.1±53.8 | 1,093.9±230.1 | <0.001 | ||
Data are expressed as mean ± SD. FF, fat fraction; SWALI, simultaneous water-fat separation and T1 mapping of the whole liver; CSE-MRI, chemical shift-encoded magnetic resonance imaging; MOLLI, modified Look-Locker inversion recovery; SD, standard deviation.
In the subset of 7 participants (5 healthy volunteers and 2 patients with fatty liver) who underwent repeated SWALI scans, the ICCs for the estimated FF and water T1 between the first and the second scan were 0.997 [95% confidence interval (CI): 0.995–0.998; P<0.001] and 0.943 (95% CI: 0.906–0.965; P<0.001), respectively.
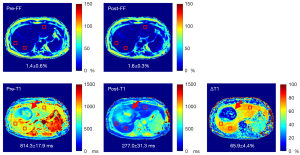
The 3 recruited patients with HCC (2 males and 1 female, age 52.7±6.7 years) completed the pre- and postcontrast SWALI acquisitions. Figure 5 shows the FF map, T1 map, and T1 map estimated from pre- and postcontrast SWALI sequences of 1 patient. The HCC region shows different T1 distributions compared to those of the liver parenchyma on pre- and postcontrast T1 maps and T1 maps (Figure 5). For all patients, the FF estimated by the pre- and postcontrast SWALI showed no significant difference (2.1%±0.8% vs. 1.8%±0.5%; P=0.08). Water T1 of the patients showed a significant reduction after contrast administration (277.7±23.6 vs. 816.7±37.7 ms; P<0.001). The mean ∆T1 of the patients was 66.0%±2.5%.
Discussion
In this study, free-breathing water-fat separation and T1 mapping of the whole liver were achieved simultaneously within 1 scan using the proposed SWALI sequence. Compared to commonly used T1 techniques in the liver, such as MOLLI and variable flip angle sequences, the advantages of the proposed sequence included free-breathing, a single scan for the whole liver coverage, and isotropic spatial resolution, and the obtained images and quantitative maps were naturally coregistered. The proposed SWALI sequence showed good FF and T1 quantification accuracy when compared with the gold standard sequences in the phantom study. The clinical feasibility of the SWALI sequence was demonstrated by the volunteer and patient study.
A free-breathing technique is valuable, especially for patients who have difficulties holding their breath. The 3D golden-angle radial trajectory used in the proposed sequence was robust to motion. The results of our study demonstrated that, even though no respiratory motion correction algorithm was applied, the proposed free-breathing technique did not show severe motion artifacts on the reconstructed images or in the estimated FF and T1 maps. In contrast to the stack-of-stars trajectory used in the MT-ME technique (30), which needs perpendicular center k-space line acquisition every 8 readouts, there was no need to acquire the additional repeated k-space data for the 3D golden-angle radial trajectory because each of the k-space lines went through the k-space center. The temporal motion information could be directly extracted from the acquired k-space data itself (30,44,45). In the future, self-respiratory motion correction algorithms can be applied to reduce the image blur caused by respiratory motion and can further improve the quantification accuracy, especially in those with large respiratory motions (30,44,45).
In addition to FS IR-SE, MOLLI was also used for T1 comparison in this study. However, fat suppression or water selection could not be applied to the MOLLI sequence; thus, MOLLI was used to measure the water-fat compound T1. Therefore, finding a significant difference between the water T1 measured by the proposed SWALI sequence and the water-fat compound T1 measured by the MOLLI sequence in patients with high liver FFs was reasonable. In this study, the TE of the MOLLI sequence was set as 0.89 ms, which was close to the out-of-phase time of water and fat at 3.0 T. Under this scan parameter, the signal recovery could be calculated by the difference between the water and fat recovery weighted by their relative fractions. Thus, the presence of fat slowed down the recovery of the water signal, resulting in a longer water-fat compound T1 obtained by MOLLI compared to water T1, as shown in the previous study and the phantom and in vivo experiments of our study (46). Although no gold-standard in vivo water T1 quantification sequence was applied in this study, the water T1 of the liver in healthy volunteers estimated by the proposed SWALI sequence was 775.9 ms, which was similar to that reported by previous studies (29,30).
This study still has some limitations. First, 3 echoes were used, and the effect of T2* was not considered in the water-fat separation of the SWALI sequence. Although T2* effect can be ignored when TE is short and in patients with normal T2*, the comparison between the SWALI sequence and the gold-standard CSE-MRI, which used 6 echoes for T2* correction, may have some underlying biases. In addition, the proposed SWALI sequence may not be suitable for patients with hepatic iron overload who have short T2* of the liver. In the future, more echoes should be applied in the SWALI sequence to estimate T2* paired with fat quantification. Second, FS IR-SE and MOLLI sequences were used in the phantom study, but only MOLLI was performed in the in vivo study because FS IR-SE has a long scan time and is sensitive to motion. T1 quantified by MOLLI will be affected by the presence of fat; therefore, other water T1 reference sequences, such as the water-fat separated variable flip angle technique, can be applied for in vivo comparison in the future. Third, the scan time of the proposed sequence was 5 min, which may still be too long for clinical application. More in vivo studies are required to explore the clinical value of the proposed technique.
Conclusions
In this study, free-breathing water-fat separation and T1 mapping quantification of the whole liver were achieved within 1 scan using the proposed SWALI sequence. The quantitative accuracy and the in vivo feasibility of the proposed sequence were demonstrated in phantom, volunteer, and patient studies.
Acknowledgments
We acknowledge the use of the International Society for Magnetic Resonance in Medicine (ISMRM) Fat-Water Toolbox (https://www.ismrm.org/workshops/FatWater12/data.htm). An abstract related to this work was presented at the ISMRM 31st Annual Meeting & Exhibition in May 2022.
Funding: This work was supported by the National Natural Science Foundation of China (No. 81930119 to Jiahong Dong) and the Beijing Municipal Natural Science Foundation (No. Z190024 to Huijun Chen).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-748/coif). Yishi Wang reports that he is an employee of Philips Healthcare. JD reports that this study was funded by the National Natural Science Foundation of China (No. 81930119). HC reports that this study was funded by the Beijing Municipal Natural Science Foundation (No. Z190024). The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of Tsinghua University, and informed consent was obtained from all individual participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Obmann VC, Mertineit N, Marx C, Berzigotti A, Ebner L, Heverhagen JT, Christe A, Huber AT. Liver MR relaxometry at 3T - segmental normal T(1) and T(2)* values in patients without focal or diffuse liver disease and in patients with increased liver fat and elevated liver stiffness. Sci Rep 2019;9:8106. [Crossref] [PubMed]
- Fahlenkamp UL, Ziegeler K, Adams LC, Böker SM, Engel G, Makowski MR. Native T1 mapping for assessment of the perilesional zone in metastases and benign lesions of the liver. Sci Rep 2020;10:12889. [Crossref] [PubMed]
- Luetkens JA, Klein S, Träber F, Schmeel FC, Sprinkart AM, Kuetting DLR, Block W, Uschner FE, Schierwagen R, Hittatiya K, Kristiansen G, Gieseke J, Schild HH, Trebicka J, Kukuk GM. Quantification of Liver Fibrosis at T1 and T2 Mapping with Extracellular Volume Fraction MRI: Preclinical Results. Radiology 2018;288:748-54. [Crossref] [PubMed]
- Li Z, Sun J, Hu X, Huang N, Han G, Chen L, Zhou Y, Bai W, Yang X. Assessment of liver fibrosis by variable flip angle T1 mapping at 3.0T. J Magn Reson Imaging 2016;43:698-703. [Crossref] [PubMed]
- Luetkens JA, Klein S, Traeber F, Schmeel FC, Sprinkart AM, Kuetting DLR, Block W, Hittatiya K, Uschner FE, Schierwagen R, Gieseke J, Schild HH, Trebicka J, Kukuk GM. Quantitative liver MRI including extracellular volume fraction for non-invasive quantification of liver fibrosis: a prospective proof-of-concept study. Gut 2018;67:593-4. [Crossref] [PubMed]
- von Ulmenstein S, Bogdanovic S, Honcharova-Biletska H, Blümel S, Deibel AR, Segna D, Jüngst C, Weber A, Kuntzen T, Gubler C, Reiner CS. Assessment of hepatic fibrosis and inflammation with look-locker T1 mapping and magnetic resonance elastography with histopathology as reference standard. Abdom Radiol (NY) 2022;47:3746-57. [Crossref] [PubMed]
- Wan Q, Peng H, Lyu J, Liu F, Cheng C, Qiao Y, Deng J, Zheng H, Wang Y, Zou C, Liu X. Water Specific MRI T1 Mapping for Evaluating Liver Inflammation Activity Grades in Rats With Methionine-Choline-Deficient Diet-Induced Nonalcoholic Fatty Liver Disease. J Magn Reson Imaging 2022;56:1429-36. [Crossref] [PubMed]
- Kim JE, Kim HO, Bae K, Choi DS, Nickel D. T1 mapping for liver function evaluation in gadoxetic acid-enhanced MR imaging: comparison of look-locker inversion recovery and B(1) inhomogeneity-corrected variable flip angle method. Eur Radiol 2019;29:3584-94. [Crossref] [PubMed]
- Zhou ZP, Long LL, Qiu WJ, Cheng G, Huang LJ, Yang TF, Huang ZK. Evaluating segmental liver function using T1 mapping on Gd-EOB-DTPA-enhanced MRI with a 3.0 Tesla. BMC Med Imaging 2017;17:20. [Crossref] [PubMed]
- Yoon JH, Lee JM, Kim E, Okuaki T, Han JK. Quantitative Liver Function Analysis: Volumetric T1 Mapping with Fast Multisection B(1) Inhomogeneity Correction in Hepatocyte-specific Contrast-enhanced Liver MR Imaging. Radiology 2017;282:408-17. [Crossref] [PubMed]
- Haase A, Frahm J, Hänicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol 1985;30:341-4. [Crossref] [PubMed]
- Meyer CH, Pauly JM, Macovski A, Nishimura DG. Simultaneous spatial and spectral selective excitation. Magn Reson Med 1990;15:287-304. [Crossref] [PubMed]
- Bydder GM, Steiner RE, Blumgart LH, Khenia S, Young IR. MR imaging of the liver using short TI inversion recovery sequences. J Comput Assist Tomogr 1985;9:1084-9. [Crossref] [PubMed]
- Cassidy FH, Yokoo T, Aganovic L, Hanna RF, Bydder M, Middleton MS, Hamilton G, Chavez AD, Schwimmer JB, Sirlin CB. Fatty liver disease: MR imaging techniques for the detection and quantification of liver steatosis. Radiographics 2009;29:231-60. [Crossref] [PubMed]
- Zhang QH, Zhao Y, Tian SF, Xie LH, Chen LH, Chen AL, Wang N, Song QW, Zhang HN, Xie LZ, Shen ZW, Liu AL. Hepatic fat quantification of magnetic resonance imaging whole-liver segmentation for assessing the severity of nonalcoholic fatty liver disease: comparison with a region of interest sampling method. Quant Imaging Med Surg 2021;11:2933-42. [Crossref] [PubMed]
- Syväri J, Junker D, Patzelt L, Kappo K, Al Sadat L, Erfanian S, Makowski MR, Hauner H, Karampinos DC. Longitudinal changes on liver proton density fat fraction differ between liver segments. Quant Imaging Med Surg 2021;11:1701-9. [Crossref] [PubMed]
- Dixon WT. Simple proton spectroscopic imaging. Radiology 1984;153:189-94. [Crossref] [PubMed]
- Glover GH. Multipoint Dixon technique for water and fat proton and susceptibility imaging. J Magn Reson Imaging 1991;1:521-30. [Crossref] [PubMed]
- Ma J. Dixon techniques for water and fat imaging. J Magn Reson Imaging 2008;28:543-58. [Crossref] [PubMed]
- Hernando D, Kellman P, Haldar JP, Liang ZP. Robust water/fat separation in the presence of large field inhomogeneities using a graph cut algorithm. Magn Reson Med 2010;63:79-90. [PubMed]
- Berglund J, Johansson L, Ahlström H, Kullberg J. Three-point Dixon method enables whole-body water and fat imaging of obese subjects. Magn Reson Med 2010;63:1659-68. [Crossref] [PubMed]
- Reeder SB, McKenzie CA, Pineda AR, Yu H, Shimakawa A, Brau AC, Hargreaves BA, Gold GE, Brittain JH. Water-fat separation with IDEAL gradient-echo imaging. J Magn Reson Imaging 2007;25:644-52. [Crossref] [PubMed]
- Yu H, McKenzie CA, Shimakawa A, Vu AT, Brau AC, Beatty PJ, Pineda AR, Brittain JH, Reeder SB. Multiecho reconstruction for simultaneous water-fat decomposition and T2* estimation. J Magn Reson Imaging 2007;26:1153-61. [Crossref] [PubMed]
- Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med 2008;60:1122-34. [Crossref] [PubMed]
- Ostenson J, Damon BM, Welch EB. MR fingerprinting with simultaneous T(1), T(2), and fat signal fraction estimation with integrated B(0) correction reduces bias in water T(1) and T(2) estimates. Magn Reson Imaging 2019;60:7-19. [Crossref] [PubMed]
- Jaubert O, Arrieta C, Cruz G, Bustin A, Schneider T, Georgiopoulos G, Masci PG, Sing-Long C, Botnar RM, Prieto C. Multi-parametric liver tissue characterization using MR fingerprinting: Simultaneous T(1), T(2), T(2) *, and fat fraction mapping. Magn Reson Med 2020;84:2625-35. [Crossref] [PubMed]
- Thompson RB, Chow K, Mager D, Pagano JJ, Grenier J. Simultaneous proton density fat-fraction and R2* imaging with water-specific T(1) mapping (PROFIT(1)): application in liver. Magn Reson Med 2021;85:223-38. [Crossref] [PubMed]
- Nezafat M, Nakamori S, Basha TA, Fahmy AS, Hauser T, Botnar RM. Imaging sequence for joint myocardial T(1) mapping and fat/water separation. Magn Reson Med 2019;81:486-94. [Crossref] [PubMed]
- Feng L, Liu F, Soultanidis G, Liu C, Benkert T, Block KT, Fayad ZA, Yang Y. Magnetization-prepared GRASP MRI for rapid 3D T1 mapping and fat/water-separated T1 mapping. Magn Reson Med 2021;86:97-114. [Crossref] [PubMed]
- Wang N, Cao T, Han F, Xie Y, Zhong X, Ma S, Kwan A, Fan Z, Han H, Bi X, Noureddin M, Deshpande V, Christodoulou AG, Li D. Free-breathing multitasking multi-echo MRI for whole-liver water-specific T(1), proton density fat fraction, and R2* quantification. Magn Reson Med 2022;87:120-37. [Crossref] [PubMed]
- Chan RW, Ramsay EA, Cunningham CH, Plewes DB. Temporal stability of adaptive 3D radial MRI using multidimensional golden means. Magn Reson Med 2009;61:354-63. [Crossref] [PubMed]
- Qi H, Sun J, Qiao H, Zhao X, Guo R, Balu N, Yuan C, Chen H. Simultaneous T(1) and T(2) mapping of the carotid plaque (SIMPLE) with T(2) and inversion recovery prepared 3D radial imaging. Magn Reson Med 2018;80:2598-608. [Crossref] [PubMed]
- Korosec FR, Frayne R, Grist TM, Mistretta CA. Time-resolved contrast-enhanced 3D MR angiography. Magn Reson Med 1996;36:345-51. [Crossref] [PubMed]
- Barger AV, Block WF, Toropov Y, Grist TM, Mistretta CA. Time-resolved contrast-enhanced imaging with isotropic resolution and broad coverage using an undersampled 3D projection trajectory. Magn Reson Med 2002;48:297-305. [Crossref] [PubMed]
- Qi H, Qiao H, Sun A, Chen S, Zhao X, Yuan C, Chen H. Highly Undersampled Kooshball reconstruction with low-rank modeling and sparsity constraints for high-resolution T1 mapping. In: Paris: Proceedings of the 27th Annual Meeting of ISMRM, 2018:abstr 248.
- Zhao B, Haldar JP, Christodoulou AG, Liang ZP. Image reconstruction from highly undersampled (k, t)-space data with joint partial separability and sparsity constraints. IEEE Trans Med Imaging 2012;31:1809-20. [Crossref] [PubMed]
- Sun A, Zhao B, Li Y, He Q, Li R, Yuan C. Real-time phase-contrast flow cardiovascular magnetic resonance with low-rank modeling and parallel imaging. J Cardiovasc Magn Reson 2017;19:19. [Crossref] [PubMed]
- Hu HH, Börnert P, Hernando D, Kellman P, Ma J, Reeder S, Sirlin C. ISMRM workshop on fat-water separation: insights, applications and progress in MRI. Magn Reson Med 2012;68:378-88. [Crossref] [PubMed]
- Qi H, Sun J, Qiao H, Chen S, Zhou Z, Pan X, Wang Y, Zhao X, Li R, Yuan C, Chen H. Carotid Intraplaque Hemorrhage Imaging with Quantitative Vessel Wall T1 Mapping: Technical Development and Initial Experience. Radiology 2018;287:276-84. [Crossref] [PubMed]
- Liu CY, McKenzie CA, Yu H, Brittain JH, Reeder SB. Fat quantification with IDEAL gradient echo imaging: correction of bias from T(1) and noise. Magn Reson Med 2007;58:354-64. [Crossref] [PubMed]
- Hines CD, Yu H, Shimakawa A, McKenzie CA, Brittain JH, Reeder SB. T1 independent, T2* corrected MRI with accurate spectral modeling for quantification of fat: validation in a fat-water-SPIO phantom. J Magn Reson Imaging 2009;30:1215-22. [Crossref] [PubMed]
- Bernard CP, Liney GP, Manton DJ, Turnbull LW, Langton CM. Comparison of fat quantification methods: a phantom study at 3.0T. J Magn Reson Imaging 2008;27:192-7. [Crossref] [PubMed]
- Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med 2004;52:141-6. [Crossref] [PubMed]
- Qi H, Jaubert O, Bustin A, Cruz G, Chen H, Botnar R, Prieto C. Free-running 3D whole heart myocardial T(1) mapping with isotropic spatial resolution. Magn Reson Med 2019;82:1331-42. [Crossref] [PubMed]
- Feng L, Axel L, Chandarana H, Block KT, Sodickson DK, Otazo R. XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing. Magn Reson Med 2016;75:775-88. [Crossref] [PubMed]
- Mozes FE, Tunnicliffe EM, Pavlides M, Robson MD. Influence of fat on liver T1 measurements using modified Look-Locker inversion recovery (MOLLI) methods at 3T. J Magn Reson Imaging 2016;44:105-11. [Crossref] [PubMed]


