
Assessment of COVID-19 lung involvement on computed tomography by deep-learning-, threshold-, and human reader-based approaches—an international, multi-center comparative study
Introduction
Coronavirus disease 2019 (COVID-19) is an infectious disease, caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which primarily manifests in the lungs (1). Chest computed tomography (CT) is regularly performed in the context of COVID pneumonia, since it has a potential role in its diagnosis and the detection of complications (1). Above all, the extent of lung involvement on chest CT is an established biomarker for the prognosis of the disease and supports clinical decision making (2-10). The originally established, semi-quantitative approach for assessment of lung involvement in COVID pneumonia was published by Pan et al. in 2020 (11). This chest CT score (CCS) considers each of the five pulmonary lobes with an individual score from 0–5, based on the extent of COVID typical findings (0: 0%; 1: <5%; 2: 5–25%; 3: 26–50%; 4: 51–75%; 5: >75%). The sum of five lobe scores equals the total CCS (range: 0–25). The semi-quantitative CCS and its later amendments have since been extensively used in clinical studies exploring the prognosis of COVID pneumonia (2,3,5,6,8).
Semi-quantitative calculation of the CCS is time-consuming and requires experienced readers (3,8). Visual estimation of the individual lobe scores demands identification of involved lung tissue and comparison to the total lobe volume, which might introduce an inter-rater bias (3). Contrarily, slice-by-slice manual segmentation of involved vs. non-involved lung tissue might be an accurate method of lung involvement assessment, but is not feasible for daily routine diagnostic of every patient during this pandemic. Several artificial intelligence-based tools for segmentation of involved lung in COVID are under development or already commercially available [e.g., syngo.via Plug-In by Siemens Healthcare, Erlangen, Germany (12,13); Thoracic VCAR by GE Healthcare, Chicago, United States of America (14); CT Lung Analysis by Canon Medical Systems, Ōtawara, Japan (15), IntelliSpace Portal CT Pulmo Auto Results by Philips Healthcare, Amsterdam, Netherlands]. Deep-learning-driven segmentation allows for fully automated, volumetric assessment of involved lung in COVID pneumonia in a fast manner. Further, numerous AI classification models of COVID pneumonia have been developed, some based on automated segmentation results (16,17). However, to the best of our knowledge, satisfactory deep-learning-based segmentation of involved lung is still limited to the commercially offered products.
Besides manual semi-quantitative and automated deep-learning-driven assessment of lung involvement, an automated threshold-based approach is a third method to evaluate the extent of lung involvement in COVID. Typical CT manifestations of COVID pneumonia include ground-glass opacifications, consolidations, and reticular patterns (1). All of those findings are apparent by an increase of lung attenuation, when compared to the well-aerated lung tissue. Notably, threshold-based COVID pneumonia segmentation is capable to achieve similar accuracy compared to the recent AI-based approaches (18). Two recent studies investigated arbitrary thresholds for identification of involved lung in COVID-19 (19,20). However, both studies examined only single-center, single-vendor data, and a robust threshold to discriminate involved vs. non-involved lung in COVID pneumonia is not yet investigated.
The aim of our study is to compare the performance of automated deep-learning- and threshold-based methods to the manual semi-quantitative lung involvement scoring by Pan et al. (11). Further, we aim to investigate an optimal threshold for quantification of COVID involved lung in chest CT, and provide an exemplary tool based on our findings. We present the following article in accordance with the GRRAS reporting checklist (available at https://qims.amegroups.com/article/view/10.21037/qims-22-175/rc).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of the University Cologne (No. 21-1426-retro) and individual consent for this retrospective analysis was waived.
Patient population
Patients were retrospectively enrolled from our institute and four publicly available datasets. This approach aimed to achieve a heterogeneous, multi-center, international dataset. If several consecutive CT exams were available for the same patient, only the initial scan was evaluated. Inclusion criteria comprised:
- Positive reverse transcription polymerase chain reaction test for SARS-CoV-2,
- Chest computed tomography without intravenous contrast administration,
- Patient age ≥18 years.
Fifty consecutive patients were included from our radiological information system, starting from 10th of March to 15th of October, 2020. Further 200 patients were randomly included from four publicly available datasets (n=50 patients each) (21-25). Three patients were excluded from the dataset by An et al., since they were obviously younger than 18 years, albeit their individual age was not reported (25). One patient was excluded from the dataset by Shakouri et al. due to severe thoracic scoliosis. CT exams were converted to a common format (.nifti) and saved to a shared folder to promote automated processing. There were no further specific steps of data curation.
Semi-quantitative scoring of lung involvement
Lung involvement was estimated semi-quantitatively by three experienced radiologists in independent reading sessions (clinical experience of >700 COVID chest CT scans each), obtaining the CCS as published by Pan et al. (8). Readers were blinded to clinical information. The readings were performed one CT scan at a time in a quiet environment on a clinically approved workstation. Readers were explicitly free to adapt window settings and adjust the time between the readings. Inter-rater reliability was assessed by the intraclass correlation coefficient (ICC) in a single rater type, two-way random-effects model (ICC2) (26).
Deep-learning-based quantification of lung involvement
To quantify lung involvement using a deep-learning-based approach, an IntelliSpace Discovery Plugin was used. This plugin is a pre-release of the CT Pulmo Auto Results application available in the CE-certified IntelliSpace Portal 11.1.6 and higher releases (Philips Healthcare) (16,27,28). No changes have been applied to the final, commercially available software; i.e., the segmentation software used in this publication fully resembles the CT Pulmo Auto Results application release. The software can identify and quantify consolidations and ground-glass opacities in adult patients in a fully automated approach, employing pre-trained neural networks.
The software operates in a cascaded fashion. First, a network is applied to detect the lungs in the CT scan. Second, the CT scan is cropped to the region identified by the first network. This cropped scan is forwarded to a second network to segment the lungs, thereby excluding the main airways (including trachea, main bronchi, and lobar bronchi), and the main vessels. Visualizations of the network can be found in Figure 1. The lung segmentation network has an architecture in line with Milletari et al. (29). Next, COVID-19 related lung infiltrations are segmented within the lungs, using the f-net architecture as proposed by Brosch et al. (30) (see Figure 2). During the last step, a second f-net is applied to the segmented lesions to perform a voxel-wise classification as either consolidation or ground-glass opacity. The percentage portion of lung involvement is then computed by adding the volumes of consolidation and ground-glass opacity, divided by the total lung volume.
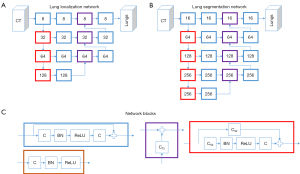

The networks were trained on more than 400 CT datasets from various public and in-house databases covering diverse cases of pneumonia (including COVID-19), cancer, and other lung pathologies. Clinical evaluation of the models was performed for product release. The estimated volume ratio errors (tolerance) of the total lesion, ground glass opacity, and consolidation as percentage portion of the whole lung were 0.638% (0.163–1.601%), 0.592% (0.320–2.961%), and 0.441% (0.115–1.092%), respectively (27).
Threshold-based quantification of lung involvement
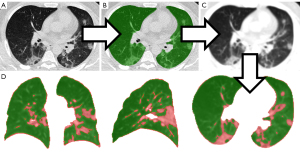
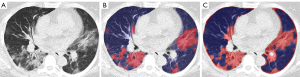
The portion of involved lung tissue was estimated by dividing the number of lung voxels above-threshold by the total lung volume. Assessment of the optimal threshold is explained below. The total lung volume was obtained by an open-source U-net, which proved robust for lung segmentation under the presence of severe COVID pneumonia (R231CovidWeb) (31). After automated lung segmentation, a Gauss filter was applied to the CT image to equalize the lung tissue attenuation. This precluded image noise from artificially generating above threshold voxels. Further, the Gauss filter equalizes smaller pulmonary vessels with the surrounding aerated lung, shifting them below the applied attenuation threshold. The Gauss filter was applied with a standard deviation (sigma) of 1.5 mm, which was rescaled to match the voxel resolution of the given image per dimension. The kernel was truncated at 4 standard deviations resulting in a kernel size of 8*1.5 mm =12 mm. The threshold-based portion of lung involvement was reported as a numerical value from 0-1. The above-reported steps were implemented in a Python script using NumPy library for array programming and in R (32,33). Threshold-based assessment of lung involvement is illustrated in Figure 3.
Assessment of optimal thresholds and five-fold cross validation
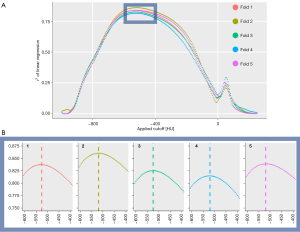
To avoid overfitting to our multi-center dataset, the optimal threshold to differentiate the involved lung was estimated using a five-fold cross validation strategy. First, our study cohort was divided into five randomized, non-overlapping folds of 50 CT scans each. Secondly, five distinct regression models were trained based on five training subsets, containing four alternating folds each, and validated on the fifth remaining fold as specified below. r2 served as a measure for the accuracy of fit of the regression model.
The optimal threshold for discrimination of involved lung vs. well-aerated lung locates between −1,000 HU (air-like attenuation) and 250 HU (soft-tissue-like attenuation). To investigate its exact location and the performance of segmentation, threshold-based assessment of lung involvement was repeated for consecutive arbitrary cutoffs from −1,000 to 250 HU by steps of 5 HU for each patient in the respective training subset. This resulted in 250 threshold-specific portions of above threshold voxels divided by the total lung volume for each training patient. A linear regression model was fitted for each threshold including the predictor variable “percentage of voxels above threshold” and the target variable “deep-learning-based lung involvement”. The threshold of the most accurate regression within each training subset was adopted for testing on the respective fifth fold. Each of five test datasets yielded one r2 alongside with the particular threshold and the regression equation, including a 95% prediction interval. Mean values of the five training and testing iterations are reported as final results.
Comparison of the manual, semi-quantitative CCS to the automated, quantitative methods
Automated, quantitative segmentation results were reviewed by an experienced radiologist. This additional sanity check, however, did not result in manual alteration of the dataset. To evaluate the relationship of the quantitative approaches (deep-learning and threshold-based) and the semi-quantitative CCS, non-linear regression models were developed.
Statistics and data analysis
Data analysis was performed using R for statistical computing (R Foundation, Vienna, Austria, version 4.0.0) (33). Intra- and inter-observer variability were reported by the intraclass correlation coefficient (ICC), using the R library irr. Variable transformations have been conducted with the R libraries car and boot. The Ramsey’s RESET test was performed using the R library lmtest. Visualization of segmentations was realized by the open source software 3D Slicer (34,35) and IntelliSpace Discovery v3.0.6 (Philips Healthcare, The Netherlands).
Results
Two-hundred-fifty non-contrast CTs of the chest were evaluated. Patient details are summarized in Table 1.
Table 1
| Dataset | Gender (F:M), age | Geographical location, time | Confirmation of COVID-19 diagnosis | Semi-quantitative lung involvement score, median of three readers [0–25] | Scanner manufacturer, reconstruction kernel (lung:soft-tissue) | Tube voltage (kV), exposure (mAs) |
|---|---|---|---|---|---|---|
| Local dataset | 26:24, 55.2±13.6 years | Germany, 03–10/2020a | Positive RT-PCR | 12 [6–15] | Philips, 50:0 | 120 kV, 29.4±9.4 mAs |
| RICORD (21) | 17:33, 57.0±15.5 years | Turkey, USA, Canada, Brazil, before 01/2021b | Positive RT-PCR | 14 [10–17] | Not reported, 0:50 | Not reported (3 low-dose CTs, 47 diagnostic CTs) |
| Shakouri et al. (22) | 31:19†, 47.2±16.3 years† | Iran, 03/2020-01/2021c | Positive RT-PCR | 11 [7–15] | Neusoft Medical Systems, 11:39 | Not reported |
| Zaffino et al. (23) | 27:23, 56 years (range, 20–83 years)† | Italy, before 12/2020d | Positive RT-PCR | 10 [6–13] | Toshiba [15] and Siemens [35], 50:0 | 110–120 kV, not reported |
| Clark et al., An et al. (24,25) | Not reported | Italy and China, 01–04/2020e | Positive RT-PCR | 7 [4–9] | Not reported | Not reported |
†, might be subject to sampling error, since patient-individual data is not reported. a, University Hospital Cologne, Cologne, Germany. b, Koç University Hospital, Koç, Turkey; San Francisco University Hospital, San Francisco, USA; Unity Health Toronto, full spectrum of care hospital network, Toronto, Canada; São Paulo University Hospital, São Paulo, Brazil. c, General University Hospital Mashhad, Mashhad, Iran. d, Azienda Ospedaliera Pugliese-Ciaccio, general hospital, Catanzaro, Italy. e, Xiangyang NO.1 People’s Hospital, Hubei University of Medicine, Hubei, China; University Hospital Milan, Milan, Italy. RICORD, The RSNA International COVID Open Radiology Database.
Semi-quantitative scoring of lung involvement
Median CCS was 10 [6–15], 10 [6–15], and 10 [6–15] for the different readers a, b, and c, respectively (Figure 4). Manual estimation of the CCS took about 2 min for each CT scan. Inter-rater reliability was excellent, yielding an ICC of 0.97 [0.97–0.98].

Deep-learning-based quantification of lung involvement
Automated assessment of lung involvement by the deep-learning-based method (IntelliSpace Discovery CT Pulmo Auto Results prototype) took about 1.5 min using a Tesla-P100 GPU card (Nvidia, Santa Carla, USA). The median portion of involved lung, as quantified by the deep-learning method, was 5.8% [0.8–15.2%].
Assessment of optimal thresholds and five-fold cross validation
After splitting the dataset into five folds, linear regressions of deep-learning vs. threshold-based lung involvement were performed for consecutive thresholds from −1,000 to 250 HU for five alternating training sets. The thresholds yielding the best fitting regression model within each training set were identified at −525, −520, −525, −520, and −520 HU (mean −522 HU, Figure 5).
For each of five test splits, an analogous linear regression was modelled using the previously identified best HU thresholds. The regression models are summarized in Table 2.
Table 2
| Fold | 1 | 2 | 3 | 4 | 5 | Mean |
|---|---|---|---|---|---|---|
| Best threshold identified in training split (HU) | −525 | −520 | −525 | −520 | −520 | −522 |
| Intercept of regression line | −0.05 | −0.05 | −0.04 | −0.04 | −0.06 | −0.05 |
| Slope of regression line | 0.93 | 0.96 | 0.89 | 0.97 | 1.03 | 0.96 |
| Width of 95% prediction interval | 0.20 | 0.30 | 0.20 | 0.18 | 0.26 | 0.23 |
| r2 | 0.82 | 0.74 | 0.88 | 0.91 | 0.83 | 0.84 |
Best thresholds were identified in five training splits [200 computed tomography (CT) scans each, second row] and consecutively tested on five non-overlapping test folds (50 CT scans each, bottom rows).
Mean values of five-fold testing yielded the final regression equation y=0.96x−0.05 for prediction of deep-learning-based lung involvement by threshold-based measurements. The regression yielded a high model fit (r2=0.84) as well as a high confidence (width of 95% prediction interval =0.23). A supplementary analysis demonstrated that introduction of an interval threshold approach did not surpass the accuracy of the arbitrary threshold model (Appendix 1). The final model is illustrated in Figure 6.
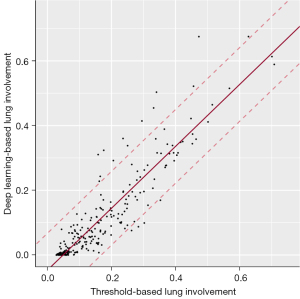
Once the optimal threshold was set, operation of the threshold-based method to determine the extent of COVID pneumonia took about 5 min per exam on a standard desktop computer (processor: Intel ® Core™ i9-9980HK CPU with 2.4 GHz clock frequency). Results of deep-learning and threshold-based quantification of lung involvement are illustrated in Figure 7.
Comparison of the manual, semi-quantitative CCS to the automated, quantitative methods
A regression model was developed to describe the outcome of the automated, quantitative lung involvement assessment methods (deep-learning and threshold-based) using the manual semi-quantitative CCS as predictor variable. Due to the bounded nature [0,1] of the deep-learning and threshold-based variables, the dependent variables were logit transformed first (36). The deep-learning-based lung involvement results were previously remapped to a range 0.025–0.975 to allow for a logit transformation. Afterwards, several models with different functional forms have been assessed with respect to their model fit. A linear model including the CCS as the predictor variable was compared against non-linear regression models (i.e., quadratic, cubic, and quartic polynomials in CCS). A Ramsey’s RESET test indicated that a linear model is an inferior approximation of the data and supports the use of non-linear regression models [(threshold-based model) P<0.05, (deep-learning model) P<0.01]. The selection of the best model was based on the Akaike information criterion (AIC) as well as the adjusted r2 as goodness-of-fit measures (37). The final models with the lowest AIC [(threshold-based model) AIC =447.6, (deep-learning model) AIC=364.5] as well as the highest adjusted r2 [(threshold-based model) r2=0.63, (deep-learning model) r2=0.80] show a moderate to strong model fit (Figure 8).
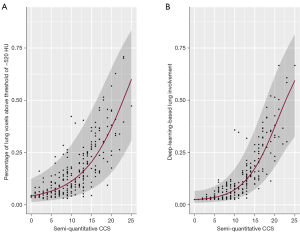
The slopes of the resulting regression curves, which predict quantitative lung involvement based on the semi-quantitative CCS, were relatively flat among lower CCS ratings, followed by a steep incline at CCS values of 10 and higher.
Discussion
Since lung involvement is a relevant predictor for the prognosis of COVID pneumonia and the obligation for intensive care treatment, we evaluated three available post-processing imaging approaches to assess the lung involvement as a valuable CT biomarker in clinical routine. Fully automated threshold- and deep-learning-based quantification of involved lung demonstrated a strong, linear relationship throughout our heterogeneous multi-center, multi-vendor dataset. Besides, the manual semi-quantitative approach correlated with the automated quantitative methods in a non-linear fashion, with larger steps of involved lung per one-point-increase of CCS rating towards higher CCS. i.e., throughout patients with severe lung infiltration, the manual CCS Pan-score underestimated the extent of lung involvement.
This limitation was consistent and independent for each of the three experienced radiologists and might restrict the value of the human-reader-based scoring in clinical routine. According to the regression equation for automated deep-learning-based lung involvement, an increase of the CCS from 5 to 10 resulted in an expanse of pulmonary infiltration by 4.0% of the total lung volume (3.5% to 7.5%); an increase of the CCS from 15 to 20, however, translated to a 26.6% increase of lung disease (19.8% to 46.4%), which was even more pronounced in the maximum CCS range (increase of the CCS from 20 to 25: further 30.0% expansion of involved lung volume). In a clinical context, a five-point CCS increase in mild COVID pneumonia might be insignificant; however, an equal five-point CCS increase in critically ill patients demonstrates a severe reduction of aerated lung tissue.
A probable explanation for the non-linear relationship of the manual CCS to the automated, quantitatively assessed fraction of involved lung tissue is the non-linear definition of the lobar CCS (11): A lobar CCS of 1 is already granted at 5% lobar involvement, and a further increase of 20% involvement raises the lobar CCS to a score of 2. For lobar CCS >2, an increase of 25% involvement is required for a raise by 1 point, which explains the flattened curve throughout mildly affected lungs. The steep curve towards extremely high CCS values might be explainable by the central tendency bias, which says that human readers tend to underestimate values larger than the average. Allred et al. recently demonstrated that the central tendency bias is particularly strong in participants under high cognitive load of their working memory, i.e., memorizing six-digit numbers during their judgements—an experimental set-up that resembles the mental estimation and calculation process of lung involvement scoring (38).
Even if doctors are aware of the non-linear characteristics of the CCS, the distortion of human judgements by exponential scaling is a well-known bias, which might promote underestimation of disease extent in critically ill COVID patients (39). Besides avoiding non-linear relationships, the abolishment of inter-rater error is another benefit of the clinical application of fully automated methods. Albeit our data suggest that inter-rater reliability of the CCS is excellent, truly quantitative, volumetric methods might reflect the biological correlate of lung infiltration more accurate than subjective, individual judgements of human raters.
Our study demonstrates that automatic quantitative evaluation and reporting of the percentage portion of aerated lung tissue could make the clinical application of lung involvement assessment more reliable. In line with this hypothesis, a recent study demonstrated that deep-learning-based quantification of lung involvement is superior to the CCS, when predicting adverse outcomes of COVID pneumonia (9). Lastly, regular implementation of an automated lung involvement assessment saves time of trained personnel, since it renders time-consuming expert scorings redundant.
Up-to-date, there is a lack of comparative data about AI-model performance of COVID pneumonia lung involvement assessment. This prohibited evidence-based identification of the best performing deep-learning model for our study and warrants further investigation. We considered the Philips IntelliSpace Portal CT Pulmo Auto Results appropriate for this study, since it achieved clinical approval for automated assessment of COVID pneumonia segmentation in patients with suspected and diagnosed COVID pneumonia. This approved scope of application includes the use case in the presented study. During clinical validation, the software proved an excellent performance with error rates <1% when calculating the percentage portion of involved lung (27). The CE-label assures that the product is not only safe to use, but also technically able to perform in a clinical context as claimed by the manufacturer (40).
For the automated threshold-based method, we found a robust cutoff at −522 HU to reproduce the deep-learning-based lung involvement quantification (r2=0.84). Yet, this threshold demonstrated a less accurate relationship to the semi-quantitative CCS, compared to the deep-learning method (r2=0.63 vs. r2=0.80, respectively). A probable explanation might be overestimation of involved lung by a threshold-based method, since it includes larger pulmonary vessels and motion blurred lung areas as above-threshold tissue. This also explains why the regression line intersects the X-axis at 0.05 in Figure 6—in contrast to the deep-learning results, the lowest estimation of involved lung by the threshold approach is 5%, and not 0%, which might correspond to the above-threshold segmented portion of lung vessels and motion blur. The slope of the regression line, however, was close to 1 (0.96), indicating that this portion of artificially above-threshold voxels was a consistent finding throughout our dataset, and did not skew the balanced relationship between deep-learning and threshold-based results. Two recent studies performed threshold-based lung involvement assessment of COVID pneumonia with arbitrary cutoffs at −700 and −500 HU, the latter in the range of our computed cutoff value of −522 HU (20,21). However, both studies only included single-center CT data with only one specific CT-scanner each. Thus, these investigated cutoffs might be of limited use for actual clinical application with different CT imaging protocols and image reconstructions. Lung attenuation measurements can be biased by the applied radiation dose, the vendor specific reconstruction filter, and depend on the quality of the CT scanner, which particularly implies accurate calibration (41). For this reason, we performed our study on an international multi-center, multi-vendor dataset with heterogeneous acquisition and reconstruction parameters, and further addressed the potential issue of overfitting by five-fold cross validation.
A further recent study by Khan et al. investigated the accuracy of seven established, advanced thresholding methods to segment the affected lung in COVID pneumonia (18). A parallel to our work is the use of the R231CovidWeb by Hofmanninger et al. to obtain the whole-lung segmentation as a starting point (31). Consecutively, however, Khan et al. transformed their data to 8-bit images (256 level greyscale) for further processing, rather than maintaining the established Hounsfield scale (18). This precludes uncomplicated adoption of their investigated thresholds for clinical use. Yet, their thresholding methods recognize advanced quantitative parameters such as histogram shape, measurement space clustering, entropy, or local gray-level surface, which might introduce a benefit compared to the absolute, arbitrary threshold in our study (42). Similar to our work, Khan et al. observed excellent reproduction of AI-based lung involvement segmentation by their threshold-based approaches (18). After all, the data is yet too scarce to allow for a definitive comparison of our methods, since Khan et al. examined only 28 patients (18).
The presented study had several limitations. First, we did not interrupt the automated processing of the quantitative assessments for manual alteration. This demonstrates feasibility of the proposed method without specific user interaction; however, clinical application should always include image inspection and a plausibility check by the reporting radiologist. Yet, we do not expect the applied U-net(R231) to limit our methodology, since it was trained on a heterogeneous dataset including severe lung pathologies, and consecutively fine-tuned to segmentation of COVID pneumonia lungs, yielding exceptional results (31,43). Additionally, marginal inclusion of highly attenuating, non-parenchymatic findings (e.g., pleural effusions) might promote the non-linear relationship between the quantitative and semi-quantitative methods. Secondly, we included only one automated deep-learning tool of one CT vendor to our study, which might affect generalizability of our method. Further, the retrospective design of our study might limit its clinical impact. The majority of included patients presented at larger university hospitals, which might hamper generalizability of our results to a non-university context. Yet, we did not observe a bias towards severely affected individuals, considering the distribution of CCS (Figure 4). Vice versa, there were relatively few severely affected patients in our population, which might imply underrepresentation of this group. Lastly, we did not obtain detailed clinical data about the severity of COVID-19 pneumonia in our study population. Nonetheless, the CCS is an established imaging biomarker for severity of COVID-19 pneumonia, and has been suggested a surrogate parameter for the clinical course of the disease by several authors (44,45).
In conclusion, our data suggest that the truly quantitative, percentage fraction of involved lung in COVID pneumonia exceeds the clinical usability of manual CCS ratings, since it indicates the extent of pulmonary pathology in an easily assessable, linear fashion. Translation into clinical daily routine seems to be warranted, but needs to be further investigated in larger prospective clinical trials. If doctors make use of the manual CCS as a support for clinical decision making, they should recognize that it operates on a non-linear scale, and an increase at higher scores translates to a much stronger expansion of pulmonary disease.
Acknowledgments
Funding: This research was supported by the German Federal Ministry of Education and Research (BMBF) as part of the University Medicine Network (Project RACOON, 01KX2021) and the German Research Foundation [Deutsche Forschungsgemeinschaft (DFG), No. 491454339]. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Footnote
Reporting Checklist: The authors have completed the GRRAS reporting checklist. Available at https://qims.amegroups.com/article/view/10.21037/qims-22-175/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-22-175/coif). DM is on the speaker’s bureau of Philips Healthcare. HC and RS are employees of Philips Healthcare. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional review board of the University Cologne (No. 21-1426-retro) and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kwee TC, Kwee RM. Chest CT in COVID-19: What the Radiologist Needs to Know. Radiographics 2020;40:1848-65. [Crossref] [PubMed]
- Feng Z, Yu Q, Yao S, Luo L, Zhou W, Mao X, et al. Early prediction of disease progression in COVID-19 pneumonia patients with chest CT and clinical characteristics. Nat Commun 2020;11:4968. [Crossref] [PubMed]
- Yuyun X, Lexi Y, Haochu W, Zhenyu S, Xiangyang G. Early Warning Information for Severe and Critical Patients With COVID-19 Based on Quantitative CT Analysis of Lung Segments. Front Public Health 2021;9:596938. [Crossref] [PubMed]
- Tabatabaei SMH, Talari H, Moghaddas F, Rajebi H. CT Features and Short-term Prognosis of COVID-19 Pneumonia: A Single-Center Study from Kashan, Iran. Radiol Cardiothorac Imaging 2020;2:e200130. [Crossref] [PubMed]
- Hu Y, Zhan C, Chen C, Ai T, Xia L. Chest CT findings related to mortality of patients with COVID-19: A retrospective case-series study. PLoS One 2020;15:e0237302. [Crossref] [PubMed]
- Li K, Chen D, Chen S, Feng Y, Chang C, Wang Z, Wang N, Zhen G. Predictors of fatality including radiographic findings in adults with COVID-19. Respir Res 2020;21:146. [Crossref] [PubMed]
- Francone M, Iafrate F, Masci GM, Coco S, Cilia F, Manganaro L, Panebianco V, Andreoli C, Colaiacomo MC, Zingaropoli MA, Ciardi MR, Mastroianni CM, Pugliese F, Alessandri F, Turriziani O, Ricci P, Catalano C. Chest CT score in COVID-19 patients: correlation with disease severity and short-term prognosis. Eur Radiol 2020;30:6808-17. [Crossref] [PubMed]
- Abdel-Tawab M, Basha MAA, Mohamed IAI, Ibrahim HM. A simple chest CT score for assessing the severity of pulmonary involvement in COVID-19. Egypt J Radiol Nucl Med 2021;52:149. [Crossref]
- Gieraerts C, Dangis A, Janssen L, Demeyere A, De Bruecker Y, De Brucker N, van Den Bergh A, Lauwerier T, Heremans A, Frans E, Laurent M, Ector B, Roosen J, Smismans A, Frans J, Gillis M, Symons R. Prognostic Value and Reproducibility of AI-assisted Analysis of Lung Involvement in COVID-19 on Low-Dose Submillisievert Chest CT: Sample Size Implications for Clinical Trials. Radiol Cardiothorac Imaging 2020;2:e200441. [Crossref] [PubMed]
- Ren HW, Wu Y, Dong JH, An WM, Yan T, Liu Y, Liu CC. Analysis of clinical features and imaging signs of COVID-19 with the assistance of artificial intelligence. Eur Rev Med Pharmacol Sci 2020;24:8210-8. [PubMed]
- Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. Time Course of Lung Changes at Chest CT during Recovery from Coronavirus Disease 2019 (COVID-19). Radiology 2020;295:715-21. [Crossref] [PubMed]
- Chaganti S, Grenier P, Balachandran A, et al. Automated Quantification of CT Patterns Associated with COVID-19 from Chest CT. Radiol Artif Intell 2020;2:e200048. [Crossref] [PubMed]
- Pulmonary Density Plug-In For syngo.via, syngo.via View&GO, and the AI-Rad Companion Chest CT 2020 [cited 2021 Jul 26]. Available online: https://cdn0.scrvt.com/39b415fb07de4d9656c7b516d8e2d907/0f0c496f6dd19ff8/2a6b31274fe7/siemens-healthineers-digital-transformation-of-radiology-ai-covid-19-pulmonary-density-plug-in-two-pager.pdf
- Thoracic VCAR | GE Healthcare (United States). [cited 2021 Jul 26]. Available online: https://www.gehealthcare.com/products/advanced-visualization/all-applications/thoracic-vcar
- CT Lung Analysis | Healthcare IT | Canon Medical Systems. [cited 2021 Jul 26]. Available online: https://global.medical.canon/products/healthcare_it/clinical_application/ct_lung_analysis
- Alsharif MH, Alsharif YH, Chaudhry SA, Albreem MA, Jahid A, Hwang E. Artificial intelligence technology for diagnosing COVID-19 cases: a review of substantial issues. Eur Rev Med Pharmacol Sci 2020;24:9226-33. [PubMed]
- Alsharif MH, Alsharif YH, Yahya K, Alomari OA, Albreem MA, Jahid A. Deep learning applications to combat the dissemination of COVID-19 disease: a review. Eur Rev Med Pharmacol Sci 2020;24:11455-60. [PubMed]
- Khan A, Garner R, Rocca M, Salehi S, Duncan D. A Novel Threshold-Based Segmentation Method for Quantification of COVID-19 Lung Abnormalities. Signal Image Video Process 2022; Epub ahead of print. [Crossref] [PubMed]
- Colombi D, Bodini FC, Petrini M, Maffi G, Morelli N, Milanese G, Silva M, Sverzellati N, Michieletti E. Well-aerated Lung on Admitting Chest CT to Predict Adverse Outcome in COVID-19 Pneumonia. Radiology 2020;296:E86-96. [Crossref] [PubMed]
- Lanza E, Muglia R, Bolengo I, Santonocito OG, Lisi C, Angelotti G, Morandini P, Savevski V, Politi LS, Balzarini L. Quantitative chest CT analysis in COVID-19 to predict the need for oxygenation support and intubation. Eur Radiol 2020;30:6770-8. [Crossref] [PubMed]
- Tsai EB, Simpson S, Lungren MP, Hershman M, Roshkovan L, Colak E, et al. The RSNA International COVID-19 Open Radiology Database (RICORD). Radiology 2021;299:E204-13. [Crossref] [PubMed]
- Shakouri S, Bakhshali MA, Layegh P, Kiani B, Masoumi F, Ataei Nakhaei S, Mostafavi SM. COVID19-CT-dataset: an open-access chest CT image repository of 1000+ patients with confirmed COVID-19 diagnosis. BMC Res Notes 2021;14:178. [Crossref] [PubMed]
- Zaffino P, Marzullo A, Moccia S, Calimeri F, De Momi E, Bertucci B, Arcuri PP, Spadea MF. An Open-Source COVID-19 CT Dataset with Automatic Lung Tissue Classification for Radiomics. Bioengineering (Basel) 2021;8:26. [Crossref] [PubMed]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L, Prior F. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 2013;26:1045-57. [Crossref] [PubMed]
- An P, Xu S, Harmon SA, Turkbey EB, Sanford TH, Amalou A, et al. CT Images in COVID-19 [Data set]. Cancer Imaging Arch 2020. Available online: https://wiki.cancerimagingarchive.net/display/Public/CT+Images+in+COVID-19
- Gamer M, Lemon J, Fellows I, Singh P. Package “irr” Title Various Coefficients of Interrater Reliability and Agreement 2019 [cited 2021 Jun 16]. Available online: https://www.r-project.org
- Instructions for Use IntelliSpace Portal, English. Available online: https://www.documents.philips.com/assets/Instruction%20for%20Use/20210412/d652a0ac1db946e8b236ad080070403c.pdf?feed=ifu_docs_feed&_gl=1*lzog9m*_ga*MTQ3ODU0MjgzMy4xNjMzNzc0NTY1*_ga_2NMXNNS6LE*MTYzMzc3NDU2NS4xLjEuMTYzMzc3NDk1OS4xNA..*_ga_7HJGB4S2PK*MTYzMzc3NDgzMy4xLjEuMTYzMzc3NTIyNC4xOQ..&_ga=2.166807357.2061606987.1633774565-1478542833.1633774565
- Si-Mohamed SA, Nasser M, Colevray M, Nempont O, Lartaud PJ, Vlachomitrou A, Broussaud T, Ahmad K, Traclet J, Cottin V, Boussel L. Automatic quantitative computed tomography measurement of longitudinal lung volume loss in interstitial lung diseases. Eur Radiol 2022;32:4292-4303. [Crossref] [PubMed]
- Milletari F, Navab N, Ahmadi SA. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. Proc - 2016 4th Int Conf 3D Vision, 3DV 2016:565-71.
- Brosch T, Saalbach A. Foveal fully convolutional nets for multi-organ segmentation. Proc. SPIE 2018;198-206. [Crossref]
- Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur Radiol Exp 2020;4:50. [Crossref] [PubMed]
- Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D, et al. Array programming with NumPy. Nature 2020;585:357-62. [Crossref] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2021. Available online: https://www.R-project.org/.
- Shetty H, Shetty S, Kakade A, Shetty A, Karobari MI, Pawar AM, Marya A, Heboyan A, Venugopal A, Nguyen TH, Rokaya D. Three-dimensional semi-automated volumetric assessment of the pulp space of teeth following regenerative dental procedures. Sci Rep 2021;11:21914. [Crossref] [PubMed]
- Kikinis R, Pieper SD, Vosburgh KG. 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In: Intraoperative Imaging and Image-Guided Therapy. Springer New York; 2014. p. 277-89.
- Baum CF. Stata Tip 63: Modeling Proportions. The Stata Journal 2008;8:299-303. [Crossref]
- Greene WH. Greene, Econometric Analysis, 8th Edition | Pearson. 8th ed. 2018 [cited 2022 Jan 10]. Available online: https://www.pearson.com/us/higher-education/program/Greene-Econometric-Analysis-8th-Edition/PGM334862.html
- Allred SR, Crawford LE, Duffy S, Smith J. Working memory and spatial judgments: Cognitive load increases the central tendency bias. Psychon Bull Rev 2016;23:1825-31. [Crossref] [PubMed]
- Ryan WH, Evers ERK. Graphs with logarithmic axes distort lay judgments. Behavioral Science & Policy 2020;6:13-23. [Crossref]
- BfArM - Placing medical devices on the market. [cited 2022 Jun 14]. Available online: https://www.bfarm.de/EN/Medical-devices/Overview/Regulatory-framework/Placing-medical-devices-on-the-market/_node.html
- Mascalchi M, Camiciottoli G, Diciotti S. Lung densitometry: why, how and when. J Thorac Dis 2017;9:3319-45. [Crossref] [PubMed]
- Sezgin M, Sankur B. Survey over image thresholding techniques and quantitative performance evaluation. Journal of Electronic Imaging 2004;13:146-65. [Crossref]
- Tilborghs S, Dirks I, Fidon L, Willems S, Eelbode T, Bertels J, et al. Comparative study of deep learning methods for the automatic segmentation of lung, lesion and lesion type in CT scans of COVID-19 patients. Available online: https://research-repository.uwa.edu.au/en/publications/comparative-study-of-deep-learning-methods-for-the-automatic-segm
- Saeed GA, Gaba W, Shah A, Al Helali AA, Raidullah E, Al Ali AB, Elghazali M, Ahmed DY, Al Kaabi SG, Almazrouei S. Correlation between Chest CT Severity Scores and the Clinical Parameters of Adult Patients with COVID-19 Pneumonia. Radiol Res Pract 2021;2021:6697677. [Crossref] [PubMed]
- Inoue A, Takahashi H, Ibe T, Ishii H, Kurata Y, Ishizuka Y, Hamamoto Y. Comparison of semiquantitative chest CT scoring systems to estimate severity in coronavirus disease 2019 (COVID-19) pneumonia. Eur Radiol 2022;32:3513-24. [Crossref] [PubMed]


